research
Despite decades of intensive cancer research by numerous biomedical scientists, cancer still holds its place as the number one cause of death in Korea. The fundamental reason behind the limitations of current cancer treatment methods is the fact that they all aim to completely destroy cancer cells, which eventually allows the cancer cells to acquire immunity. In other words, recurrences and side-effects caused by the destruction of healthy cells are inevitable. To this end, some have suggested anticancer treatment methods based on cancer reversion, which can revert cancer cells back to normal or near-normal cells under certain conditions. However, the practical development of this idea has not yet been attempted.
On June 8, a KAIST research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering reported to have successfully identified the fundamental principle of a process that can revert cancer cells back to normal cells without killing the cells.
Professor Cho’s team focused on the fact that unlike normal cells, which react according to external stimuli, cancer cells tend to ignore such stimuli and only undergo uncontrolled cell division. Through computer simulation analysis, the team discovered that the input-output (I/O) relationships that were distorted by genetic mutations could be reverted back to normal I/O relationships under certain conditions. The team then demonstrated through molecular cell experiments that such I/O relationship recovery also occurred in real cancer cells.
The results of this study, written by Dr. Jae Il Joo and Dr. Hwa-Jeong Park, were published in Wiley’s Advanced Science online on June 2 under the title, "Normalizing input-output relationships of cancer networks for reversion therapy."
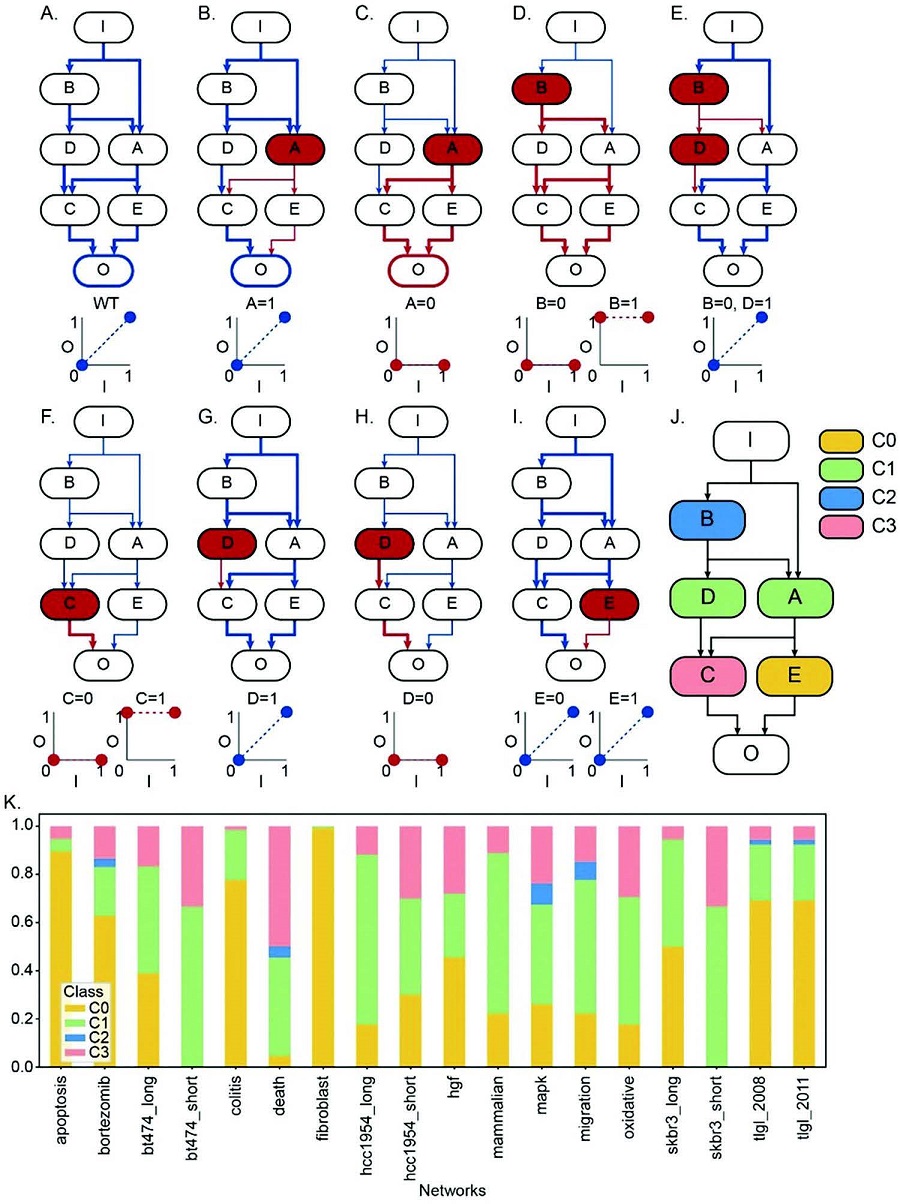
< Image 1. Input-output (I/O) relationships in gene regulatory networks >
Professor Kwang-Hyun Cho's research team classified genes into four types by simulation-analyzing the effect of gene mutations on the I/O relationship of gene regulatory networks. (Figure A-J) In addition, by analyzing 18 genes of the cancer-related gene regulatory network, it was confirmed that when mutations occur in more than half of the genes constituting each network, reversibility is possible through appropriate control. (Figure K)
Professor Cho’s team uncovered that the reason the distorted I/O relationships of cancer cells could be reverted back to normal ones was the robustness and redundancy of intracellular gene control networks that developed over the course of evolution. In addition, they found that some genes were more promising as targets for cancer reversion than others, and showed through molecular cell experiments that controlling such genes could revert the distorted I/O relationships of cancer cells back to normal ones.

