Bio-Inspired+Engineering
-
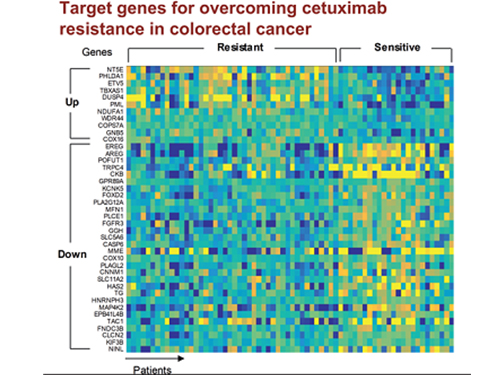 5 Biomarkers for Overcoming Colorectal Cancer Drug Resistance Identified
< Professor Kwang-Hyun Cho's Team >
KAIST researchers have identified five biomarkers that will help them address resistance to cancer-targeting therapeutics. This new treatment strategy will bring us one step closer to precision medicine for patients who showed resistance.
Colorectal cancer is one of the most common types of cancer worldwide. The number of patients has surpassed 1 million, and its five-year survival rate significantly drops to about 20 percent when metastasized. In Korea, the surge of colorectal cancer has been the highest in the last 10 years due to increasing Westernized dietary patterns and obesity. It is expected that the number and mortality rates of colorectal cancer patients will increase sharply as the nation is rapidly facing an increase in its aging population.
Recently, anticancer agents targeting only specific molecules of colon cancer cells have been developed. Unlike conventional anticancer medications, these selectively treat only specific target factors, so they can significantly reduce some of the side-effects of anticancer therapy while enhancing drug efficacy.
Cetuximab is the most well-known FDA approved anticancer medication. It is a biomarker that predicts drug reactivity and utilizes the presence of the ‘KRAS’ gene mutation. Cetuximab is prescribed to patients who don’t carry the KRAS gene mutation.
However, even in patients without the KRAS gene mutation, the response rate of Cetuximab is only about fifty percent, and there is also resistance to drugs after targeted chemotherapy. Compared with conventional chemotherapy alone, the life expectancy only lasts five months on average.
In research featured in the FEBS Journal as the cover paper for the April 7 edition, the KAIST research team led by Professor Kwang-Hyun Cho at the Department of Bio and Brain Engineering presented five additional biomarkers that could increase Cetuximab responsiveness using systems biology approach that combines genomic data analysis, mathematical modeling, and cell experiments. The experimental inhibition of newly discovered biomarkers DUSP4, ETV5, GNB5, NT5E, and PHLDA1 in colorectal cancer cells has been shown to overcome Cetuximab resistance in KRAS-normal genes. The research team confirmed that when suppressing GNB5, one of the new biomarkers, it was shown to overcome resistance to Cetuximab regardless of having a mutation in the KRAS gene.
Professor Cho said, “There has not been an example of colorectal cancer treatment involving regulation of the GNB5 gene.” He continued, “Identifying the principle of drug resistance in cancer cells through systems biology and discovering new biomarkers that could be a new molecular target to overcome drug resistance suggest real potential to actualize precision medicine.”
This study was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT (2017R1A2A1A17069642 and 2015M3A9A7067220).
Image 1. The cover of FEBS Journal for April 2019
2019.05.27 View 60989
5 Biomarkers for Overcoming Colorectal Cancer Drug Resistance Identified
< Professor Kwang-Hyun Cho's Team >
KAIST researchers have identified five biomarkers that will help them address resistance to cancer-targeting therapeutics. This new treatment strategy will bring us one step closer to precision medicine for patients who showed resistance.
Colorectal cancer is one of the most common types of cancer worldwide. The number of patients has surpassed 1 million, and its five-year survival rate significantly drops to about 20 percent when metastasized. In Korea, the surge of colorectal cancer has been the highest in the last 10 years due to increasing Westernized dietary patterns and obesity. It is expected that the number and mortality rates of colorectal cancer patients will increase sharply as the nation is rapidly facing an increase in its aging population.
Recently, anticancer agents targeting only specific molecules of colon cancer cells have been developed. Unlike conventional anticancer medications, these selectively treat only specific target factors, so they can significantly reduce some of the side-effects of anticancer therapy while enhancing drug efficacy.
Cetuximab is the most well-known FDA approved anticancer medication. It is a biomarker that predicts drug reactivity and utilizes the presence of the ‘KRAS’ gene mutation. Cetuximab is prescribed to patients who don’t carry the KRAS gene mutation.
However, even in patients without the KRAS gene mutation, the response rate of Cetuximab is only about fifty percent, and there is also resistance to drugs after targeted chemotherapy. Compared with conventional chemotherapy alone, the life expectancy only lasts five months on average.
