Laboratory+for+Systems+Biology+and+Bio-Inspired+Engineering
-
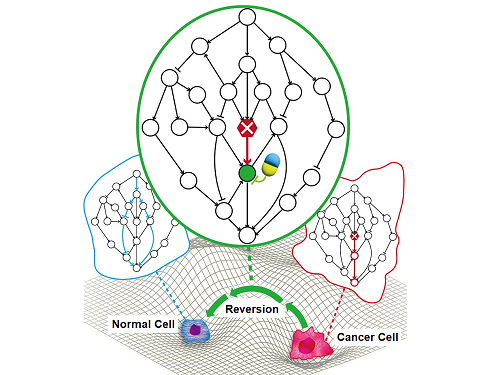 A KAIST Research Team Identifies a Cancer Reversion Mechanism
Despite decades of intensive cancer research by numerous biomedical scientists, cancer still holds its place as the number one cause of death in Korea. The fundamental reason behind the limitations of current cancer treatment methods is the fact that they all aim to completely destroy cancer cells, which eventually allows the cancer cells to acquire immunity. In other words, recurrences and side-effects caused by the destruction of healthy cells are inevitable. To this end, some have suggested anticancer treatment methods based on cancer reversion, which can revert cancer cells back to normal or near-normal cells under certain conditions. However, the practical development of this idea has not yet been attempted.
On June 8, a KAIST research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering reported to have successfully identified the fundamental principle of a process that can revert cancer cells back to normal cells without killing the cells.
Professor Cho’s team focused on the fact that unlike normal cells, which react according to external stimuli, cancer cells tend to ignore such stimuli and only undergo uncontrolled cell division. Through computer simulation analysis, the team discovered that the input-output (I/O) relationships that were distorted by genetic mutations could be reverted back to normal I/O relationships under certain conditions. The team then demonstrated through molecular cell experiments that such I/O relationship recovery also occurred in real cancer cells.
The results of this study, written by Dr. Jae Il Joo and Dr. Hwa-Jeong Park, were published in Wiley’s Advanced Science online on June 2 under the title, "Normalizing input-output relationships of cancer networks for reversion therapy."
< Image 1. Input-output (I/O) relationships in gene regulatory networks >
Professor Kwang-Hyun Cho's research team classified genes into four types by simulation-analyzing the effect of gene mutations on the I/O relationship of gene regulatory networks. (Figure A-J) In addition, by analyzing 18 genes of the cancer-related gene regulatory network, it was confirmed that when mutations occur in more than half of the genes constituting each network, reversibility is possible through appropriate control. (Figure K)
Professor Cho’s team uncovered that the reason the distorted I/O relationships of cancer cells could be reverted back to normal ones was the robustness and redundancy of intracellular gene control networks that developed over the course of evolution. In addition, they found that some genes were more promising as targets for cancer reversion than others, and showed through molecular cell experiments that controlling such genes could revert the distorted I/O relationships of cancer cells back to normal ones.
< Image 2. Simulation results of restoration of bladder cancer gene regulation network and I/O relationship of bladder cancer cells. >
The research team classified the effects of gene mutations on the I/O relationship in the bladder cancer gene regulation network by simulation analysis and classified them into 4 types. (Figure A) Through this, it was found that the distorted input-output relationship between bladder cancer cell lines KU-1919 and HCT-1197 could be restored to normal. (Figure B)
< Image 3. Analysis of survival of bladder cancer patients according to reversible gene mutation and I/O recovery experiment of bladder cancer cells. >
As predicted through network simulation analysis, Professor Kwang-Hyun Cho's research team confirmed through molecular cell experiments that the response to TGF-b was normally restored when AKT and MAP3K1 were inhibited in the bladder cancer cell line KU-1919. (Figure A-G) In addition, it was confirmed that there is a difference in the survival rate of bladder cancer patients depending on the presence or absence of a reversible gene mutation. (Figure H)
The results of this research show that the reversion of real cancer cells does not happen by chance, and that it is possible to systematically explore targets that can induce this phenomenon, thereby creating the potential for the development of innovative anticancer drugs that can control such target genes.
< Image 4. Cancer cell reversibility principle >
The research team analyzed the reversibility, redundancy, and robustness of various networks and found that there was a positive correlation between them. From this, it was found that reversibility was additionally inherent in the process of evolution in which the gene regulatory network acquired redundancy and consistency.
Professor Cho said, “By uncovering the fundamental principles of a new cancer reversion treatment strategy that may overcome the unresolved limitations of existing chemotherapy, we have increased the possibility of developing new and innovative drugs that can improve both the prognosis and quality of life of cancer patients.”
< Image 5. Conceptual diagram of research results >
The research team identified the fundamental control principle of cancer cell reversibility through systems biology research. When the I/O relationship of the intracellular gene regulatory network is distorted by mutation, the distorted I/O relationship can be restored to a normal state by identifying and adjusting the reversible gene target based on the redundancy of the molecular circuit inherent in the complex network.
After Professor Cho’s team first suggested the concept of reversion treatment, they published their results for reverting colorectal cancer in January 2020, and in January 2022 they successfully re-programmed malignant breast cancer cells back into hormone-treatable ones. In January 2023, the team successfully removed the metastasis ability from lung cancer cells and reverted them back to a state that allowed improved drug reactivity. However, these results were case studies of specific types of cancer and did not reveal what common principle allowed cancer reversion across all cancer types, making this the first revelation of the general principle of cancer reversion and its evolutionary origins.
This research was funded by the Ministry of Science and ICT of the Republic of Korea and the National Research Foundation of Korea.
2023.06.20 View 4989
A KAIST Research Team Identifies a Cancer Reversion Mechanism
Despite decades of intensive cancer research by numerous biomedical scientists, cancer still holds its place as the number one cause of death in Korea. The fundamental reason behind the limitations of current cancer treatment methods is the fact that they all aim to completely destroy cancer cells, which eventually allows the cancer cells to acquire immunity. In other words, recurrences and side-effects caused by the destruction of healthy cells are inevitable. To this end, some have suggested anticancer treatment methods based on cancer reversion, which can revert cancer cells back to normal or near-normal cells under certain conditions. However, the practical development of this idea has not yet been attempted.
On June 8, a KAIST research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering reported to have successfully identified the fundamental principle of a process that can revert cancer cells back to normal cells without killing the cells.
Professor Cho’s team focused on the fact that unlike normal cells, which react according to external stimuli, cancer cells tend to ignore such stimuli and only undergo uncontrolled cell division. Through computer simulation analysis, the team discovered that the input-output (I/O) relationships that were distorted by genetic mutations could be reverted back to normal I/O relationships under certain conditions. The team then demonstrated through molecular cell experiments that such I/O relationship recovery also occurred in real cancer cells.
