Department+of+Chemical
-
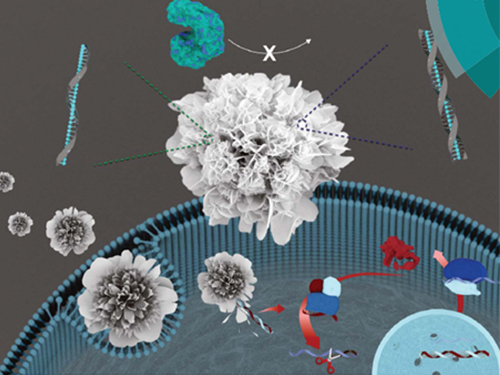 Nuclease-Resistant Hybrid Nanoflowers
An eco-friendly method to synthesize DNA-copper nanoflowers with high load efficiencies, low cytotoxicity, and strong resistance against nucleases has been developed by Professor Hyun Gyu Park in the Department of Chemical and Biomolecular Engineering and his collaborators.
The research team successfully formed a flower-shaped nanostructure in an eco-friendly condition by using interactions between copper ions and DNA containing amide and amine groups. The resulting nanoflowers exhibit high DNA loading capacities in addition to low cytotoxicity.
Flower-shaped nanocrystals called nanoflowers have gained attention for their distinct features of high surface roughness and high surface area to volume ratios. The nanoflowers have been used in many areas including catalysis, electronics, and analytical chemistry.
Of late, research breakthroughs were made in the generation of hybrid inorganic-organic nanoflowers containing various enzymes as organic components. The hybridization with inorganic materials greatly enhanced enzymatic activity, stability, and durability compared to the corresponding free enzymes.
Generally, the formation of protein nanocrystals requires high heat treatment so it has limitations for achieving the high loading capacities of intact DNA.
The research team addressed the issue, focusing on the fact that nucleic acids with well-defined structures and selective recognition properties also contain amide and amine groups in their nucleobases. They proved that flower-like structures could be formed by using nucleic acids as a synthetic template, which paved the way to synthesize the hybrid nanoflowers containing DNA as an organic component in an eco-friendly condition.
The team also confirmed that this synthetic method can be universally applied to any DNA sequences containing amide and amine groups. They said their approach is quite unique considering that the majority of previous works focused on the utilization of DNA as a linker to assemble the nanomaterials. They said the method has several advantageous features. First, the ‘green’ synthetic procedure doesn’t involve any toxic chemicals, and shows low cytotoxicity and strong resistance against nucleases. Second, the obtained nanoflowers exhibit exceptionally high DNA loading capacities.
Above all, such superior features of hybrid nanoflowers enabled the sensitive detection of various molecules including phenol, hydrogen peroxide, and glucose. DNA-copper nanoflowers showed even higher peroxidase activity than those of protein-copper nanoflowers, which may be due to the larger surface area of the flower- shaped structures, creating a greater chance for applying them in the field of sensing of detection of hydrogen peroxide.
The research team expects that their research will create diverse applications in many areas including biosensors and will be further applied into therapeutic applications.
Professor Park said, “The inorganic component in the hybrid nanoflowers not only exhibits low cytotoxicity, but also protects the encapsulated DNA from being cleaved by endonuclease enzymes. Using this feature, the nanostructure will be applied into developing gene therapeutic carriers.”
This research was co-led by Professor Moon Il Kim at Gachon University and KAIST graduate Ki Soo Park, currently a professor at Konkuk University, is the first author. The research was featured as the front cover article of the Journal of Materials Chemistry B on March 28, Issue 12, published by the Royal Society of Chemistry.
The research was funded by the Mid-Career Researcher Support Program of the National Research Foundation of Korea and the Global Frontier Project of the Ministry of Science, ICT & Future Planning.
(Figure: (A) Schematic illustration of the formation of nuclease-resistant DNA–inorganic nanoflowers. (B) SEM images showing time-dependent growth of DNA-nanoflowers. The concentration of A-rich ssDNA (Table S1, ESI†) was 0.25 mM.)