In research featured in the FEBS Journal as the cover paper for the April 7 edition, the KAIST research team led by Professor Kwang-Hyun Cho at the Department of Bio and Brain Engineering presented five additional biomarkers that could increase Cetuximab responsiveness using systems biology approach that combines genomic data analysis, mathematical modeling, and cell experiments. The experimental inhibition of newly discovered biomarkers DUSP4, ETV5, GNB5, NT5E, and PHLDA1 in colorectal cancer cells has been shown to overcome Cetuximab resistance in KRAS-normal genes. The research team confirmed that when suppressing GNB5, one of the new biomarkers, it was shown to overcome resistance to Cetuximab regardless of having a mutation in the KRAS gene.
Professor Cho said, “There has not been an example of colorectal cancer treatment involving regulation of the GNB5 gene.” He continued, “Identifying the principle of drug resistance in cancer cells through systems biology and discovering new biomarkers that could be a new molecular target to overcome drug resistance suggest real potential to actualize precision medicine.”
This study was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT (2017R1A2A1A17069642 and 2015M3A9A7067220).
Image 1. The cover of FEBS Journal for April 2019
2019.05.27 View 60989 -
 KAIST Unveils the Hidden Control Architecture of Brain Networks
(Professor Kwang-Hyun Cho and his team)
A KAIST research team identified the intrinsic control architecture of brain networks. The control properties will contribute to providing a fundamental basis for the exogenous control of brain networks and, therefore, has broad implications in cognitive and clinical neuroscience.
Although efficiency and robustness are often regarded as having a trade-off relationship, the human brain usually exhibits both attributes when it performs complex cognitive functions. Such optimality must be rooted in a specific coordinated control of interconnected brain regions, but the understanding of the intrinsic control architecture of brain networks is lacking.
Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering and his team investigated the intrinsic control architecture of brain networks. They employed an interdisciplinary approach that spans connectomics, neuroscience, control engineering, network science, and systems biology to examine the structural brain networks of various species and compared them with the control architecture of other biological networks, as well as man-made ones, such as social, infrastructural and technological networks.
In particular, the team reconstructed the structural brain networks of 100 healthy human adults by performing brain parcellation and tractography with structural and diffusion imaging data obtained from the Human Connectome Project database of the US National Institutes of Health.
The team developed a framework for analyzing the control architecture of brain networks based on the minimum dominating set (MDSet), which refers to a minimal subset of nodes (MD-nodes) that control the remaining nodes with a one-step direct interaction. MD-nodes play a crucial role in various complex networks including biomolecular networks, but they have not been investigated in brain networks.
By exploring and comparing the structural principles underlying the composition of MDSets of various complex networks, the team delineated their distinct control architectures. Interestingly, the team found that the proportion of MDSets in brain networks is remarkably small compared to those of other complex networks. This finding implies that brain networks may have been optimized for minimizing the cost required for controlling networks. Furthermore, the team found that the MDSets of brain networks are not solely determined by the degree of nodes, but rather strategically placed to form a particular control architecture.
Consequently, the team revealed the hidden control architecture of brain networks, namely, the distributed and overlapping control architecture that is distinct from other complex networks. The team found that such a particular control architecture brings about robustness against targeted attacks (i.e., preferential attacks on high-degree nodes) which might be a fundamental basis of robust brain functions against preferential damage of high-degree nodes (i.e., brain regions).
Moreover, the team found that the particular control architecture of brain networks also enables high efficiency in switching from one network state, defined by a set of node activities, to another – a capability that is crucial for traversing diverse cognitive states.
Professor Cho said, “This study is the first attempt to make a quantitative comparison between brain networks and other real-world complex networks. Understanding of intrinsic control architecture underlying brain networks may enable the development of optimal interventions for therapeutic purposes or cognitive enhancement.”
This research, led by Byeongwook Lee, Uiryong Kang and Hongjun Chang, was published in iScience (10.1016/j.isci.2019.02.017) on March 29, 2019.
Figure 1. Schematic of identification of control architecture of brain networks.
Figure 2. Identified control architectures of brain networks and other real-world complex networks.
2019.04.23 View 39117
KAIST Unveils the Hidden Control Architecture of Brain Networks
(Professor Kwang-Hyun Cho and his team)
A KAIST research team identified the intrinsic control architecture of brain networks. The control properties will contribute to providing a fundamental basis for the exogenous control of brain networks and, therefore, has broad implications in cognitive and clinical neuroscience.
Although efficiency and robustness are often regarded as having a trade-off relationship, the human brain usually exhibits both attributes when it performs complex cognitive functions. Such optimality must be rooted in a specific coordinated control of interconnected brain regions, but the understanding of the intrinsic control architecture of brain networks is lacking.
Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering and his team investigated the intrinsic control architecture of brain networks. They employed an interdisciplinary approach that spans connectomics, neuroscience, control engineering, network science, and systems biology to examine the structural brain networks of various species and compared them with the control architecture of other biological networks, as well as man-made ones, such as social, infrastructural and technological networks.
In particular, the team reconstructed the structural brain networks of 100 healthy human adults by performing brain parcellation and tractography with structural and diffusion imaging data obtained from the Human Connectome Project database of the US National Institutes of Health.