The results of this study, written by Dr. Jae Il Joo and Dr. Hwa-Jeong Park, were published in Wiley’s Advanced Science online on June 2 under the title, "Normalizing input-output relationships of cancer networks for reversion therapy."
< Image 1. Input-output (I/O) relationships in gene regulatory networks >
Professor Kwang-Hyun Cho's research team classified genes into four types by simulation-analyzing the effect of gene mutations on the I/O relationship of gene regulatory networks. (Figure A-J) In addition, by analyzing 18 genes of the cancer-related gene regulatory network, it was confirmed that when mutations occur in more than half of the genes constituting each network, reversibility is possible through appropriate control. (Figure K)
Professor Cho’s team uncovered that the reason the distorted I/O relationships of cancer cells could be reverted back to normal ones was the robustness and redundancy of intracellular gene control networks that developed over the course of evolution. In addition, they found that some genes were more promising as targets for cancer reversion than others, and showed through molecular cell experiments that controlling such genes could revert the distorted I/O relationships of cancer cells back to normal ones.
< Image 2. Simulation results of restoration of bladder cancer gene regulation network and I/O relationship of bladder cancer cells. >
The research team classified the effects of gene mutations on the I/O relationship in the bladder cancer gene regulation network by simulation analysis and classified them into 4 types. (Figure A) Through this, it was found that the distorted input-output relationship between bladder cancer cell lines KU-1919 and HCT-1197 could be restored to normal. (Figure B)
< Image 3. Analysis of survival of bladder cancer patients according to reversible gene mutation and I/O recovery experiment of bladder cancer cells. >
As predicted through network simulation analysis, Professor Kwang-Hyun Cho's research team confirmed through molecular cell experiments that the response to TGF-b was normally restored when AKT and MAP3K1 were inhibited in the bladder cancer cell line KU-1919. (Figure A-G) In addition, it was confirmed that there is a difference in the survival rate of bladder cancer patients depending on the presence or absence of a reversible gene mutation. (Figure H)
The results of this research show that the reversion of real cancer cells does not happen by chance, and that it is possible to systematically explore targets that can induce this phenomenon, thereby creating the potential for the development of innovative anticancer drugs that can control such target genes.
< Image 4. Cancer cell reversibility principle >
The research team analyzed the reversibility, redundancy, and robustness of various networks and found that there was a positive correlation between them. From this, it was found that reversibility was additionally inherent in the process of evolution in which the gene regulatory network acquired redundancy and consistency.
Professor Cho said, “By uncovering the fundamental principles of a new cancer reversion treatment strategy that may overcome the unresolved limitations of existing chemotherapy, we have increased the possibility of developing new and innovative drugs that can improve both the prognosis and quality of life of cancer patients.”
< Image 5. Conceptual diagram of research results >
The research team identified the fundamental control principle of cancer cell reversibility through systems biology research. When the I/O relationship of the intracellular gene regulatory network is distorted by mutation, the distorted I/O relationship can be restored to a normal state by identifying and adjusting the reversible gene target based on the redundancy of the molecular circuit inherent in the complex network.
After Professor Cho’s team first suggested the concept of reversion treatment, they published their results for reverting colorectal cancer in January 2020, and in January 2022 they successfully re-programmed malignant breast cancer cells back into hormone-treatable ones. In January 2023, the team successfully removed the metastasis ability from lung cancer cells and reverted them back to a state that allowed improved drug reactivity. However, these results were case studies of specific types of cancer and did not reveal what common principle allowed cancer reversion across all cancer types, making this the first revelation of the general principle of cancer reversion and its evolutionary origins.
This research was funded by the Ministry of Science and ICT of the Republic of Korea and the National Research Foundation of Korea.
2023.06.20 View 4989 -
 KAIST presents a fundamental technology to remove metastatic traits from lung cancer cells
KAIST (President Kwang Hyung Lee) announced on January 30th that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering succeeded in using systems biology research to change the properties of carcinogenic cells in the lungs and eliminate both drug resistance and their ability to proliferate out to other areas of the body.
As the incidences of cancer increase within aging populations, cancer has become the most lethal disease threatening healthy life. Fatality rates are especially high when early detection does not happen in time and metastasis has occurred in various organs. In order to resolve this problem, a series of attempts were made to remove or lower the ability of cancer cells to spread, but they resulted in cancer cells in the intermediate state becoming more unstable and even more malignant, which created serious treatment challenges.
Professor Kwang-Hyun Cho's research team simulated various cancer cell states in the Epithelial-to-Mesenchymal Transition (EMT) of lung cancer cells, between epithelial cells without metastatic ability and mesenchymal cells with metastatic ability. A mathematical model of molecular network was established, and key regulators that could reverse the state of invasive and drug resistant mesenchymal cells back to the epithelial state were discovered through computer simulation analysis and molecular cell experiments. In particular, this process succeeded in properly reverting the mesenchymal lung cancer cells to a state where they were sensitive to chemotherapy treatment while avoiding the unstable EMT hybrid cell state in the middle process, which had remained a difficult problem.
The results of this research, in which KAIST Ph.D. student Namhee Kim, Dr. Chae Young Hwang, Researcher Taeyoung Kim, and Ph.D. student Hyunjin Kim participated, were published as an online paper in the international journal “Cancer Research” published by the American Association for Cancer Research (AACR) on January 30th. (Paper title: A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states)
Cells in an EMT hybrid state, which are caused by incomplete transitions during the EMT process in cancer cells, have the characteristics of both epithelial cells and mesenchymal cells, and are known to have high drug resistance and metastatic potential by acquiring high stem cell capacity. In particular, EMT is further enhanced through factors such as transforming growth factor-beta (TGF-β) secreted from the tumor microenvironment (TME) and, as a result, various cell states with high plasticity appear. Due to the complexity of EMT, it has been very difficult to completely reverse the transitional process of the mesenchymal cancer cells to an epithelial cell state in which metastatic ability and drug resistance are eliminated while avoiding the EMT hybrid cell state with high metastatic ability and drug resistance.
Professor Kwang-Hyun Cho's research team established a mathematical model of the gene regulation network that governs the complex process of EMT, and then applied large-scale computer simulation analysis and complex system network control technology to identify and verify 'p53', 'SMAD4', and 'ERK1' and 'ERK 2' (collectively ERKs) through molecular cell experiments as the three key molecular targets that can transform lung cancer cells in the mesenchymal cell state, reversed back to an epithelial cell state that no longer demonstrates the ability to metastasize, while avoiding the EMT hybrid cell state.
