research
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences.
Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.
Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
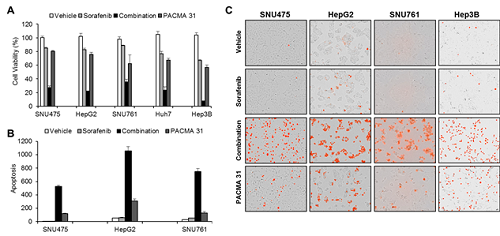
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)

(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)

(Figure 3. ER stress network model)
-
research What Fuels a “Domino Effect” in Cancer Drug Resistance?
KAIST researchers have identified mechanisms that relay prior acquired resistance to the first-line chemotherapy to the second-line targeted therapy, fueling a “domino effect” in cancer drug resistance. Their study featured in the February 7 edition of Science Advances suggests a new strategy for improving the second-line setting of cancer treatment for patients who showed resistance to anti-cancer drugs. Resistance to cancer drugs is often managed in the clinic by chemotherap
2020-02-10 -
research Novel Material Properties of Hybrid Perovskite Nanostructures for Next-generation Non-linear Electronic Devices
(from left: Juho Lee, Dr. Muhammad Ejaz Khan and Professor Yong-Hoon Kim) A KAIST research team reported a novel non-linear device with the founding property coming from perovskite nanowires. They showed that hybrid perovskite-derived, inorganic-framework nanowires can acquire semi-metallicity, and proposed negative differential resistance (NDR) devices with excellent NDR characteristics that resulted from a novel quantum-hybridization NDR mechanism, implying the potential of perovskite nano
2019-02-22 -
research Hierarchical Porous Titanium Nitride Synthesized by Multiscale Phase Separation for LSBs
(from left: Professor Jinwoo Lee and PhD candidate Won-Gwang Lim) A KAIST research team developed ultra-stable, high-rate lithium-sulfur batteries (LSBs) by using hierarchical porous titanium nitride as a sulfur host, and achieved superior cycle stability and high rate performance for LSBs. The control of large amounts of energy is required for use in an electric vehicle or smart grid system. In this sense, the development of next-generation secondary batteries is in high
2019-01-28 -
research Highly Scalable Process to Obtain Stable 2D Nanosheet Dispersion
(Professor Do Hyun Kim and his team) A KAIST team developed technology that allows the mass production of two-dimensional (2D) nanomaterial dispersion by utilizing the characteristic shearing force of hydraulic power. The 2D nanosheet dispersion can be directly applied to solution-based processes to manufacture devices for electronics as well as energy storage and conversion. It is expected to be used in these devices with improved performance. There have been numero
2018-12-19 -
research Characteristics of Submesoscale Geophysical Turbulence Reported
A KAIST research team has reported some of unique characteristics and driving forces behind submesoscale geophysical turbulence. Using big data analysis on ocean surface currents and chlorophyll concentrations observed using coastal radars and satellites has brought better understanding of oceanic processes in space and time scales of O(1) kilometer and O(1) hour. The outcomes of this work will lead to improved tracking of water-borne materials and performance in global and regional climate
2018-12-13