The team developed a framework for analyzing the control architecture of brain networks based on the minimum dominating set (MDSet), which refers to a minimal subset of nodes (MD-nodes) that control the remaining nodes with a one-step direct interaction. MD-nodes play a crucial role in various complex networks including biomolecular networks, but they have not been investigated in brain networks.
By exploring and comparing the structural principles underlying the composition of MDSets of various complex networks, the team delineated their distinct control architectures. Interestingly, the team found that the proportion of MDSets in brain networks is remarkably small compared to those of other complex networks. This finding implies that brain networks may have been optimized for minimizing the cost required for controlling networks. Furthermore, the team found that the MDSets of brain networks are not solely determined by the degree of nodes, but rather strategically placed to form a particular control architecture.
Consequently, the team revealed the hidden control architecture of brain networks, namely, the distributed and overlapping control architecture that is distinct from other complex networks. The team found that such a particular control architecture brings about robustness against targeted attacks (i.e., preferential attacks on high-degree nodes) which might be a fundamental basis of robust brain functions against preferential damage of high-degree nodes (i.e., brain regions).
Moreover, the team found that the particular control architecture of brain networks also enables high efficiency in switching from one network state, defined by a set of node activities, to another – a capability that is crucial for traversing diverse cognitive states.
Professor Cho said, “This study is the first attempt to make a quantitative comparison between brain networks and other real-world complex networks. Understanding of intrinsic control architecture underlying brain networks may enable the development of optimal interventions for therapeutic purposes or cognitive enhancement.”
This research, led by Byeongwook Lee, Uiryong Kang and Hongjun Chang, was published in iScience (10.1016/j.isci.2019.02.017) on March 29, 2019.
Figure 1. Schematic of identification of control architecture of brain networks.
Figure 2. Identified control architectures of brain networks and other real-world complex networks.
2019.04.23 View 39117 -
 Trigger of the Hyperactivation of Fibrosis Identified
(Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering)
Scientists have been investigating the negative effects that the hyperactivation of fibrosis has on fibrotic diseases and cancer. A KAIST research team unveiled a positive feedback loop that bi-stably activates fibroblasts in collaboration with Samsung Medical Center. This finding will contribute to developing therapeutic targets for both fibrosis and cancer.
Human fibroblasts are dormant in normal tissue, but show radical activation during wound healing. However, the principle that induces their explosive activation has not yet been identified.
Here, Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering, in collaboration with Professor Seok-Hyung Kim from Samsung Medical Center, discovered the principle of a circuit that continuously activates fibroblasts.
They constructed a positive feedback loops (PFLs) where Twist1, Prrx1, and Tenascin-C (TNC) molecules consecutively activate fibroblasts. They confirmed that these are the main inducers of fibroblast activation by conducting various experiments, including molecular biological tests, mathematical modeling, animal testing, and computer simulations to conclude that they are the main inducers of fibroblast activation.
According to their research, Twist 1 is a key regulator of cancer-associated fibroblasts, which directly upregulates Prrx1 and then triggers TNC, which also increases Twist1 expression. This circuit consequently forms a Twist-Prrx1-TNC positive feedback loop.
Activated fibroblasts need to be deactivated after wounds are healed. However, if the PFLs continue, the fibroblasts become the major cause of worsening fibrotic diseases and cancers.
Therefore, the team expects that Twist1-Prrx1-TNC positive PFLs will be applied for novel and effective therapeutic targeting of fibrotic diseases and cancers.
This research was published in Nature Communications on August 1, 2018.
Figure 1. Twist1 increases tenascin-c expression in cancer-associated fibroblasts. Twist1 is a potent but indirect inducer of tenascin-c (TNC), which is essential for maintaining Twist1 expression in cancer-associated fibroblasts (CAFs).
Figure 2. Summary of the study. The Twist1-Prrx1-TNC positive feedback regulation provides clues for understanding the activation of fibroblasts during wound healing under normal conditions, as well as abnormally activated fibroblasts in pathological conditions such as cancerous and fibrotic diseases. Under normal conditions, the PFL acts as a reversible bistable switch by which the activation of fibroblasts is “ON" above a sufficient level of stimulation and “OFF" for the withdrawal of the stimulus. However, this switch can be permanently turned on under pathologic conditions by continued activation of the PFL, resulting in sustained proliferation of fibroblasts.
2018.10.11 View 7002
Trigger of the Hyperactivation of Fibrosis Identified
(Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering)
Scientists have been investigating the negative effects that the hyperactivation of fibrosis has on fibrotic diseases and cancer. A KAIST research team unveiled a positive feedback loop that bi-stably activates fibroblasts in collaboration with Samsung Medical Center. This finding will contribute to developing therapeutic targets for both fibrosis and cancer.
Human fibroblasts are dormant in normal tissue, but show radical activation during wound healing. However, the principle that induces their explosive activation has not yet been identified.