In particular, by analyzing the molecular regulatory mechanism of the complex EMT process at the system level, the key pathways were identified that were linked to the positive feedback that plays an important role in completely returning cancer cells to an epithelial cell state in which metastatic ability and drug resistance are removed.
This discovery is significant in that it proved that mesenchymal cells can be reverted to the state of epithelial cells under conditions where TGF-β stimulation are present, like they are in the actual environment where cancer tissue forms in the human body.
Abnormal EMT in cancer cells leads to various malignant traits such as the migration and invasion of cancer cells, changes in responsiveness to chemotherapy treatment, enhanced stem cell function, and the dissemination of cancer. In particular, the acquisition of the metastatic ability of cancer cells is a key determinant factor for the prognosis of cancer patients. The EMT reversal technology in lung cancer cells developed in this research is a new anti-cancer treatment strategy that reprograms cancer cells to eliminate their high plasticity and metastatic potential and increase their responsiveness to chemotherapy.
Professor Kwang-Hyun Cho said, "By succeeding in reversing the state of lung cancer cells that acquired high metastatic traits and resistance to drugs and reverting them to a treatable epithelial cell state with renewed sensitivity to chemotherapy, the research findings propose a new strategy for treatments that can improve the prognosis of cancer patients.”
Professor Kwang-Hyun Cho's research team was the first to present the principle of reversal treatment to revert cancer cells to normal cells, following through with the announcement of the results of their study that reverted colon cancer cells to normal colon cells in January of 2020, and also presenting successful re-programming research where the most malignant basal type breast cancer cells turned into less-malignant luminal type breast cancer cells that were treatable with hormonal therapies in January of 2022. This latest research result is the third in the development of reversal technology where lung cancer cells that had acquired metastatic traits returned to a state in which their metastatic ability was removed and drug sensitivity was enhanced.
This research was carried out with support from the Ministry of Science and ICT and the National Research Foundation of Korea's Basic Research in Science & Engineering Program for Mid-Career Researchers.
< Figure 1. Construction of the mathematical model of the regulatory network to represent the EMT phenotype based on the interaction between various molecules related to EMT.
(A) Professor Kwang-Hyun Cho's research team investigated numerous literatures and databases related to complex EMT, and based on comparative analysis of cell line data showing epithelial and mesenchymal cell conditions, they extracted key signaling pathways related to EMT and built a mathematical model of regulatory network (B) By comparing the results of computer simulation analysis and the molecular cell experiments, it was verified how well the constructed mathematical model simulated the actual cellular phenomena. >
< Figure 2. Understanding of various EMT phenotypes through large-scale computer simulation analysis and complex system network control technology.
(A) Through computer simulation analysis and experiments, Professor Kwang-Hyun Cho's research team found that complete control of EMT is impossible with single-molecule control alone. In particular, through comparison of the relative stability of attractors, it was revealed that the cell state exhibiting EMT hybrid characteristics has unstable properties. (B), (C) Based on these results, Prof. Cho’s team identified two feedbacks (positive feedback consisting of Snail-miR-34 and ZEB1-miR-200) that play an important role in avoiding the EMT hybrid state that appeared in the TGF-β-ON state. It was found through computer simulation analysis that the two feedbacks restore relatively high stability when the excavated p53 and SMAD4 are regulated. In addition, molecular cell experiments demonstrated that the expression levels of E-cad and ZEB1, which are representative phenotypic markers of EMT, changed similarly to the expression profile in the epithelial cell state, despite the TGF-β-ON state. >
< Figure 3. Complex molecular network analysis and discovery of reprogramming molecular targets for intact elimination of EMT hybrid features.
(A) Controlling the expression of p53 and SMAD4 in lung cancer cell lines was expected to overcome drug resistance, but contrary to expectations, chemotherapy responsiveness was not restored. (B) Professor Kwang-Hyun Cho's research team additionally analyzed computer simulations, genome data, and experimental results and found that high expression levels of TWIST1 and EPCAM were related to drug resistance. (C) Prof. Cho’s team identified three key molecular targets: p53, SMAD4 and ERK1 & ERK2. (D), (E) Furthermore, they identified a key pathway that plays an important role in completely reversing into epithelial cells while avoiding EMT hybrid characteristics, and confirmed through network analysis and attractor analysis that high stability of the key pathway was restored when the proposed molecular target was controlled. >
< Figure 4. Verification through experiments with lung cancer cell lines.
When p53 was activated and SMAD4 and ERK1/2 were inhibited in lung cancer cell lines, (A), (B) E-cad protein expression increased and ZEB1 protein expression decreased, and (C) mesenchymal cell status including TWIST1 and EPCAM and gene expression of markers related to stem cell potential characteristics were completely inhibited. In addition, (D) it was confirmed that resistance to chemotherapy treatment was also overcome as the cell state was reversed by the regulated target. >
< Figure 5. A schematic representation of the research results.
Prof. Cho’s research team identified key molecular regulatory pathways to avoid high plasticity formed by abnormal EMT of cancer cells and reverse it to an epithelial cell state through systems biology research. From this analysis, a reprogramming molecular target that can reverse the state of mesenchymal cells with acquired invasiveness and drug resistance to the state of epithelial cells with restored drug responsiveness was discovered.
For lung cancer cells, when a drug that enhances the expression of p53, one of the molecular targets discovered, and inhibits the expression of SMAD4 and ERK1 & ERK2 is administered, the molecular network of genes in the state of mesenchymal cells is modified, eventually eliminating metastatic ability and it is reprogrammed to turn into epithelial cells without the resistance to chemotherapy treatments. >
2023.01.30 View 11809
KAIST presents a fundamental technology to remove metastatic traits from lung cancer cells
KAIST (President Kwang Hyung Lee) announced on January 30th that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering succeeded in using systems biology research to change the properties of carcinogenic cells in the lungs and eliminate both drug resistance and their ability to proliferate out to other areas of the body.
As the incidences of cancer increase within aging populations, cancer has become the most lethal disease threatening healthy life. Fatality rates are especially high when early detection does not happen in time and metastasis has occurred in various organs. In order to resolve this problem, a series of attempts were made to remove or lower the ability of cancer cells to spread, but they resulted in cancer cells in the intermediate state becoming more unstable and even more malignant, which created serious treatment challenges.
