research
Our bodies have a variety of biological clocks that follow rhythms or oscillations with periods ranging from seconds to days. For example, our hearts beat every second, and cells divide periodically. The circadian clock located in the hypothalamus generates twenty-four hour rhythms, timing our sleep and hormone release. How do these biological clocks or circuits generate and sustain the stable rhythms that are essential to life?
Jae Kyoung Kim, who is an assistant professor in the Department of Mathematical Sciences at KAIST, has predicted how these biological circuits generate rhythms and control their robustness, utilizing mathematical modeling based on differential equations and stochastic parameter sampling. Based on his prediction, using synthetic biology, a research team headed by Matthew Bennett of Rice University constructed a novel biological circuit that spans two genetically engineered strains of bacteria, one serves as an activator and the other as a repressor to regulate gene expression across multiple cell types, and found that the circuit generates surprisingly robust rhythms under various conditions.
The results of the research conducted in collaboration with KAIST (Korea Institute of Science and Technology), Rice University, and the University of Houston were published in Science (August 28, 2015 issue). The title of the paper is "Emergent Genetic Oscillations in a Synthetic Microbial Consortium" .
The top-down research approach, which focuses on identifying the components of biological circuits, limits our understanding of the mechanisms in which the circuits generate rhythms. Synthetic biology, a rapidly growing field at the interface of biosciences and engineering, however, uses a bottom-up approach.
Synthetic biologists can create complex circuits out of simpler components, and some of these new genetic circuits are capable of fluctuation to regulate gene production. In the same way that electrical engineers understand how an electrical circuit works as they construct batteries, resistors, and wires, synthetic biologists can understand better about biological circuits if they put them together using genes and proteins. However, due to the complexity of biological systems, both experiments and mathematical modeling need to be applied hand in hand to design these biological circuits and understand their function.
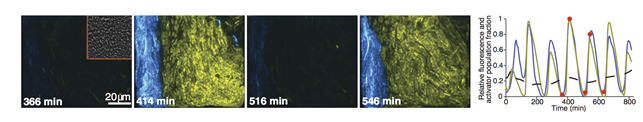
In this research, an interdisciplinary approach proved that a synthetic intercellular singling circuit generates robust rhythms to create a cooperative microbial system. Specifically, Kim's mathematical analysis suggested, and experiments confirmed, that the presence of negative feedback loops in addition to a core transcriptional negative feedback loop can explain the robustness of rhythms in this system. This result provides important clues about the fundamental mechanism of robust rhythm generation in biological systems.
Furthermore, rather than constructing the entire circuit inside a single bacterial strain, the circuit was split among two strains of Escherichia coli bacterium. When the strains were grown together, the bacteria exchanged information, completing the circuit. Thus, this research also shows how, by regulating individual cells within the system, complex biological systems can be controlled, which in turn influences each other (e.g., the gut microbiome in humans).
###
Ye Chen, a graduate student in Bennett's laboratory at Rice University, and Jae Kyoung Kim, an assistant professor at KAIST and a former postdoctoral fellow at Ohio State University, are the lead authors of the paper. The co-authors are Rice graduate student Andrew Hirning and Krešimir Josic?, a professor of mathematics at the University of Houston. Bennett is the Assistant Professor of the Biochemistry and Cell Biology Department at Rice University.
About the researcher: While Jae Kyoung Kim is a mathematician, he has also solved various biological puzzles in collaboration with various experimental laboratories of Matthew Bennett at Rice University, David Virshup at Duke and the National University of Singapore, Carla Finkielstein at Virginia Polytechnic Institute and State University, Choo-Gon Lee at the Florida State University, Seung-Hee Yoo at the Medical School of the University of Texas, Toru Takumi at RIKEN Brain Science Institute, and Travis Wager at Pfizer Inc. He has used non-linear dynamics and stochastic analysis to understand the function of biochemical networks in biological systems. In particular, he is interested in mechanisms generating and regulating biological rhythms.
Picture 1: This schematic image is the design of a biological circuit between two strains of bacteria and the part of differential equations used to understand the function of the biological circuit.

Picture 2: The core transcriptional negative feedback loop and additional negative feedback loop in the biological circuit (picture 1) generate robust rhythms. The snapshots correspond the red dots in the time series graph.
- No Data