Here, Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering, in collaboration with Professor Seok-Hyung Kim from Samsung Medical Center, discovered the principle of a circuit that continuously activates fibroblasts.
They constructed a positive feedback loops (PFLs) where Twist1, Prrx1, and Tenascin-C (TNC) molecules consecutively activate fibroblasts. They confirmed that these are the main inducers of fibroblast activation by conducting various experiments, including molecular biological tests, mathematical modeling, animal testing, and computer simulations to conclude that they are the main inducers of fibroblast activation.
According to their research, Twist 1 is a key regulator of cancer-associated fibroblasts, which directly upregulates Prrx1 and then triggers TNC, which also increases Twist1 expression. This circuit consequently forms a Twist-Prrx1-TNC positive feedback loop.
Activated fibroblasts need to be deactivated after wounds are healed. However, if the PFLs continue, the fibroblasts become the major cause of worsening fibrotic diseases and cancers.
Therefore, the team expects that Twist1-Prrx1-TNC positive PFLs will be applied for novel and effective therapeutic targeting of fibrotic diseases and cancers.
This research was published in Nature Communications on August 1, 2018.
Figure 1. Twist1 increases tenascin-c expression in cancer-associated fibroblasts. Twist1 is a potent but indirect inducer of tenascin-c (TNC), which is essential for maintaining Twist1 expression in cancer-associated fibroblasts (CAFs).
Figure 2. Summary of the study. The Twist1-Prrx1-TNC positive feedback regulation provides clues for understanding the activation of fibroblasts during wound healing under normal conditions, as well as abnormally activated fibroblasts in pathological conditions such as cancerous and fibrotic diseases. Under normal conditions, the PFL acts as a reversible bistable switch by which the activation of fibroblasts is “ON" above a sufficient level of stimulation and “OFF" for the withdrawal of the stimulus. However, this switch can be permanently turned on under pathologic conditions by continued activation of the PFL, resulting in sustained proliferation of fibroblasts.
2018.10.11 View 7002 -
 Technology to Find Optimum Drug Target for Cancer Developed
(Professor Kwang-Hyun Cho (right) and lead author Dr. Minsoo Choi)
A KAIST research team led by Professor Kwang-Hyun Cho of the Department of Bio and Brain Engineering developed technology to find the optimum drug target according to the type of cancer cell. The team used systems biology to analyze molecular network dynamics that reflect genetic mutations in cancer cells and to predict drug response. The technology could contribute greatly to future anti-cancer drug development.
There are many types of genetic variations found in cancer cells, including gene mutations and copy number variations. These variations differ in cancer cells even within the same type of cancer, and thus the drug response varies cell by cell. Cancer researchers worked towards identifying frequently occurring genetic variations in cancer patients and, in particular, the mutations that can be used as an index for specific drugs. Previous studies focused on identifying a single genetic mutation or creating an analysis of the structural characteristics of a gene network. However, this approach was limited in its inability to explain the biological properties of cancer which are induced by various gene and protein interactions in cancer cells, which result in differences in drug response.
Gene mutations in cancer cells not only affect the function of the affected gene, but also other genes that interact with the mutated gene and proteins. As a consequence, one mutation could lead to changes in the dynamical properties of the molecular network. Therefore, the responses to anti-cancer drugs by cancer cells differ. The current treatment approach that ignores molecular network dynamics and targets a few cancer-related genes is only effective on a fraction of patients, while many other patients exhibit resistance to the drug.
Professor Cho’s team integrated a large-scale computer simulation using super-computing and cellular experiments to analyze changes in molecular network dynamics in cancer cells.
This led to development of technology to find the optimum drug target according to the type of cancer cells by predicting drug response. This technology was applied to the molecular network of known tumor suppressor p53. The team used large-scale cancer cell genomic data available from The Cancer Cell Line Encyclopedia (CCLE) to construct different molecular networks specific to the characteristics of genetic variations.
Perturbation analysis on drug response in each molecular network was used to quantify changes in cancer cells from drug response and similar networks were clustered. Then, computer simulations were used to analyze the synergetic effects in terms of efficacy and combination to predict the level of drug response. Based on the simulation results from various cancer cell lines including lung, breast, bone, skin, kidney, and ovary cancers were used in drug response experiments for compare analysis.
This technique can be applied in any molecular network to identify the optimum drug target for personalized medicine.
The research team suggests that the technology can analyze varying drug response due to the heterogeneity of cancer cells by considering the overall modulatory interactions rather than focusing only on a specific gene or protein. Further, the technology aids the prediction of causes of drug resistance and thus the identification of the optimum drug target to inhibit the resistance. This could be core source technology that can be used in drug repositioning, a process of applying existing drugs to new disease targets.
Professor Cho said, “Genetic variations in cancer cells are the cause of diverse drug response, but a complete analysis had not yet been made.” He continued, “Systems biology allowed the simulation of drug responses by cancer cell molecular networks to identify fundamental principles of drug response and optimum drug targets using a new conceptual approach.”