2017.04.14 View 10056
Nuclease-Resistant Hybrid Nanoflowers
An eco-friendly method to synthesize DNA-copper nanoflowers with high load efficiencies, low cytotoxicity, and strong resistance against nucleases has been developed by Professor Hyun Gyu Park in the Department of Chemical and Biomolecular Engineering and his collaborators.
The research team successfully formed a flower-shaped nanostructure in an eco-friendly condition by using interactions between copper ions and DNA containing amide and amine groups. The resulting nanoflowers exhibit high DNA loading capacities in addition to low cytotoxicity.
Flower-shaped nanocrystals called nanoflowers have gained attention for their distinct features of high surface roughness and high surface area to volume ratios. The nanoflowers have been used in many areas including catalysis, electronics, and analytical chemistry.
Of late, research breakthroughs were made in the generation of hybrid inorganic-organic nanoflowers containing various enzymes as organic components. The hybridization with inorganic materials greatly enhanced enzymatic activity, stability, and durability compared to the corresponding free enzymes.
Generally, the formation of protein nanocrystals requires high heat treatment so it has limitations for achieving the high loading capacities of intact DNA.
The research team addressed the issue, focusing on the fact that nucleic acids with well-defined structures and selective recognition properties also contain amide and amine groups in their nucleobases. They proved that flower-like structures could be formed by using nucleic acids as a synthetic template, which paved the way to synthesize the hybrid nanoflowers containing DNA as an organic component in an eco-friendly condition.
The team also confirmed that this synthetic method can be universally applied to any DNA sequences containing amide and amine groups. They said their approach is quite unique considering that the majority of previous works focused on the utilization of DNA as a linker to assemble the nanomaterials. They said the method has several advantageous features. First, the ‘green’ synthetic procedure doesn’t involve any toxic chemicals, and shows low cytotoxicity and strong resistance against nucleases. Second, the obtained nanoflowers exhibit exceptionally high DNA loading capacities.
Above all, such superior features of hybrid nanoflowers enabled the sensitive detection of various molecules including phenol, hydrogen peroxide, and glucose. DNA-copper nanoflowers showed even higher peroxidase activity than those of protein-copper nanoflowers, which may be due to the larger surface area of the flower- shaped structures, creating a greater chance for applying them in the field of sensing of detection of hydrogen peroxide.
The research team expects that their research will create diverse applications in many areas including biosensors and will be further applied into therapeutic applications.
Professor Park said, “The inorganic component in the hybrid nanoflowers not only exhibits low cytotoxicity, but also protects the encapsulated DNA from being cleaved by endonuclease enzymes. Using this feature, the nanostructure will be applied into developing gene therapeutic carriers.”
This research was co-led by Professor Moon Il Kim at Gachon University and KAIST graduate Ki Soo Park, currently a professor at Konkuk University, is the first author. The research was featured as the front cover article of the Journal of Materials Chemistry B on March 28, Issue 12, published by the Royal Society of Chemistry.
The research was funded by the Mid-Career Researcher Support Program of the National Research Foundation of Korea and the Global Frontier Project of the Ministry of Science, ICT & Future Planning.
(Figure: (A) Schematic illustration of the formation of nuclease-resistant DNA–inorganic nanoflowers. (B) SEM images showing time-dependent growth of DNA-nanoflowers. The concentration of A-rich ssDNA (Table S1, ESI†) was 0.25 mM.)
2017.04.14 View 10056 -
 Mystery of Biological Plastic Synthesis Machinery Unveiled
Plastics and other polymers are used every day. These polymers are mostly made from fossil resources by refining petrochemicals. On the other hand, many microorganisms naturally synthesize polyesters known as polyhydroxyalkanoates (PHAs) as distinct granules inside cells.
