research

Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
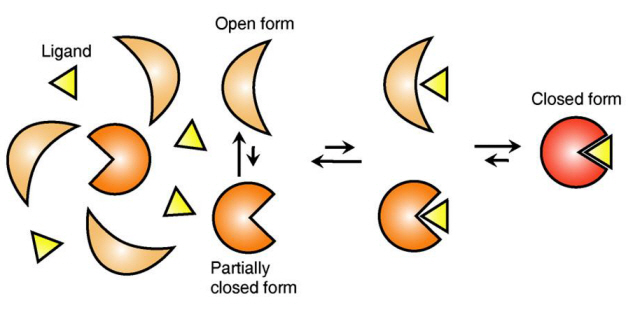
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
-
people Professor Won-Ki Cho Selected as the 2020 SUHF Young Investigator
Professor Won-Ki Cho from the Department of Biological Sciences was named one of three recipients of the 2020 Suh Kyung-Bae Science Foundation (SUHF) Young Investigator Award. The SUHF is a non-profit organization established in 2016 and funded by a personal donation of 300 billion KRW in shares from Chairman and CEO Kyung-Bae Suh of the Amorepacific Group. The primary purpose of the foundation is to serve as a platform to nurture and provide comprehensive long-term support for creative and p
2020-10-15 -
research Breastfeeding Helps Prevent Mothers from Developing Diabetes after Childbirth
A team of South Korean researchers found that lactation can lower the incidence and reduce the risk of maternal postpartum diabetes. The researchers identified that lactation increases the mass and function of pancreatic beta cells through serotonin production. The team suggested that sustained improvements in pancreatic beta cells, which can last for years even after the cessation of lactation, improve mothers’ metabolic health in addition to providing health benefits for infants. Pre
2020-04-29 -
research Highly Efficient and Stable Double Layer Solar Cell Developed
Solar cells convert light into energy, but they can be inefficient and vulnerable to the environment, degrading with, ironically, too much light or other factors, including moisture and low temperature. An international research team has developed a new type of solar cell that can both withstand environmental hazards and is 26.7% efficient in power conversion. They published their results on March 26 in Science. The researchers, led by Byungha Shin, a professor from the Department of Materia
2020-03-27 -
policy KAIST GSAI and SNUBH Join Hands for AI in Healthcare
< Dean Song Chong (left) and Director Chang Wan Oh (right) at the KAIST GSAI - SNUBH MOU Signing Ceremony > The Graduate School of AI (GSAI) at KAIST and the Seoul National University Bundang Hospital (SNUBH) signed a memorandum of understanding (MOU) to cooperate in AI education and research in the field of healthcare last month. The two institutions have agreed to collaborate on research and technology development through the implementation of academic and personnel exchange progr
2019-12-27 -
research Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke (Professor Young Seok Ju at the Graduate School of Medical Science and Engineering) Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancer
2019-05-31