This research was published in Nature Communications on December 5 and was funded by Ministry of Science and ICT and National Research Foundation of Korea.
(Figure 1. Drug response prediction for each cancer cell type from computer simulation and cellular experiment verification for comparison)
(Figure 2. Drug response prediction based on cancer cell molecular network dynamics and clustering of cancer cells by their molecular networks)
(Figure 3. Identification of drug target for each cancer cell type by cellular molecular network analysis and establishment for personalized medicine strategy for each cancer patient)
2017.12.15 View 7584
Technology to Find Optimum Drug Target for Cancer Developed
(Professor Kwang-Hyun Cho (right) and lead author Dr. Minsoo Choi)
A KAIST research team led by Professor Kwang-Hyun Cho of the Department of Bio and Brain Engineering developed technology to find the optimum drug target according to the type of cancer cell. The team used systems biology to analyze molecular network dynamics that reflect genetic mutations in cancer cells and to predict drug response. The technology could contribute greatly to future anti-cancer drug development.
There are many types of genetic variations found in cancer cells, including gene mutations and copy number variations. These variations differ in cancer cells even within the same type of cancer, and thus the drug response varies cell by cell. Cancer researchers worked towards identifying frequently occurring genetic variations in cancer patients and, in particular, the mutations that can be used as an index for specific drugs. Previous studies focused on identifying a single genetic mutation or creating an analysis of the structural characteristics of a gene network. However, this approach was limited in its inability to explain the biological properties of cancer which are induced by various gene and protein interactions in cancer cells, which result in differences in drug response.
Gene mutations in cancer cells not only affect the function of the affected gene, but also other genes that interact with the mutated gene and proteins. As a consequence, one mutation could lead to changes in the dynamical properties of the molecular network. Therefore, the responses to anti-cancer drugs by cancer cells differ. The current treatment approach that ignores molecular network dynamics and targets a few cancer-related genes is only effective on a fraction of patients, while many other patients exhibit resistance to the drug.
Professor Cho’s team integrated a large-scale computer simulation using super-computing and cellular experiments to analyze changes in molecular network dynamics in cancer cells.
This led to development of technology to find the optimum drug target according to the type of cancer cells by predicting drug response. This technology was applied to the molecular network of known tumor suppressor p53. The team used large-scale cancer cell genomic data available from The Cancer Cell Line Encyclopedia (CCLE) to construct different molecular networks specific to the characteristics of genetic variations.
Perturbation analysis on drug response in each molecular network was used to quantify changes in cancer cells from drug response and similar networks were clustered. Then, computer simulations were used to analyze the synergetic effects in terms of efficacy and combination to predict the level of drug response. Based on the simulation results from various cancer cell lines including lung, breast, bone, skin, kidney, and ovary cancers were used in drug response experiments for compare analysis.
This technique can be applied in any molecular network to identify the optimum drug target for personalized medicine.
The research team suggests that the technology can analyze varying drug response due to the heterogeneity of cancer cells by considering the overall modulatory interactions rather than focusing only on a specific gene or protein. Further, the technology aids the prediction of causes of drug resistance and thus the identification of the optimum drug target to inhibit the resistance. This could be core source technology that can be used in drug repositioning, a process of applying existing drugs to new disease targets.
Professor Cho said, “Genetic variations in cancer cells are the cause of diverse drug response, but a complete analysis had not yet been made.” He continued, “Systems biology allowed the simulation of drug responses by cancer cell molecular networks to identify fundamental principles of drug response and optimum drug targets using a new conceptual approach.”
This research was published in Nature Communications on December 5 and was funded by Ministry of Science and ICT and National Research Foundation of Korea.
(Figure 1. Drug response prediction for each cancer cell type from computer simulation and cellular experiment verification for comparison)
(Figure 2. Drug response prediction based on cancer cell molecular network dynamics and clustering of cancer cells by their molecular networks)
(Figure 3. Identification of drug target for each cancer cell type by cellular molecular network analysis and establishment for personalized medicine strategy for each cancer patient)
2017.12.15 View 7584 -
 Mutant Gene Network in Colon Cancer Identified
The principles of the gene network for colon tumorigenesis have been identified by a KAIST research team. The principles will be used to find the molecular target for effective anti-cancer drugs in the future. Further, this research gained attention for using a systems biology approach, which is an integrated research area of IT and BT.
The KAIST research team led by Professor Kwang-Hyun Cho for the Department of Bio and Brain Engineering succeeded in the identification. Conducted by Dr. Dongkwan Shin and student researchers Jonghoon Lee and Jeong-Ryeol Gong, the research was published in Nature Communications online on November 2.
Human cancer is caused by genetic mutations. The frequency of the mutations differs by the type of cancer; for example, only around 10 mutations are found in leukemia and childhood cancer, but an average of 50 mutations are found in adult solid cancers and even hundreds of mutations are found in cancers due to external factors, such as with lung cancer.