PHAs are a family of microbial polyesters that have attracted much attention as biodegradable and biocompatible plastics and elastomers that can substitute petrochemical counterparts. There have been numerous papers and patents on gene cloning and metabolic engineering of PHA biosynthetic machineries, biochemical studies, and production of PHAs; simple Google search with “polyhydroxyalkanoates” yielded returns of 223,000 document pages. PHAs have always been considered amazing examples of biological polymer synthesis. It is astounding to see PHAs of 500 kDa to sometimes as high as 10,000 kDa can be synthesized in vivo by PHA synthase, the key polymerizing enzyme in PHA biosynthesis. They have attracted great interest in determining the crystal structure of PHA synthase over the last 30 years, but unfortunately without success. Thus, the characteristics and molecular mechanisms of PHA synthase were under a dark veil.
In two papers published back-to-back in Biotechnology Journal online on November 30, 2016, a Korean research team led by Professor Kyung-Jin Kim at Kyungpook National University and Distinguished Professor Sang Yup Lee at the Korea Advanced Institute of Science and Technology (KAIST) described the crystal structure of PHA synthase from Ralstonia eutropha, the best studied bacterium for PHA production, and reported the structural basis for the detailed molecular mechanisms of PHA biosynthesis. The crystal structure has been deposited to Protein Data Bank in February 2016. After deciphering the crystal structure of the catalytic domain of PHA synthase, in addition to other structural studies on whole enzyme and related proteins, the research team also performed experiments to elucidate the mechanisms of the enzyme reaction, validating detailed structures, enzyme engineering, and also N-terminal domain studies among others.
Through several biochemical studies based on crystal structure, the authors show that PHA synthase exists as a dimer and is divided into two distinct domains, the N-terminal domain (RePhaC1ND) and the C-terminal domain (RePhaC1CD). The RePhaC1CD catalyzes the polymerization reaction via a non-processive ping-pong mechanism using a Cys-His-Asp catalytic triad. The two catalytic sites of the RePhaC1CD dimer are positioned 33.4 Å apart, suggesting that the polymerization reaction occurs independently at each site. This study also presents the structure-based mechanisms for substrate specificities of various PHA synthases from different classes.
Professor Sang Yup Lee, who has worked on this topic for more than 20 years, said,
“The results and information presented in these two papers have long been awaited not only in the PHA community, but also metabolic engineering, bacteriology/microbiology, and in general biological sciences communities. The structural information on PHA synthase together with the recently deciphered reaction mechanisms will be valuable for understanding the detailed mechanisms of biosynthesizing this important energy/redox storage material, and also for the rational engineering of PHA synthases to produce designer bioplastics from various monomers more efficiently.”
Indeed, these two papers published in Biotechnology Journal finally reveal the 30-year mystery of machinery of biological polyester synthesis, and will serve as the essential compass in creating designer and more efficient bioplastic machineries.
References:
Jieun Kim, Yeo-Jin Kim, So Young Choi, Sang Yup Lee and Kyung-Jin Kim. “Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms” Biotechnology Journal DOI: 10.1002/biot.201600648
http://onlinelibrary.wiley.com/doi/10.1002/biot.201600648/abstract
Yeo-Jin Kim, So Young Choi, Jieun Kim, Kyeong Sik Jin, Sang Yup Lee and Kyung-Jin Kim. “Structure and function of the N-terminal domain of Ralstonia eutropha polyhydroxyalkanoate synthase, and the proposed structure and mechanisms of the whole enzyme” Biotechnology Journal DOI: 10.1002/biot.201600649
http://onlinelibrary.wiley.com/doi/10.1002/biot.201600649/abstract
2016.12.02 View 11429
Mystery of Biological Plastic Synthesis Machinery Unveiled
Plastics and other polymers are used every day. These polymers are mostly made from fossil resources by refining petrochemicals. On the other hand, many microorganisms naturally synthesize polyesters known as polyhydroxyalkanoates (PHAs) as distinct granules inside cells.