Professor Kwang-Hyun Cho's research team simulated various cancer cell states in the Epithelial-to-Mesenchymal Transition (EMT) of lung cancer cells, between epithelial cells without metastatic ability and mesenchymal cells with metastatic ability. A mathematical model of molecular network was established, and key regulators that could reverse the state of invasive and drug resistant mesenchymal cells back to the epithelial state were discovered through computer simulation analysis and molecular cell experiments. In particular, this process succeeded in properly reverting the mesenchymal lung cancer cells to a state where they were sensitive to chemotherapy treatment while avoiding the unstable EMT hybrid cell state in the middle process, which had remained a difficult problem.
The results of this research, in which KAIST Ph.D. student Namhee Kim, Dr. Chae Young Hwang, Researcher Taeyoung Kim, and Ph.D. student Hyunjin Kim participated, were published as an online paper in the international journal “Cancer Research” published by the American Association for Cancer Research (AACR) on January 30th. (Paper title: A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states)
Cells in an EMT hybrid state, which are caused by incomplete transitions during the EMT process in cancer cells, have the characteristics of both epithelial cells and mesenchymal cells, and are known to have high drug resistance and metastatic potential by acquiring high stem cell capacity. In particular, EMT is further enhanced through factors such as transforming growth factor-beta (TGF-β) secreted from the tumor microenvironment (TME) and, as a result, various cell states with high plasticity appear. Due to the complexity of EMT, it has been very difficult to completely reverse the transitional process of the mesenchymal cancer cells to an epithelial cell state in which metastatic ability and drug resistance are eliminated while avoiding the EMT hybrid cell state with high metastatic ability and drug resistance.
Professor Kwang-Hyun Cho's research team established a mathematical model of the gene regulation network that governs the complex process of EMT, and then applied large-scale computer simulation analysis and complex system network control technology to identify and verify 'p53', 'SMAD4', and 'ERK1' and 'ERK 2' (collectively ERKs) through molecular cell experiments as the three key molecular targets that can transform lung cancer cells in the mesenchymal cell state, reversed back to an epithelial cell state that no longer demonstrates the ability to metastasize, while avoiding the EMT hybrid cell state.
In particular, by analyzing the molecular regulatory mechanism of the complex EMT process at the system level, the key pathways were identified that were linked to the positive feedback that plays an important role in completely returning cancer cells to an epithelial cell state in which metastatic ability and drug resistance are removed.
This discovery is significant in that it proved that mesenchymal cells can be reverted to the state of epithelial cells under conditions where TGF-β stimulation are present, like they are in the actual environment where cancer tissue forms in the human body.
Abnormal EMT in cancer cells leads to various malignant traits such as the migration and invasion of cancer cells, changes in responsiveness to chemotherapy treatment, enhanced stem cell function, and the dissemination of cancer. In particular, the acquisition of the metastatic ability of cancer cells is a key determinant factor for the prognosis of cancer patients. The EMT reversal technology in lung cancer cells developed in this research is a new anti-cancer treatment strategy that reprograms cancer cells to eliminate their high plasticity and metastatic potential and increase their responsiveness to chemotherapy.
Professor Kwang-Hyun Cho said, "By succeeding in reversing the state of lung cancer cells that acquired high metastatic traits and resistance to drugs and reverting them to a treatable epithelial cell state with renewed sensitivity to chemotherapy, the research findings propose a new strategy for treatments that can improve the prognosis of cancer patients.”
Professor Kwang-Hyun Cho's research team was the first to present the principle of reversal treatment to revert cancer cells to normal cells, following through with the announcement of the results of their study that reverted colon cancer cells to normal colon cells in January of 2020, and also presenting successful re-programming research where the most malignant basal type breast cancer cells turned into less-malignant luminal type breast cancer cells that were treatable with hormonal therapies in January of 2022. This latest research result is the third in the development of reversal technology where lung cancer cells that had acquired metastatic traits returned to a state in which their metastatic ability was removed and drug sensitivity was enhanced.
This research was carried out with support from the Ministry of Science and ICT and the National Research Foundation of Korea's Basic Research in Science & Engineering Program for Mid-Career Researchers.
< Figure 1. Construction of the mathematical model of the regulatory network to represent the EMT phenotype based on the interaction between various molecules related to EMT.
(A) Professor Kwang-Hyun Cho's research team investigated numerous literatures and databases related to complex EMT, and based on comparative analysis of cell line data showing epithelial and mesenchymal cell conditions, they extracted key signaling pathways related to EMT and built a mathematical model of regulatory network (B) By comparing the results of computer simulation analysis and the molecular cell experiments, it was verified how well the constructed mathematical model simulated the actual cellular phenomena. >
< Figure 2. Understanding of various EMT phenotypes through large-scale computer simulation analysis and complex system network control technology.
(A) Through computer simulation analysis and experiments, Professor Kwang-Hyun Cho's research team found that complete control of EMT is impossible with single-molecule control alone. In particular, through comparison of the relative stability of attractors, it was revealed that the cell state exhibiting EMT hybrid characteristics has unstable properties. (B), (C) Based on these results, Prof. Cho’s team identified two feedbacks (positive feedback consisting of Snail-miR-34 and ZEB1-miR-200) that play an important role in avoiding the EMT hybrid state that appeared in the TGF-β-ON state. It was found through computer simulation analysis that the two feedbacks restore relatively high stability when the excavated p53 and SMAD4 are regulated. In addition, molecular cell experiments demonstrated that the expression levels of E-cad and ZEB1, which are representative phenotypic markers of EMT, changed similarly to the expression profile in the epithelial cell state, despite the TGF-β-ON state. >
< Figure 3. Complex molecular network analysis and discovery of reprogramming molecular targets for intact elimination of EMT hybrid features.
(A) Controlling the expression of p53 and SMAD4 in lung cancer cell lines was expected to overcome drug resistance, but contrary to expectations, chemotherapy responsiveness was not restored. (B) Professor Kwang-Hyun Cho's research team additionally analyzed computer simulations, genome data, and experimental results and found that high expression levels of TWIST1 and EPCAM were related to drug resistance. (C) Prof. Cho’s team identified three key molecular targets: p53, SMAD4 and ERK1 & ERK2. (D), (E) Furthermore, they identified a key pathway that plays an important role in completely reversing into epithelial cells while avoiding EMT hybrid characteristics, and confirmed through network analysis and attractor analysis that high stability of the key pathway was restored when the proposed molecular target was controlled. >
< Figure 4. Verification through experiments with lung cancer cell lines.
When p53 was activated and SMAD4 and ERK1/2 were inhibited in lung cancer cell lines, (A), (B) E-cad protein expression increased and ZEB1 protein expression decreased, and (C) mesenchymal cell status including TWIST1 and EPCAM and gene expression of markers related to stem cell potential characteristics were completely inhibited. In addition, (D) it was confirmed that resistance to chemotherapy treatment was also overcome as the cell state was reversed by the regulated target. >
< Figure 5. A schematic representation of the research results.
