research
Professor Tae-Young Yoon’s research team from the Department of Physics at KAIST has developed a new method to form DNA nanostructures by using magnetic tweezers to observe and to induce the formation of the structure in real time.
Unlike traditional designs of "DNA origami" which relies on thermal or chemical annealing methods, the new technology utilizes a completely different dynamic in DNA folding. This allows the folding to be done within only ten minutes.
Developed in 2006, the "DNA origami" allows a long skeleton of DNA to be folded into an arbitrary structure by using small stapler DNA pieces. This has been a prominent method in DNA nanotechnology.
However, the traditional technology which adopts thermal processes could not control the DNA formation during the folding because every interaction among DNAs occurs simultaneously. Thus, the thermal processes, which take dozens of hours to complete, had to be repeated multiple times in order to find the optimal condition.
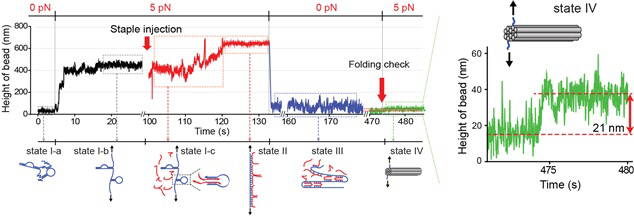
The research team designed a DNA folding using uni-molecular magnetic tweezers that applied force to a single DNA molecule while measuring the state of the DNA. Through this technology, they were able to induce the formation of DNA nanostructure and observe it at the same time.
During high temperature heat treatment, the first stage of conventional thermal processes, the internal structure of the long skeleton DNA untangles. To induce such state, after attaching one side of the skeleton DNA to the surface of glass and the other side to a magnetic material, the team unfolded the internal structure of the DNA by pulling the two sides apart with magnetic force.
Unlike the conventional thermal processes, this method lets the stapler DNA swiftly adhere to the skeleton DNA within a minute because the sites are revealed at room temperature.
After the stapler pieces connected to the skeleton, the team removed the magnetic force. Next, the structure folded through self-assembly as the stapler DNAs stuck to different sites on the skeleton DNA.
Professor Yoon said, “With the existing thermal methods, we could not differentiate the reactions of the DNA because the response of each DNA pieces mutually interacted with each other.” He added that “Using the magnetic tweezers, we were able to sort the process of DNA nanostructure formation into a series of reactions of DNA molecules that are well known, and shorten the time taken for formation in only ten minutes.”
He commented, “This nanostructure formation method will enable us to create more intricate and desirable DNA nanostructures by programming the folding of DNA origami structures.”
Conducted by Dr. Woori Bae under the guidance of Professor Yoon, the research findings were published online in the December 4th issue of Nature Communications.
Figure 1: Uni-molecular magnetic tweezers orchestrating the DNA nanostructure formation

Figure 2: The evolution of DNA nanostructure formation using magnetic tweezers. The DNA nanostructure with a 21-nanometer size was formed in about eight minutes.
-
research A Hole in One for Holographic Display
(Professor YongKeun Park) Researchers have designed an ultrathin display that can project dynamic, multi-coloured, 3D holographic images, according to a study published in Nature Communications. The system’s critical component is a thin film of titanium filled with tiny holes that precisely correspond with each pixel in a liquid crystal display (LCD) panel. This film acts as a ‘photon sieve’ – each pinhole diffracts light emerging from them widely, resulting in a
2019-04-18 -
research Unravelling Inherent Electrocatalysis to Improve the Performance of Hydrogen Fuel Cells
(Figure 1. Electrode structure for the precise evaluation of the metal nanoparticles’ electrochemical catalytic characteristics at a high temperature.) A KAIST team presented an ideal electrode design to enhance the performance of high-temperature fuel cells. The new analytical platform with advanced nanoscale patterning method quantitatively revealed the electrochemical value of metal nanoparticles dispersed on the oxide electrode, thus leading to electrode design directions that c
2019-03-28 -
research New Catalyst for Synthesizing Chiral Molecules Selectively
(from left: Dr. Yoonsu Park and Professor Sukbok Chang from the Department of Chemistry) Molecules in nature often have “twin” molecules that look identical. In particular, the twin molecules that look like mirror images to each other are called enantiomers. However, even though they have the same type and number of elements, these twin molecules exhibit completely different properties. Professor Sukbok Chang and Dr. Yoonsu Park from the Department of Chemistry
2019-03-05 -
research New LSB with Theoretical Capacity over 90%
(Professor Hee-Tak Kim and Hyunwon Chu) A KAIST research team has developed a lithium sulfur battery (LSB) that realizes 92% of the theoretical capacity and an areal capacity of 4mAh/cm2. LSBs are gaining a great deal of attention as an alternative for lithium ion batteries (LIBs) because they have a theoretical energy density up to six to seven times higher than that of LIBs, and can be manufactured in a more cost-effective way. However, LSBs face the obstacle of
2019-02-11 -
research Noninvasive Light-Sensitive Recombinase for Deep Brain Genetic Manipulation
A KAIST team presented a noninvasive light-sensitive photoactivatable recombinase suitable for genetic manipulation in vivo. The highly light-sensitive property of photoactivatable Flp recombinase will be ideal for controlling genetic manipulation in deep mouse brain regions by illumination with a noninvasive light-emitting diode. This easy-to-use optogenetic module made by Professor Won Do Heo and his team will provide a side-effect free and expandable genetic manipulation tool for neurosci
2019-01-22