PHAs are a family of microbial polyesters that have attracted much attention as biodegradable and biocompatible plastics and elastomers that can substitute petrochemical counterparts. There have been numerous papers and patents on gene cloning and metabolic engineering of PHA biosynthetic machineries, biochemical studies, and production of PHAs; simple Google search with “polyhydroxyalkanoates” yielded returns of 223,000 document pages. PHAs have always been considered amazing examples of biological polymer synthesis. It is astounding to see PHAs of 500 kDa to sometimes as high as 10,000 kDa can be synthesized in vivo by PHA synthase, the key polymerizing enzyme in PHA biosynthesis. They have attracted great interest in determining the crystal structure of PHA synthase over the last 30 years, but unfortunately without success. Thus, the characteristics and molecular mechanisms of PHA synthase were under a dark veil.
In two papers published back-to-back in Biotechnology Journal online on November 30, 2016, a Korean research team led by Professor Kyung-Jin Kim at Kyungpook National University and Distinguished Professor Sang Yup Lee at the Korea Advanced Institute of Science and Technology (KAIST) described the crystal structure of PHA synthase from Ralstonia eutropha, the best studied bacterium for PHA production, and reported the structural basis for the detailed molecular mechanisms of PHA biosynthesis. The crystal structure has been deposited to Protein Data Bank in February 2016. After deciphering the crystal structure of the catalytic domain of PHA synthase, in addition to other structural studies on whole enzyme and related proteins, the research team also performed experiments to elucidate the mechanisms of the enzyme reaction, validating detailed structures, enzyme engineering, and also N-terminal domain studies among others.
Through several biochemical studies based on crystal structure, the authors show that PHA synthase exists as a dimer and is divided into two distinct domains, the N-terminal domain (RePhaC1ND) and the C-terminal domain (RePhaC1CD). The RePhaC1CD catalyzes the polymerization reaction via a non-processive ping-pong mechanism using a Cys-His-Asp catalytic triad. The two catalytic sites of the RePhaC1CD dimer are positioned 33.4 Å apart, suggesting that the polymerization reaction occurs independently at each site. This study also presents the structure-based mechanisms for substrate specificities of various PHA synthases from different classes.
Professor Sang Yup Lee, who has worked on this topic for more than 20 years, said,
“The results and information presented in these two papers have long been awaited not only in the PHA community, but also metabolic engineering, bacteriology/microbiology, and in general biological sciences communities. The structural information on PHA synthase together with the recently deciphered reaction mechanisms will be valuable for understanding the detailed mechanisms of biosynthesizing this important energy/redox storage material, and also for the rational engineering of PHA synthases to produce designer bioplastics from various monomers more efficiently.”
Indeed, these two papers published in Biotechnology Journal finally reveal the 30-year mystery of machinery of biological polyester synthesis, and will serve as the essential compass in creating designer and more efficient bioplastic machineries.
References:
Jieun Kim, Yeo-Jin Kim, So Young Choi, Sang Yup Lee and Kyung-Jin Kim. “Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms” Biotechnology Journal DOI: 10.1002/biot.201600648
http://onlinelibrary.wiley.com/doi/10.1002/biot.201600648/abstract
Yeo-Jin Kim, So Young Choi, Jieun Kim, Kyeong Sik Jin, Sang Yup Lee and Kyung-Jin Kim. “Structure and function of the N-terminal domain of Ralstonia eutropha polyhydroxyalkanoate synthase, and the proposed structure and mechanisms of the whole enzyme” Biotechnology Journal DOI: 10.1002/biot.201600649
http://onlinelibrary.wiley.com/doi/10.1002/biot.201600649/abstract
2016.12.02 View 11429 -
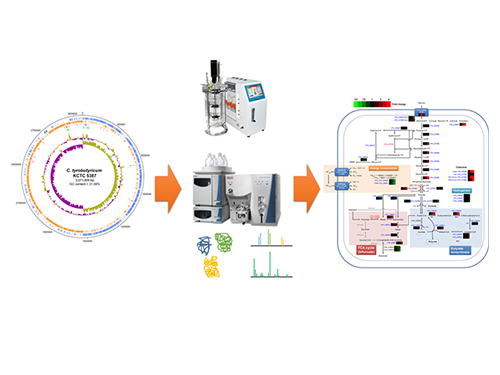 Unveiling the Distinctive Features of Industrial Microorganism
KAIST researchers have sequenced the whole genome of Clostridium tyrobutyricum, which has a higher tolerance to toxic chemicals, such as 1-butanol, compared to other clostridial bacterial strains.