Prof. Cho’s research team identified key molecular regulatory pathways to avoid high plasticity formed by abnormal EMT of cancer cells and reverse it to an epithelial cell state through systems biology research. From this analysis, a reprogramming molecular target that can reverse the state of mesenchymal cells with acquired invasiveness and drug resistance to the state of epithelial cells with restored drug responsiveness was discovered.
For lung cancer cells, when a drug that enhances the expression of p53, one of the molecular targets discovered, and inhibits the expression of SMAD4 and ERK1 & ERK2 is administered, the molecular network of genes in the state of mesenchymal cells is modified, eventually eliminating metastatic ability and it is reprogrammed to turn into epithelial cells without the resistance to chemotherapy treatments. >
2023.01.30 View 11809 -
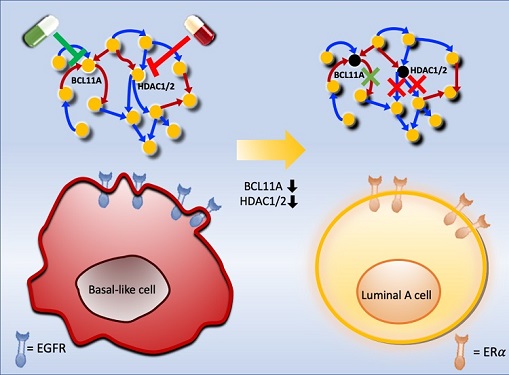 Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 7057
Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 7057 -
 Simulations Open a New Way to Reverse Cell Aging
Turning off a newly identified enzyme could reverse a natural aging process in cells.
Research findings by a KAIST team provide insight into the complex mechanism of cellular senescence and present a potential therapeutic strategy for reducing age-related diseases associated with the accumulation of senescent cells.
Simulations that model molecular interactions have identified an enzyme that could be targeted to reverse a natural aging process called cellular senescence. The findings were validated with laboratory experiments on skin cells and skin equivalent tissues, and published in the Proceedings of the National Academy of Sciences (PNAS).
“Our research opens the door for a new generation that perceives aging as a reversible biological phenomenon,” says Professor Kwang-Hyun Cho of the Department of Bio and Brain engineering at the Korea Advanced Institute of Science and Technology (KAIST), who led the research with colleagues from KAIST and Amorepacific Corporation in Korea.
Cells respond to a variety of factors, such as oxidative stress, DNA damage, and shortening of the telomeres capping the ends of chromosomes, by entering a stable and persistent exit from the cell cycle. This process, called cellular senescence, is important, as it prevents damaged cells from proliferating and turning into cancer cells. But it is also a natural process that contributes to aging and age-related diseases. Recent research has shown that cellular senescence can be reversed. But the laboratory approaches used thus far also impair tissue regeneration or have the potential to trigger malignant transformations.
Professor Cho and his colleagues used an innovative strategy to identify molecules that could be targeted for reversing cellular senescence. The team pooled together information from the literature and databases about the molecular processes involved in cellular senescence. To this, they added results from their own research on the molecular processes involved in the proliferation, quiescence (a non-dividing cell that can re-enter the cell cycle) and senescence of skin fibroblasts, a cell type well known for repairing wounds. Using algorithms, they developed a model that simulates the interactions between these molecules. Their analyses allowed them to predict which molecules could be targeted to reverse cell senescence.
They then investigated one of the molecules, an enzyme called PDK1, in incubated senescent skin fibroblasts and three-dimensional skin equivalent tissue models. They found that blocking PDK1 led to the inhibition of two downstream signalling molecules, which in turn restored the cells’ ability to enter back into the cell cycle. Notably, the cells retained their capacity to regenerate wounded skin without proliferating in a way that could lead to malignant transformation.
The scientists recommend investigations are next done in organs and organisms to determine the full effect of PDK1 inhibition. Since the gene that codes for PDK1 is overexpressed in some cancers, the scientists expect that inhibiting it will have both anti-aging and anti-cancer effects.
-Profile
Professor Kwang-Hyun Cho
Laboratory for Systems Biology and Bio-Inspired Engineering
http://sbie.kaist.ac.kr
Department of Bio and Brain Engineering
KAIST
2020.11.26 View 10989
Simulations Open a New Way to Reverse Cell Aging
Turning off a newly identified enzyme could reverse a natural aging process in cells.
Research findings by a KAIST team provide insight into the complex mechanism of cellular senescence and present a potential therapeutic strategy for reducing age-related diseases associated with the accumulation of senescent cells.
Simulations that model molecular interactions have identified an enzyme that could be targeted to reverse a natural aging process called cellular senescence. The findings were validated with laboratory experiments on skin cells and skin equivalent tissues, and published in the Proceedings of the National Academy of Sciences (PNAS).
“Our research opens the door for a new generation that perceives aging as a reversible biological phenomenon,” says Professor Kwang-Hyun Cho of the Department of Bio and Brain engineering at the Korea Advanced Institute of Science and Technology (KAIST), who led the research with colleagues from KAIST and Amorepacific Corporation in Korea.
Cells respond to a variety of factors, such as oxidative stress, DNA damage, and shortening of the telomeres capping the ends of chromosomes, by entering a stable and persistent exit from the cell cycle. This process, called cellular senescence, is important, as it prevents damaged cells from proliferating and turning into cancer cells. But it is also a natural process that contributes to aging and age-related diseases. Recent research has shown that cellular senescence can be reversed. But the laboratory approaches used thus far also impair tissue regeneration or have the potential to trigger malignant transformations.
Professor Cho and his colleagues used an innovative strategy to identify molecules that could be targeted for reversing cellular senescence. The team pooled together information from the literature and databases about the molecular processes involved in cellular senescence. To this, they added results from their own research on the molecular processes involved in the proliferation, quiescence (a non-dividing cell that can re-enter the cell cycle) and senescence of skin fibroblasts, a cell type well known for repairing wounds. Using algorithms, they developed a model that simulates the interactions between these molecules. Their analyses allowed them to predict which molecules could be targeted to reverse cell senescence.
They then investigated one of the molecules, an enzyme called PDK1, in incubated senescent skin fibroblasts and three-dimensional skin equivalent tissue models. They found that blocking PDK1 led to the inhibition of two downstream signalling molecules, which in turn restored the cells’ ability to enter back into the cell cycle. Notably, the cells retained their capacity to regenerate wounded skin without proliferating in a way that could lead to malignant transformation.
The scientists recommend investigations are next done in organs and organisms to determine the full effect of PDK1 inhibition. Since the gene that codes for PDK1 is overexpressed in some cancers, the scientists expect that inhibiting it will have both anti-aging and anti-cancer effects.
