research
Binding Regulatory Mechanism of Protein Biomolecules Revealed
View : 9289
Date : 2014-05-02
Writer : ed_news

Professor Hak-Sung Kim
A research team led by Professor Hak-Sung Kim of Biological Sciences, KAIST, and Dr. Mun-Hyeong Seo, KAIST, has revealed a regulatory mechanism that controls the binding affinity of protein’s biomolecules, which is crucial for the protein to recognize molecules and carry out functions within the body.
The research results were published in the April 24th online edition of Nature Communications.
The protein, represented by enzyme, antibody, or hormones, specifically recognizes a variety of biomolecules in all organisms and implements signaling or immune response to precisely adjust and maintain important biological processes. The protein binding affinity of biomolecules plays a crucial role in determining the duration of the bond between two molecules, and hence to determine and control the in-vivo function of proteins.
The researchers have noted that, during the process of proteins’ recognizing biomolecules, the protein binding affinity of biomolecules is closely linked not only to the size of non-covalent interaction between two molecules, but also to the unique kinetic properties of proteins.
To identify the basic mechanism that determines the protein binding affinity of biomolecules, Professor Kim and his research team have made mutation in the allosteric site of protein to create a variety of mutant proteins with the same chemical binding surface, but with the binding affinity vastly differing from 10 to 100 times. The allosteric site of the protein refers to a region which does not directly bind with biomolecules, but crucially influences the biomolecule recognition site.
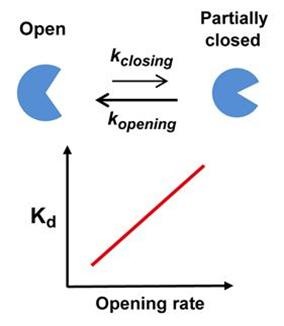
Using real-time analysis at the single-molecule level of unique kinetic properties of the produced mutant proteins, the researchers were able to identify that the protein binding affinity of biomolecules is directly associated with the protein’s specific kinetic characteristics, its structure opening rate.
Also, by proving that unique characteristics of the protein can be changed at the allosteric site, instead of protein’s direct binding site with biomolecules, the researchers have demonstrated a new methodology of regulating the in-vivo function of proteins.
The researchers expect that these results will contribute greatly to a deeper understanding of protein’s nature that governs various life phenomena and help evaluate the proof of interpreting protein binding affinity of biomolecules from the perspective of protein kinetics.
Professor Kim said, “Until now, the protein binding affinity of biomolecules was determined by a direct interaction between two molecules. Our research has identified an important fact that the structure opening rate of proteins also plays a crucial role in determining their binding affinity.”
[Picture]
A correlation graph of opening rate (kopening) and binding affinity (kd) between protein’s stable, open state and its unstable, partially closed state.
Releated news
- No Data