Clostridium tyrobutyricum, a Gram-positive, anaerobic spore-forming bacterium, is considered a promising industrial host strain for the production of various chemicals including butyric acid which has many applications in different industries such as a precursor to biofuels. Despite such potential, C. tyrobutyricum has received little attention, mainly due to a limited understanding of its genotypic and metabolic characteristics at the genome level.
A Korean research team headed by Distinguished Professor Sang Yup Lee of the Chemical and Biomolecular Engineering Department at the Korea Advanced Institute of Science and Technology (KAIST) deciphered the genome sequence of C. tyrobutyricum and its proteome profiles during the course of batch fermentation. As a result, the research team learned that the bacterium is not only capable of producing a large amount of butyric acid but also can tolerate toxic compounds such as 1-butanol. The research results were published in mBio on June 14, 2016.
The team adopted a genoproteomic approach, combining genomics and proteomics, to investigate the metabolic features of C. tyrobutyricum. Unlike Clostridium acetobutylicum, the most widely used organism for 1-butanol production, C. tyrobutyricum has a novel butyrate-producing pathway and various mechanisms for energy conservation under anaerobic conditions. The expression of various metabolic genes, including those involved in butyrate formation, was analyzed using the “shotgun” proteome approach.
To date, the bio-based production of 1-butanol, a next-generation biofuel, has relied on several clostridial hosts including C. acetobutylicum and C. beijerinckii. However, these organisms have a low tolerance against 1-butanol even though they are naturally capable of producing it. C. tyrobutyricum cannot produce 1-butanol itself, but has a higher 1-butanol-tolerance and rapid uptake of monosaccharides, compared to those two species.
The team identified most of the genes involved in the central metabolism of C. tyrobutyricum from the whole-genome and shotgun proteome data, and this study will accelerate the bacterium’s engineering to produce useful chemicals including butyric acid and 1-butanol, replacing traditional bacterial hosts.
Professor Lee said,
“The unique metabolic features and energy conservation mechanisms of C. tyrobutyricum can be employed in the various microbial hosts we have previously developed to further improve their productivity and yield. Moreover, findings on C. tyrobutyricum revealed by this study will be the first step to directly engineer this bacterium.”
Director Jin-Woo Kim at the Platform Technology Division of the Ministry of Science, ICT and Future Planning of Korea, who oversees the Technology Development Program to Solve Climate Change, said,
“Over the years, Professor Lee’s team has researched the development of a bio-refinery system to produce natural and non-natural chemicals with the systems metabolic engineering of microorganisms. They were able to design strategies for the development of diverse industrial microbial strains to produce useful chemicals from inedible biomass-based carbon dioxide fixation. We believe the efficient production of butyric acid using a metabolic engineering approach will play an important role in the establishment of a bioprocess for chemical production.”
The title of the research paper is “Deciphering Clostridium tyrobutyricum Metabolism Based on the Who-Genome Sequence and Proteome Analyses.” (DOI: 10.1128/mBio.00743-16)
The lead authors are Joungmin Lee, a post-doctoral fellow in the BioProcess Research Center at KAIST, currently working in CJ CheilJedang Research Institute; Yu-Sin Jang, a research fellow in the BioProcess Research Center at KAIST, currently working at Gyeongsang National University as an assistant professor; and Mee-Jung Han, an assistant professor in the Environmental Engineering and Energy Department at Dongyang University. Jin Young Kim, a senior researcher at the Korea Basic Science Institute, also participated in the research.