-Profile
Professor Kwang-Hyun Cho
Laboratory for Systems Biology and Bio-Inspired Engineering
http://sbie.kaist.ac.kr
Department of Bio and Brain Engineering
KAIST
2020.11.26 View 10989 -
 Cancer cell reversion may offer a new approach to colorectal cancer treatment
A novel approach to reverse the progression of healthy cells to malignant ones may offer a more effective way to eradicate colorectal cancer cells with far fewer side effects, according to a team of researchers based in South Korea.
Colorectal cancer, or cancer of the colon, is the third most common cancer in men and the second most common in women worldwide. South Korea has the second highest incident rate of colorectal cancer in the world, topped only by Hungary, according to the World Cancer Research Fund.
Their results were published as a featured cover article on January 2 in Molecular Cancer Research, a journal of the American Association for Cancer Research.
Led by Kwang-Hyun Cho, a professor and associate vice president of research at KAIST , the researchers used a computational framework to analyze healthy colon cells and colorectal cancer cells. They found that some master regulator proteins involved in cellular replication helped healthy colon cells mature, or differentiate into their specific cell type, and remain healthy. One particular protein, called SETDB1, suppressed the helpful proteins, forcing new cells to remain in a state of immaturity with the potential to become cancerous.
“This suggests that differentiated cells have an inherent resistance mechanism against malignant transformation and indicates that cellular reprogramming is indispensable for malignancy,” said Cho. “We speculated that malignant properties might be eradicated if the tissue-specific gene expression is reinstated — if we repress SETDB1 and allow the colon cells to mature and differentiate as they would normally.”
Image credit: Kwang-Hyun Cho, KAIST
Image restriction: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Using human-derived cells, Cho and his team targeted the tissue-specific gene expression programs identified in their computational analysis. These are the blueprints for the proteins that eventually help immature cells differentiate into tissue-specific cell types, such as colon cells. When a person has a genetic mutation, or has exposure to certain environmental factors, this process can go awry, leading to an overexpression of unhelpful proteins, such as SEDTB1.
The researchers specifically reduced the amount of SEDTB1 in these tissue-specific gene expression programs, which allowed the cells to mature and fully differentiate into colon cells.
“Our experiment also shows that SETDB1 depletion combined with cytotoxic drugs might be potentially beneficial to anticancer treatment,” Cho said. Cytotoxic drugs are often used for cancer treatment because the type of medicine contains chemicals that are toxic to cancer cells which can prevent them from replicating or growing. He noted that this combination could be more effective in treating cancer by transforming the cancer cell state into a less malignant or resistant state. He eventually pursues a cancer reversion therapy alone instead of conventional cytotoxic drug therapy since the cancer reversion therapy can provide a much less painful experience for patients with cancer who often have severe side effects from treatments intended to kill off cancerous cells, such as chemotherapy.
The researchers plan to continue studying how to return cancer cells to healthier states, with the ultimate goal of translating their work to therapeutic treatment for patients with colorectal cancer.
“I think our study of cancer reversion would eventually change the current medical practice of treating cancer toward the direction of keeping the patient’s quality of life while minimizing the side effects of current anti-cancer therapies,” Cho said.
###
This work was funded by KAIST and the National Research Foundation of Korea grants funded by the Korean government, the Ministry of Science and Information and Communication Technology.
Other authors include Soobeom Lee, Chae Young Hwang and Dongsan Kim, all of whom are affiliated with the Laboratory for Systems Biology and Bio-Inspired Engineering in the Department of Bio and Brain Engineering at KAIST; Chansu Lee and Sung Noh Hong, both with the Department of Medicine, and Seok-Hyung Kim of the Department of Pathology in the Samsung Medical Center at the Sungkyunkwan University School of Medicine.
-Profile
Professor Kwang-Hyun Cho
ckh@kaist.ac.kr
http://sbie.kaist.ac.kr/
Department of Bio and Brain Engineering
KAIST
https://www.kaist.ac.kr
2020.01.31 View 5866
Cancer cell reversion may offer a new approach to colorectal cancer treatment
A novel approach to reverse the progression of healthy cells to malignant ones may offer a more effective way to eradicate colorectal cancer cells with far fewer side effects, according to a team of researchers based in South Korea.
Colorectal cancer, or cancer of the colon, is the third most common cancer in men and the second most common in women worldwide. South Korea has the second highest incident rate of colorectal cancer in the world, topped only by Hungary, according to the World Cancer Research Fund.
Their results were published as a featured cover article on January 2 in Molecular Cancer Research, a journal of the American Association for Cancer Research.
Led by Kwang-Hyun Cho, a professor and associate vice president of research at KAIST , the researchers used a computational framework to analyze healthy colon cells and colorectal cancer cells. They found that some master regulator proteins involved in cellular replication helped healthy colon cells mature, or differentiate into their specific cell type, and remain healthy. One particular protein, called SETDB1, suppressed the helpful proteins, forcing new cells to remain in a state of immaturity with the potential to become cancerous.
“This suggests that differentiated cells have an inherent resistance mechanism against malignant transformation and indicates that cellular reprogramming is indispensable for malignancy,” said Cho. “We speculated that malignant properties might be eradicated if the tissue-specific gene expression is reinstated — if we repress SETDB1 and allow the colon cells to mature and differentiate as they would normally.”
Image credit: Kwang-Hyun Cho, KAIST
Image restriction: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Using human-derived cells, Cho and his team targeted the tissue-specific gene expression programs identified in their computational analysis. These are the blueprints for the proteins that eventually help immature cells differentiate into tissue-specific cell types, such as colon cells. When a person has a genetic mutation, or has exposure to certain environmental factors, this process can go awry, leading to an overexpression of unhelpful proteins, such as SEDTB1.
The researchers specifically reduced the amount of SEDTB1 in these tissue-specific gene expression programs, which allowed the cells to mature and fully differentiate into colon cells.
“Our experiment also shows that SETDB1 depletion combined with cytotoxic drugs might be potentially beneficial to anticancer treatment,” Cho said. Cytotoxic drugs are often used for cancer treatment because the type of medicine contains chemicals that are toxic to cancer cells which can prevent them from replicating or growing. He noted that this combination could be more effective in treating cancer by transforming the cancer cell state into a less malignant or resistant state. He eventually pursues a cancer reversion therapy alone instead of conventional cytotoxic drug therapy since the cancer reversion therapy can provide a much less painful experience for patients with cancer who often have severe side effects from treatments intended to kill off cancerous cells, such as chemotherapy.
