research
Research led by Korean medical scientists has discovered that a specific type of drug resistance mechanism to EGFR inhibitor therapy in lung cancer is predisposed by mutations in two canonical cancer-related genes: RB1 and TP53. Published in Journal of Clinical Oncology on May 12, the study also found those mutations can be detectable in patients' tumors at the point of clinical diagnosis. Therefore, it can be used as strong markers in clinic for predicting poor outcome for the targeted treatment for lung adenocarcinoma.
Lung adenocarcinoma is the most common type of lung cancer, and about 15% of patients in Western countries and 50% of patients in Asian countries have mutations in the EGFR gene, which is critical for the development of lung cancer. Patients with lung adenocarcinoma harboring the EGFR mutation show favorable responses to EGFR inhibitors such as erlotinib (Tarceva) or gefitinib (Iressa), but ultimately relapse with drug-resistant tumors. Since the initial report in 2006, it has been known that in about 5~15% of patients, the lung adenocarcinoma cells undergo a mysterious transformation into a very different cancer cell type called “small cell lung cancer,” a much more aggressive lung cancer subtype, common in cigarette smokers.
To find out the genetic basis of this process, the researchers compared the genome sequences of multiple cancer tissues acquired during the treatment courses of patients whose tumors underwent small-cell transformation. They reconstructed the cancer cell pedigree by comparing mutations between cancer tissues, and identified that RB1 and TP53 genes are completely inactivated by mutations already in their lung adenocarcinoma tissues.
"We tried to compare the somatic mutational profile of pre-EGFR inhibitor treatment lung adenocarcinomas and post-treatment small cell carcinomas and to reconstruct the pedigrees of the cancer evolution in each patient. Strikingly, both copies of RB1 and TP53 genes were already inactivated at the stage of lung adenocarcinomas in all sequenced cases," said Dr. Jake June-Koo Lee, the first author from KAIST.
They further pursued the clinical implications of RB1 and TP53 inactivation by investigating 75 EGFR-mutated lung adenocarcinoma tissues from patients who received EGFR inhibitor therapy, including patients with small-cell transformation. In this analysis, the lung adenocarcinomas with a complete inactivation of both RB1 and TP53 genes tended to have a 43-times greater risk of transformation into small cell lung cancer during their EGFR inhibitor treatment courses.
Dr. Young Seok Ju, the co-last author from KAIST, explained, "This study shows the power of entire genome analyses to better understand the mechanisms underlying mysterious phenomenon encountered in clinic. Upon accurate bioinformatics, we are finding cancer-specific somatic mutations from the whole-genomes of patients’ cancer cells. These mutations allow us to track the evolution of cancer cells throughout the extraordinary clinical course of a special set of lung cancers."
The complete inactivation of both RB1 and TP53 tumor suppressor genes is found in a minor (<10%) subset of lung adenocarcinoma. This study suggests that the clinical course against targeted therapy is endogenously different for the cancers in the subgroup, and specific drug-resistance mechanisms are predisposed by the two genetic mutations. Indeed, RB1 and TP53 double inactivation is a genetic hallmark of primary small cell lung cancer, observed in nearly all cases.
"We are actively investigating patient tumor tissues to develop optimal surveillance plans and treatment options for patients with lung adenocarcinomas more prone to small-cell transformation," said Dr. Tae Min Kim, the co-last author from Seoul National University Hospital.
The researchers are implementing their findings into lung cancer clinics by screening the RB1 and TP53 mutational status in lung adenocarcinoma patients receiving EGFR inhibitor treatment, and following their treatment courses to develop a treatment strategy for those patients.
This research (doi.org/10.1200/JCO.2016.71.9096) was funded by the National Research Foundation of Korea (NRF-2013H1A2A1032691 to J.-K.L., NRF-2014R1A2A2A05003665 to Y.T.K.); Korea Institute of Science and Technology Information (K-16-L03-C02-S02 to J.L.); and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, which was funded by the Ministry of Health and Welfare (HI14C1234 to T.M.K., HI16C2387 to Y.S.J.)