Cancer researchers around the world are working to identify frequently found genetic mutations in patients, and in turn identify important cancer-inducing genes (called ‘driver genes’) to develop targets for anti-cancer drugs. However, gene mutations not only affect their own functions but also affect other genes through interactions. Therefore, there are limitations in current treatments targeting a few cancer-inducing genes without further knowledge on gene networks, hence current drugs are only effective in a few patients and often induce drug resistance.
Professor Cho’s team used large-scale genomic data from cancer patients to construct a mathematical model on the cooperative effects of multiple genetic mutations found in gene interaction networks. The basis of the model construction was The Cancer Genome Atlas (TCGA) presented at the International Cancer Genome Consortium. The team successfully quantified the effects of mutations in gene networks to group colon cancer patients by clinical characteristics.
Further, the critical transition phenomenon that occurs in tumorigenesis was identified using large-scale computer simulation analysis, which was the first hidden gene network principle to be identified. Critical transition is the phenomenon in which the state of matter is suddenly changed through phase transition. It was not possible to identify the presence of transition phenomenon in the past, as it was difficult to track the sequence of gene mutations during tumorigenesis.
The research team used a systems biology-based research method to find that colon cancer tumorigenesis shows a critical transition phenomenon if the known driver gene mutations follow sequentially. Using the developed mathematical model, it can be possible to develop a new anti-cancer targeting drug that most effectively inhibits the effects of many gene mutations found in cancer patients. In particular, not only driver genes, but also other passenger genes affected by the gene mutations, could be evaluated to find the most effective drug targets.
Professor Cho said, “Little was known about the contribution of many gene mutations during tumorigenesis.” He continued, “In this research, a systems biology approach identified the principle of gene networks for the first time to suggest the possibility of anti-cancer drug target identification from a new perspective.”
This research was funded by the Ministry of Science and ICT and the National Research Foundation of Korea.
Figure1. Formation of giant clusters via mutation propagation
Figure2. Critical transition phenomenon by cooperative effect of mutations in tumorigenesis
2017.11.10 View 8655
Mutant Gene Network in Colon Cancer Identified
The principles of the gene network for colon tumorigenesis have been identified by a KAIST research team. The principles will be used to find the molecular target for effective anti-cancer drugs in the future. Further, this research gained attention for using a systems biology approach, which is an integrated research area of IT and BT.
The KAIST research team led by Professor Kwang-Hyun Cho for the Department of Bio and Brain Engineering succeeded in the identification. Conducted by Dr. Dongkwan Shin and student researchers Jonghoon Lee and Jeong-Ryeol Gong, the research was published in Nature Communications online on November 2.
Human cancer is caused by genetic mutations. The frequency of the mutations differs by the type of cancer; for example, only around 10 mutations are found in leukemia and childhood cancer, but an average of 50 mutations are found in adult solid cancers and even hundreds of mutations are found in cancers due to external factors, such as with lung cancer.
Cancer researchers around the world are working to identify frequently found genetic mutations in patients, and in turn identify important cancer-inducing genes (called ‘driver genes’) to develop targets for anti-cancer drugs. However, gene mutations not only affect their own functions but also affect other genes through interactions. Therefore, there are limitations in current treatments targeting a few cancer-inducing genes without further knowledge on gene networks, hence current drugs are only effective in a few patients and often induce drug resistance.
Professor Cho’s team used large-scale genomic data from cancer patients to construct a mathematical model on the cooperative effects of multiple genetic mutations found in gene interaction networks. The basis of the model construction was The Cancer Genome Atlas (TCGA) presented at the International Cancer Genome Consortium. The team successfully quantified the effects of mutations in gene networks to group colon cancer patients by clinical characteristics.
Further, the critical transition phenomenon that occurs in tumorigenesis was identified using large-scale computer simulation analysis, which was the first hidden gene network principle to be identified. Critical transition is the phenomenon in which the state of matter is suddenly changed through phase transition. It was not possible to identify the presence of transition phenomenon in the past, as it was difficult to track the sequence of gene mutations during tumorigenesis.
The research team used a systems biology-based research method to find that colon cancer tumorigenesis shows a critical transition phenomenon if the known driver gene mutations follow sequentially. Using the developed mathematical model, it can be possible to develop a new anti-cancer targeting drug that most effectively inhibits the effects of many gene mutations found in cancer patients. In particular, not only driver genes, but also other passenger genes affected by the gene mutations, could be evaluated to find the most effective drug targets.
Professor Cho said, “Little was known about the contribution of many gene mutations during tumorigenesis.” He continued, “In this research, a systems biology approach identified the principle of gene networks for the first time to suggest the possibility of anti-cancer drug target identification from a new perspective.”
This research was funded by the Ministry of Science and ICT and the National Research Foundation of Korea.