The researchers plan to continue studying how to return cancer cells to healthier states, with the ultimate goal of translating their work to therapeutic treatment for patients with colorectal cancer.
“I think our study of cancer reversion would eventually change the current medical practice of treating cancer toward the direction of keeping the patient’s quality of life while minimizing the side effects of current anti-cancer therapies,” Cho said.
###
This work was funded by KAIST and the National Research Foundation of Korea grants funded by the Korean government, the Ministry of Science and Information and Communication Technology.
Other authors include Soobeom Lee, Chae Young Hwang and Dongsan Kim, all of whom are affiliated with the Laboratory for Systems Biology and Bio-Inspired Engineering in the Department of Bio and Brain Engineering at KAIST; Chansu Lee and Sung Noh Hong, both with the Department of Medicine, and Seok-Hyung Kim of the Department of Pathology in the Samsung Medical Center at the Sungkyunkwan University School of Medicine.
-Profile
Professor Kwang-Hyun Cho
ckh@kaist.ac.kr
http://sbie.kaist.ac.kr/
Department of Bio and Brain Engineering
KAIST
https://www.kaist.ac.kr
2020.01.31 View 5866 -
 Hidden Mechanism for the Suppression of Colon Cancer Identified
Published in Cell Reports : cells at the risk of causing colorectal cancer due to genetic mutation are discharged outside the colon tissue
Korean researchers have successfully identified the cancer inhibitory mechanism of the colon tissue. The discovery of the inherent defense mechanism of the colon tissues is expected to provide understanding of the cause of colorectal cancer.
The research was led by Kwang-Hyun Cho, a professor of Bio and Brain Engineering at KAIST (corresponding author) and participated by Dr. Jehun Song (the first author), as well as Dr. Owen Sansom, David Huels, and Rachel Ridgway from the Beatson Institute for Cancer Research in the UK and Dr. Walter Kolch from Conway Institute in Ireland.
The research was funded by the Ministry of Science, ICT and Future Planning and the National Research Foundation of Korea, and its results were published in the 28th March online edition of Cell Reports under the title of “The APC network regulates the removal of mutated cells from colonic crypts.”
The organism can repair damaged tissues by itself, but genetic mutations, which may cause cancer, can occur in the process of cell division s for the repair. The rapid cell division s and toxic substances from the digestive process cause a problem especially in colon crypt that has a high probability for genetic mutation.
The research team was able to find out that the colon tissues prevent cancer by rapidly discharging carcinogenic cells with genetic mutations from the colon crypt durin ga frequent tissue repair process.
This defense mechanism, which inhibits abnormal cell division s by reducing the time mutated cells reside in the crypt, is inherent in the colon.
Extensive mathematical simulation results show that the mutated cells with enhanced Wnt signaling acquire increased adhesion in comparison to the normal cells, which therefore move rapidly toward the upper part of the crypt and are discharged more easily.
If beta-catenine, the key factor in Wnt signal transduction pathway, is not degraded due to genetic mutation, the accumulated beta-catenine activates cell proliferation and increases cell adhesion. The special environment of crypt tissue and the tendency of the cells with similar adhesion to aggregate will therefore discharge the mutated cell, hence maintaining the tissue homeostasis.
In vivo experiment with a mouse model confirms the simulation results that, in the case of abnormal crypt, the cells with high proliferation in fact move slower.
Professor Cho said, “This research has identified that multicellular organism is exquisitely designed to maintain the tissue homeostasis despite abnormal cell mutation. This also proves the systems biology research, which is a convergence of information technology and bio-technology , can discover hidden mechanisms behind complex biological phenomena.”
Crypt: Epithelium, consisting of approximately 2,000 cells, forms a colon surface in the shape of a cave.
Wnt Signaling: A signal transduction pathway involved in the proliferation and differentiation of cells that are particularly important for the embryonic development and management of adult tissue homeostasis.
2014.04.17 View 11223
Hidden Mechanism for the Suppression of Colon Cancer Identified
Published in Cell Reports : cells at the risk of causing colorectal cancer due to genetic mutation are discharged outside the colon tissue
Korean researchers have successfully identified the cancer inhibitory mechanism of the colon tissue. The discovery of the inherent defense mechanism of the colon tissues is expected to provide understanding of the cause of colorectal cancer.
The research was led by Kwang-Hyun Cho, a professor of Bio and Brain Engineering at KAIST (corresponding author) and participated by Dr. Jehun Song (the first author), as well as Dr. Owen Sansom, David Huels, and Rachel Ridgway from the Beatson Institute for Cancer Research in the UK and Dr. Walter Kolch from Conway Institute in Ireland.
The research was funded by the Ministry of Science, ICT and Future Planning and the National Research Foundation of Korea, and its results were published in the 28th March online edition of Cell Reports under the title of “The APC network regulates the removal of mutated cells from colonic crypts.”
The organism can repair damaged tissues by itself, but genetic mutations, which may cause cancer, can occur in the process of cell division s for the repair. The rapid cell division s and toxic substances from the digestive process cause a problem especially in colon crypt that has a high probability for genetic mutation.
The research team was able to find out that the colon tissues prevent cancer by rapidly discharging carcinogenic cells with genetic mutations from the colon crypt durin ga frequent tissue repair process.
This defense mechanism, which inhibits abnormal cell division s by reducing the time mutated cells reside in the crypt, is inherent in the colon.
Extensive mathematical simulation results show that the mutated cells with enhanced Wnt signaling acquire increased adhesion in comparison to the normal cells, which therefore move rapidly toward the upper part of the crypt and are discharged more easily.
If beta-catenine, the key factor in Wnt signal transduction pathway, is not degraded due to genetic mutation, the accumulated beta-catenine activates cell proliferation and increases cell adhesion. The special environment of crypt tissue and the tendency of the cells with similar adhesion to aggregate will therefore discharge the mutated cell, hence maintaining the tissue homeostasis.
In vivo experiment with a mouse model confirms the simulation results that, in the case of abnormal crypt, the cells with high proliferation in fact move slower.
Professor Cho said, “This research has identified that multicellular organism is exquisitely designed to maintain the tissue homeostasis despite abnormal cell mutation. This also proves the systems biology research, which is a convergence of information technology and bio-technology , can discover hidden mechanisms behind complex biological phenomena.”
Crypt: Epithelium, consisting of approximately 2,000 cells, forms a colon surface in the shape of a cave.
Wnt Signaling: A signal transduction pathway involved in the proliferation and differentiation of cells that are particularly important for the embryonic development and management of adult tissue homeostasis.