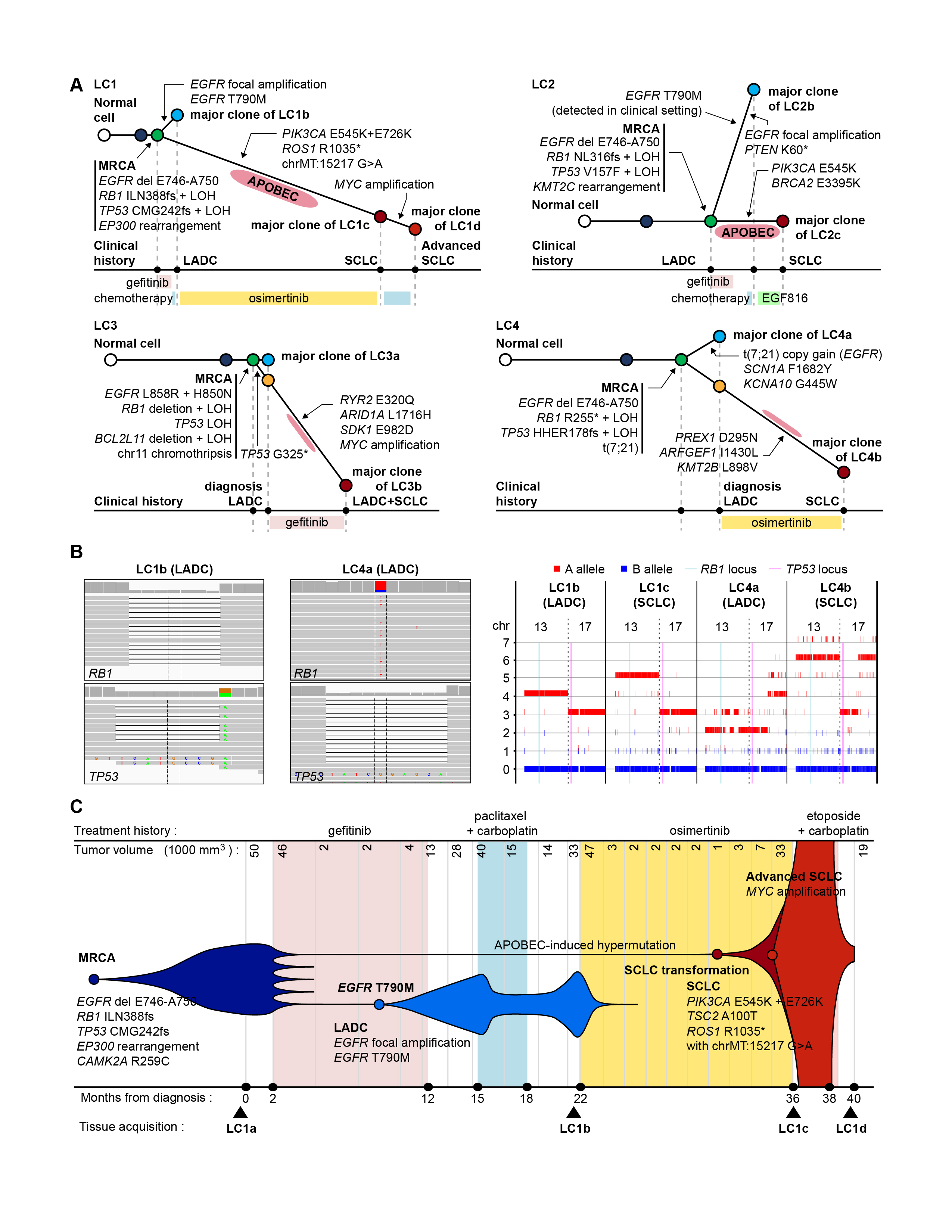
A. Phylogeny trees of sequenced cases (LC1−LC4) are reconstructed from the WGS data. Conceptual illustrations are depicted with grey color. Circles indicate major clones of the tumors. The length of each branch is proportional to the number of mutations that occurred in the branch. Mutations of cancer-related genes in each branch are indicated with arrows. The time points of relevant treatments are summarized below the trees.
B. Mutations of RB1 and TP53 in two early LADCs (LC1b and LC4a) are visualized using Integrative Genomics Viewer (left panel). Allele-specific copy number analysis shows loss of heterozygosity of chromosomes 13 and 17 in both early LADCs and EGFR TKI-resistant SCLCs (right panel).
C. Clonal evolution of LC1 is described with clinical history and tumor volumes. The horizontal axis represents the time from the diagnosis (0), and the vertical axis indicates the volume of tumors calculated from the computed tomography images.
Abbreviations: LADC, lung adenocarcinoma; SCLC, small cell lung cancer
-
event KAIST ISSS Research Session Captivates 150↑ International Scholars, Achieve Major Success
< Photo. Scholars gatheres for NRF Information Session at Chung Keun Mo Hall > KAIST’s International Office, headed by Vice President Soyoung Kim, successfully organized the ‘NRF Information Session for International Scholars’ on September 11, 2024, in collaboration with the National Research Foundation of Korea (NRF). The event was held at KAIST’s main campus to enourage the international scholar’s active participation in research projects and suppor
2024-09-13 -
research KAIST Research Team Proves How a Neurotransmitter may be the Key in Controlling Alzheimer’s Toxicity
With nearly 50 million dementia patients worldwide, and Alzheimers’s disease is the most common neurodegenerative disease. Its main symptom is the impairment of general cognitive abilities, including the ability to speak or to remember. The importance of finding a cure is widely understood with increasingly aging population and the life expectancy being ever-extended. However, even the cause of the grim disease is yet to be given a clear definition. A KAIST research team in the Departme
2022-07-29 -
people Professor Byungha Shin Named Scientist of the Month
Professor Byungha Shin from the Department of Materials Science and Engineering won the Scientist of the Month Award presented by the Ministry of Science and ICT (MSIT) and the National Research Foundation of Korea (NRF) on May 4. Professor Shin was recognized for his research in the field of next-generation perovskite solar cells and received 10 million won in prize money. To achieve ‘carbon neutrality,’ which many countries across the globe including Korea hope to realize, the
2021-05-07 -
people Professor Bumjoon Kim Named Scientist of the Month
Professor Bumjoon Kim from the Department of Chemical and Biomolecular Engineering won January’s Scientist of the Month Award presented by the Ministry of Science and ICT (MSIT) and the National Research Foundation of Korea (NRF) on January 6. Professor Kim also received 10 million won in prize money. Professor Kim was recognized for his research in the field of fuel cells. Since the first paper on fuel cells was published in 1839 by the German chemist Friedrich Schonbein, there has bee
2021-01-22 -
people Scientist of October: Professor Jungwon Kim
Professor Jungwon Kim from the Department of Mechanical Engineering was selected as the ‘Scientist of the Month’ for October 2020 by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Kim was recognized for his contributions to expanding the horizons of the basics of precision engineering through his research on multifunctional ultrahigh-speed, high-resolution sensors. He received 10 million KRW in prize money. Professor Kim was selected as th
2020-10-15