< Image 2. Simulation results of restoration of bladder cancer gene regulation network and I/O relationship of bladder cancer cells. >
The research team classified the effects of gene mutations on the I/O relationship in the bladder cancer gene regulation network by simulation analysis and classified them into 4 types. (Figure A) Through this, it was found that the distorted input-output relationship between bladder cancer cell lines KU-1919 and HCT-1197 could be restored to normal. (Figure B)
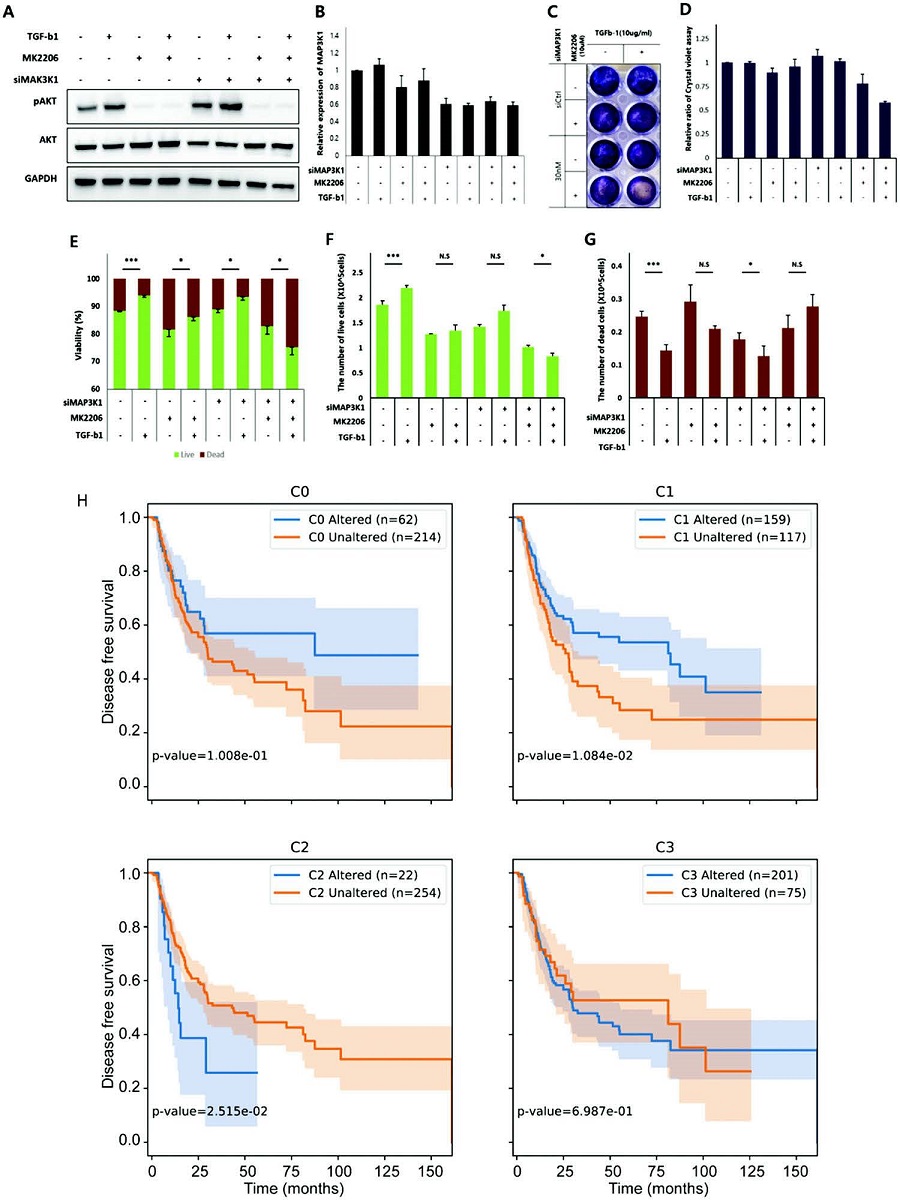
< Image 3. Analysis of survival of bladder cancer patients according to reversible gene mutation and I/O recovery experiment of bladder cancer cells. >
As predicted through network simulation analysis, Professor Kwang-Hyun Cho's research team confirmed through molecular cell experiments that the response to TGF-b was normally restored when AKT and MAP3K1 were inhibited in the bladder cancer cell line KU-1919. (Figure A-G) In addition, it was confirmed that there is a difference in the survival rate of bladder cancer patients depending on the presence or absence of a reversible gene mutation. (Figure H)
The results of this research show that the reversion of real cancer cells does not happen by chance, and that it is possible to systematically explore targets that can induce this phenomenon, thereby creating the potential for the development of innovative anticancer drugs that can control such target genes.