Figure1. Formation of giant clusters via mutation propagation
Figure2. Critical transition phenomenon by cooperative effect of mutations in tumorigenesis
2017.11.10 View 8655 -
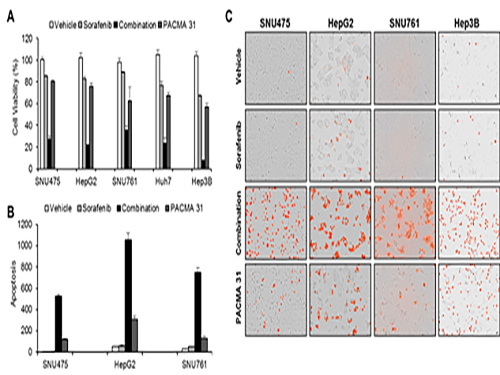 Discovery of an Optimal Drug Combination: Overcoming Resistance to Targeted Drugs for Liver Cancer
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences. Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)
(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)
(Figure 3. ER stress network model)
2017.08.30 View 13088
Discovery of an Optimal Drug Combination: Overcoming Resistance to Targeted Drugs for Liver Cancer
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences. Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)
(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)
(Figure 3. ER stress network model)
2017.08.30 View 13088 -
 A mechanism for how reactive oxygen species cause cell responses studied
A research team led by Professor Kwang-Hyun Cho of the Department of Biology and Brain Engineering, KAIST, and Dr. Gi-Sun Kwon of the Korea Research Institute of Bioscience and Biotechnology succeeded in proving the mechanism behind the determination of cell life in relation to reactive oxygen species.
The results of the venture were published in the June 3rd edition of Science Signaling. The title of the research paper is “MLK3 is part of a feedback mechanism that regulates different cellular responses to reactive oxygen species.”
The research team discovered that the molecular switch that determines the division of apoptosis of a cell was based on MLK3 feedback mechanism. MLK stands for mixed-lineage kinase.
Under sufficient stress, the mechanism instructs the cell to undergo the division but in an overly stressful environment, the mechanism stops the cell division and instead, induces apoptosis. This discovery is expected to be a breakthrough in illnesses related to the concentration of the reactive oxygen species (ROS).
At low concentration of ROS, the protein associated with cell division, ERK (extracellular-signal-regulated kinase), is activated while as the ROS concentration increases, JNK (c-Jun N-terminal protein kinases), responsible for apoptosis, becomes activated. Furthermore, through computer simulation analysis and mathematical modeling, in tandem with molecular cell biology experiments, the MLK3 based feedback mechanism was the fundamental molecular switch that determines the balance between ERK and JNK, and ultimately the cell’s responses.
Professor Cho commented that “the contradicting cell responses to ROS had remained a mystery, but with the system biology, an approach in which information technology and biotechnology converge, such riddles can be resolved. We expect that the proven mechanism will be used to overcome aging or cancer growth as a result of ROS in the near future.”
Picture shows the process of identifying cell responses caused by reactive oxygen species.
2014.06.13 View 10431
A mechanism for how reactive oxygen species cause cell responses studied
A research team led by Professor Kwang-Hyun Cho of the Department of Biology and Brain Engineering, KAIST, and Dr. Gi-Sun Kwon of the Korea Research Institute of Bioscience and Biotechnology succeeded in proving the mechanism behind the determination of cell life in relation to reactive oxygen species.
The results of the venture were published in the June 3rd edition of Science Signaling. The title of the research paper is “MLK3 is part of a feedback mechanism that regulates different cellular responses to reactive oxygen species.”
The research team discovered that the molecular switch that determines the division of apoptosis of a cell was based on MLK3 feedback mechanism. MLK stands for mixed-lineage kinase.
Under sufficient stress, the mechanism instructs the cell to undergo the division but in an overly stressful environment, the mechanism stops the cell division and instead, induces apoptosis. This discovery is expected to be a breakthrough in illnesses related to the concentration of the reactive oxygen species (ROS).
At low concentration of ROS, the protein associated with cell division, ERK (extracellular-signal-regulated kinase), is activated while as the ROS concentration increases, JNK (c-Jun N-terminal protein kinases), responsible for apoptosis, becomes activated. Furthermore, through computer simulation analysis and mathematical modeling, in tandem with molecular cell biology experiments, the MLK3 based feedback mechanism was the fundamental molecular switch that determines the balance between ERK and JNK, and ultimately the cell’s responses.
Professor Cho commented that “the contradicting cell responses to ROS had remained a mystery, but with the system biology, an approach in which information technology and biotechnology converge, such riddles can be resolved. We expect that the proven mechanism will be used to overcome aging or cancer growth as a result of ROS in the near future.”
Picture shows the process of identifying cell responses caused by reactive oxygen species.
2014.06.13 View 10431 -
 Systems biology demystifies the resistance mechanism of targeted cancer medication
Korean researchers have found the fundamental resistance mechanism of the MEK inhibitor, a recently highlighted chemotherapy method, laying the foundation for future research on overcoming cancer drug resistance and improving cancer survival rates. This research is meaningful because it was conducted through systems biology, a fusion of IT and biotechnology.