2014.04.17 View 11223 -
 Professor Kwang-Hyun Cho publishes Encyclopaedia of Systems Biology
Professor Kwang-Hyun Cho
KAIST Biological and Brain Engineering Department’s Professor Kwang-Hyun Cho edited the Encyclopaedia of Systems Biology with three scholars, all experts of Systems Biology in England, Germany and the United States. It is rare that a Korean scientist edits a world renowned academic science encyclopaedia.
The Encyclopaedia, published by the New York office of Springer Verlag, was a grand international project five years in the making by 28 editors and 391 scientists with expertise in Systems Biology from around the world. The Encyclopaedia compiles various research areas of Systems Biology, the new academic paradigm of the 21st century through the integration of IT and BT, comprehensively on 3,000 pages in 4 four volumes.
Professor Kwang-Hyun Cho, who led this international project, majored in electrical engineering and pioneered the field of Systems Biology, the integrated study of biological sciences and engineering, as a new integrated field of IT since the 1990s. The professor has achieved various innovative research results since then. Recently he has investigated “kernel,” an evolutionary core structure in complex biological networks and developed a new cancer treatment through the state space analysis of the molecular network of cancer cells. His work was published in Science Signalling, a sister journal of Science, as a cover story several times, and contributed to foundational research as well as commercialisation of the integrated fields of IT and BT.
2013.08.27 View 8776
Professor Kwang-Hyun Cho publishes Encyclopaedia of Systems Biology
Professor Kwang-Hyun Cho
KAIST Biological and Brain Engineering Department’s Professor Kwang-Hyun Cho edited the Encyclopaedia of Systems Biology with three scholars, all experts of Systems Biology in England, Germany and the United States. It is rare that a Korean scientist edits a world renowned academic science encyclopaedia.
The Encyclopaedia, published by the New York office of Springer Verlag, was a grand international project five years in the making by 28 editors and 391 scientists with expertise in Systems Biology from around the world. The Encyclopaedia compiles various research areas of Systems Biology, the new academic paradigm of the 21st century through the integration of IT and BT, comprehensively on 3,000 pages in 4 four volumes.
Professor Kwang-Hyun Cho, who led this international project, majored in electrical engineering and pioneered the field of Systems Biology, the integrated study of biological sciences and engineering, as a new integrated field of IT since the 1990s. The professor has achieved various innovative research results since then. Recently he has investigated “kernel,” an evolutionary core structure in complex biological networks and developed a new cancer treatment through the state space analysis of the molecular network of cancer cells. His work was published in Science Signalling, a sister journal of Science, as a cover story several times, and contributed to foundational research as well as commercialisation of the integrated fields of IT and BT.
2013.08.27 View 8776 -
 Prof. Cho Elected Editor-in-Chief of Systems Biology
Prof. Kwang-Hyun Cho of Department of Bio and Brain Engineering at KAIST has been recently elected editor-in-chief of the Systems Biology, an international journal published by the London-based Institution of Engineering and Technology (IET), the university authorities said on Wednesday (Sept. 23)
By the year 2012, Cho will oversee the editorial process of the journal covering intra- and inter-cellular dynamics, using systems- and signal-oriented approaches. IET, one of the world"s leading professional societies for the engineering and technology community, has a worldwide membership of more than 150,000.
Prof. Cho"s research interests cover the areas of systems science with bio-medical applications including systems biology and bio-inspired engineering based on molecular systems biology. He is currently an editorial board member of Systems and Synthetic Biology (Springer, Netherlands, from 2006), BMC Systems Biology (BMC, London, U.K., from 2007), Gene Regulation and Systems Biology (Libertas Academica, New Zealand, from 2007), and Bulletin of Mathematical Biology (Springer, New York, from 2008), and an editorial advisory board member of Molecular BioSystems (The Royal Society of Chemistry, U.K.).
2009.09.24 View 14663
Prof. Cho Elected Editor-in-Chief of Systems Biology
Prof. Kwang-Hyun Cho of Department of Bio and Brain Engineering at KAIST has been recently elected editor-in-chief of the Systems Biology, an international journal published by the London-based Institution of Engineering and Technology (IET), the university authorities said on Wednesday (Sept. 23)
By the year 2012, Cho will oversee the editorial process of the journal covering intra- and inter-cellular dynamics, using systems- and signal-oriented approaches. IET, one of the world"s leading professional societies for the engineering and technology community, has a worldwide membership of more than 150,000.
Prof. Cho"s research interests cover the areas of systems science with bio-medical applications including systems biology and bio-inspired engineering based on molecular systems biology. He is currently an editorial board member of Systems and Synthetic Biology (Springer, Netherlands, from 2006), BMC Systems Biology (BMC, London, U.K., from 2007), Gene Regulation and Systems Biology (Libertas Academica, New Zealand, from 2007), and Bulletin of Mathematical Biology (Springer, New York, from 2008), and an editorial advisory board member of Molecular BioSystems (The Royal Society of Chemistry, U.K.).
2009.09.24 View 14663 -
 Prof. Cho Appointed Editor-in-Chief of Systems Biology Encyclopedia
Prof. Kwang-Hyun Cho of the Department of Bio and Brain Engineering, KAIST, has been appointed as the editor-in-chief of the Encyclopedia of Systems Biology which is currently in development by Springer, a New York-based publishing company, university authorities said on Monday (July 6).
Prof. Cho will share the position with three other eminent scholars from Britain, Germany and the United States. Cho will be responsible for selecting editorial members for each section of the Encyclopedia and overseeing the overall editorial process.
The Encyclopedia of Systems Biology is a multi-volume reference compilation of the research outcomes in the field of systems biology all over the world. The ESB will consist of alphabetically ordered description of systems biology concepts and is envisaged to ultimately comprise 6-12 volumes. Publication of the Encyclopedia is scheduled for 2011.
2009.07.08 View 12217
Prof. Cho Appointed Editor-in-Chief of Systems Biology Encyclopedia
Prof. Kwang-Hyun Cho of the Department of Bio and Brain Engineering, KAIST, has been appointed as the editor-in-chief of the Encyclopedia of Systems Biology which is currently in development by Springer, a New York-based publishing company, university authorities said on Monday (July 6).
Prof. Cho will share the position with three other eminent scholars from Britain, Germany and the United States. Cho will be responsible for selecting editorial members for each section of the Encyclopedia and overseeing the overall editorial process.
The Encyclopedia of Systems Biology is a multi-volume reference compilation of the research outcomes in the field of systems biology all over the world. The ESB will consist of alphabetically ordered description of systems biology concepts and is envisaged to ultimately comprise 6-12 volumes. Publication of the Encyclopedia is scheduled for 2011.
2009.07.08 View 12217