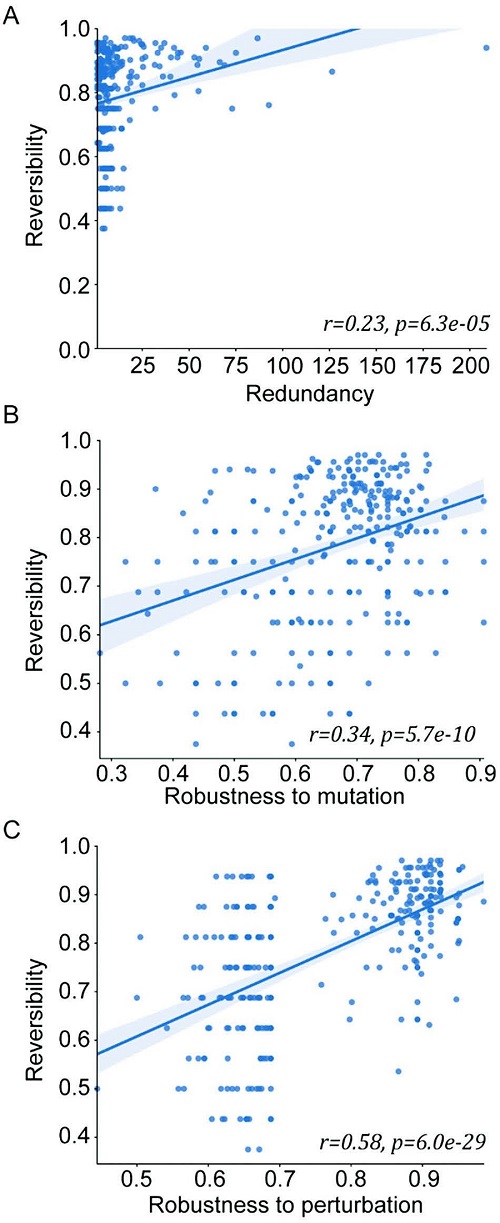
< Image 4. Cancer cell reversibility principle >
The research team analyzed the reversibility, redundancy, and robustness of various networks and found that there was a positive correlation between them. From this, it was found that reversibility was additionally inherent in the process of evolution in which the gene regulatory network acquired redundancy and consistency.
Professor Cho said, “By uncovering the fundamental principles of a new cancer reversion treatment strategy that may overcome the unresolved limitations of existing chemotherapy, we have increased the possibility of developing new and innovative drugs that can improve both the prognosis and quality of life of cancer patients.”

< Image 5. Conceptual diagram of research results >
The research team identified the fundamental control principle of cancer cell reversibility through systems biology research. When the I/O relationship of the intracellular gene regulatory network is distorted by mutation, the distorted I/O relationship can be restored to a normal state by identifying and adjusting the reversible gene target based on the redundancy of the molecular circuit inherent in the complex network.
After Professor Cho’s team first suggested the concept of reversion treatment, they published their results for reverting colorectal cancer in January 2020, and in January 2022 they successfully re-programmed malignant breast cancer cells back into hormone-treatable ones. In January 2023, the team successfully removed the metastasis ability from lung cancer cells and reverted them back to a state that allowed improved drug reactivity. However, these results were case studies of specific types of cancer and did not reveal what common principle allowed cancer reversion across all cancer types, making this the first revelation of the general principle of cancer reversion and its evolutionary origins.
This research was funded by the Ministry of Science and ICT of the Republic of Korea and the National Research Foundation of Korea.
- No Data