The research was conducted by Professor Gwang hyun Cho’s team from the Department of Biology at KAIST and was supported by the Ministry of Education, Science and Technology and the National Research Foundation of Korea.
The research was published as the cover paper for the June edition of the Journal of Molecular Cell Biology (Title: The cross regulation between ERK and PI3K signaling pathways determines the tumoricidal efficacy of MEK inhibitor).
Targeted anticancer medication targets certain molecules in the signaling pathway of the tumor cell and not only has fewer side effects than pre-existing anticancer medication, but also has high clinical efficacy. The technology also allows the creation of personalized medication and has been widely praised by scientists worldwide.
However, resistances to the targeted medication have often been found before or during the clinical stage, eventually causing the medications to fail to reach the drug development stage. Moreover, even if the drug is effective, the survival rate is low and the redevelopment rate is high.
An active pathway in most tumor cells is the ERK (Extracellular signal-regulated kinases) signaling pathway. This pathway is especially important in the development of skin cancer or thyroid cancer, which are developed by the mutation of the BRAF gene inside the path. In these cases, the MEK (Extracellular signal-regulated kinases) inhibitor is an effective treatment because it targets the pathway itself. However, the built-up resistance to the inhibitor commonly leads to the redevelopment of cancer.
Professor Cho’s research team used large scale computer simulations to analyze the fundamental resistance mechanism of the MEK inhibitor and used molecular cell biological experiments as well as bio-imaging* techniques to verify the results.
* Bio-imaging: Checking biological phenomena at the cellular and molecular levels using imagery
The research team used different mutational variables, which revealed that the use of the MEK inhibitor reduced the transmission of the ERK signal but led to the activation of another signaling pathway (the PI3K signaling pathway), reducing the effectiveness of the medication. Professor Cho’s team also found that this response originated from the complex interaction between the signaling matter as well as the feedback network structure, suggesting that the mix of the MEK inhibitor with other drugs could improve the effects of the targeted anticancer medication.
Professor Cho stated that this research was the first of its kind to examine the drug resistivity against the MEK inhibitor at the systematic dimension and showed how the effects of drugs on the signaling pathways of cells could be predicted using computer simulation. It also showed how basic research on signaling networks can be applied to clinical drug use, successfully suggesting a new research platform on overcoming resistance to targeting medication using its fundamental mechanism.
2012.07.06 View 13374
Systems biology demystifies the resistance mechanism of targeted cancer medication
Korean researchers have found the fundamental resistance mechanism of the MEK inhibitor, a recently highlighted chemotherapy method, laying the foundation for future research on overcoming cancer drug resistance and improving cancer survival rates. This research is meaningful because it was conducted through systems biology, a fusion of IT and biotechnology.
The research was conducted by Professor Gwang hyun Cho’s team from the Department of Biology at KAIST and was supported by the Ministry of Education, Science and Technology and the National Research Foundation of Korea.
The research was published as the cover paper for the June edition of the Journal of Molecular Cell Biology (Title: The cross regulation between ERK and PI3K signaling pathways determines the tumoricidal efficacy of MEK inhibitor).
Targeted anticancer medication targets certain molecules in the signaling pathway of the tumor cell and not only has fewer side effects than pre-existing anticancer medication, but also has high clinical efficacy. The technology also allows the creation of personalized medication and has been widely praised by scientists worldwide.
However, resistances to the targeted medication have often been found before or during the clinical stage, eventually causing the medications to fail to reach the drug development stage. Moreover, even if the drug is effective, the survival rate is low and the redevelopment rate is high.
An active pathway in most tumor cells is the ERK (Extracellular signal-regulated kinases) signaling pathway. This pathway is especially important in the development of skin cancer or thyroid cancer, which are developed by the mutation of the BRAF gene inside the path. In these cases, the MEK (Extracellular signal-regulated kinases) inhibitor is an effective treatment because it targets the pathway itself. However, the built-up resistance to the inhibitor commonly leads to the redevelopment of cancer.
Professor Cho’s research team used large scale computer simulations to analyze the fundamental resistance mechanism of the MEK inhibitor and used molecular cell biological experiments as well as bio-imaging* techniques to verify the results.
* Bio-imaging: Checking biological phenomena at the cellular and molecular levels using imagery
The research team used different mutational variables, which revealed that the use of the MEK inhibitor reduced the transmission of the ERK signal but led to the activation of another signaling pathway (the PI3K signaling pathway), reducing the effectiveness of the medication. Professor Cho’s team also found that this response originated from the complex interaction between the signaling matter as well as the feedback network structure, suggesting that the mix of the MEK inhibitor with other drugs could improve the effects of the targeted anticancer medication.
Professor Cho stated that this research was the first of its kind to examine the drug resistivity against the MEK inhibitor at the systematic dimension and showed how the effects of drugs on the signaling pathways of cells could be predicted using computer simulation. It also showed how basic research on signaling networks can be applied to clinical drug use, successfully suggesting a new research platform on overcoming resistance to targeting medication using its fundamental mechanism.
2012.07.06 View 13374