This research was supported by the Technology Development Program to Solve Climate Change’s research project entitled “Systems Metabolic Engineering for Biorefineries” from the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea (NRF-2012M1A2A2026556).
Schematic Diagram of C. tyrobutyricum’s Genome Sequence and Its Proteome Profiles
The picture below shows the complete genome sequence, global protein expression profiles, and the genome-based metabolic characteristics during batch fermentation of C. tyrobutyricum.
2016.06.20 View 11684
Unveiling the Distinctive Features of Industrial Microorganism
KAIST researchers have sequenced the whole genome of Clostridium tyrobutyricum, which has a higher tolerance to toxic chemicals, such as 1-butanol, compared to other clostridial bacterial strains.
Clostridium tyrobutyricum, a Gram-positive, anaerobic spore-forming bacterium, is considered a promising industrial host strain for the production of various chemicals including butyric acid which has many applications in different industries such as a precursor to biofuels. Despite such potential, C. tyrobutyricum has received little attention, mainly due to a limited understanding of its genotypic and metabolic characteristics at the genome level.
A Korean research team headed by Distinguished Professor Sang Yup Lee of the Chemical and Biomolecular Engineering Department at the Korea Advanced Institute of Science and Technology (KAIST) deciphered the genome sequence of C. tyrobutyricum and its proteome profiles during the course of batch fermentation. As a result, the research team learned that the bacterium is not only capable of producing a large amount of butyric acid but also can tolerate toxic compounds such as 1-butanol. The research results were published in mBio on June 14, 2016.
The team adopted a genoproteomic approach, combining genomics and proteomics, to investigate the metabolic features of C. tyrobutyricum. Unlike Clostridium acetobutylicum, the most widely used organism for 1-butanol production, C. tyrobutyricum has a novel butyrate-producing pathway and various mechanisms for energy conservation under anaerobic conditions. The expression of various metabolic genes, including those involved in butyrate formation, was analyzed using the “shotgun” proteome approach.
To date, the bio-based production of 1-butanol, a next-generation biofuel, has relied on several clostridial hosts including C. acetobutylicum and C. beijerinckii. However, these organisms have a low tolerance against 1-butanol even though they are naturally capable of producing it. C. tyrobutyricum cannot produce 1-butanol itself, but has a higher 1-butanol-tolerance and rapid uptake of monosaccharides, compared to those two species.
The team identified most of the genes involved in the central metabolism of C. tyrobutyricum from the whole-genome and shotgun proteome data, and this study will accelerate the bacterium’s engineering to produce useful chemicals including butyric acid and 1-butanol, replacing traditional bacterial hosts.
Professor Lee said,
“The unique metabolic features and energy conservation mechanisms of C. tyrobutyricum can be employed in the various microbial hosts we have previously developed to further improve their productivity and yield. Moreover, findings on C. tyrobutyricum revealed by this study will be the first step to directly engineer this bacterium.”
Director Jin-Woo Kim at the Platform Technology Division of the Ministry of Science, ICT and Future Planning of Korea, who oversees the Technology Development Program to Solve Climate Change, said,
“Over the years, Professor Lee’s team has researched the development of a bio-refinery system to produce natural and non-natural chemicals with the systems metabolic engineering of microorganisms. They were able to design strategies for the development of diverse industrial microbial strains to produce useful chemicals from inedible biomass-based carbon dioxide fixation. We believe the efficient production of butyric acid using a metabolic engineering approach will play an important role in the establishment of a bioprocess for chemical production.”
The title of the research paper is “Deciphering Clostridium tyrobutyricum Metabolism Based on the Who-Genome Sequence and Proteome Analyses.” (DOI: 10.1128/mBio.00743-16)
The lead authors are Joungmin Lee, a post-doctoral fellow in the BioProcess Research Center at KAIST, currently working in CJ CheilJedang Research Institute; Yu-Sin Jang, a research fellow in the BioProcess Research Center at KAIST, currently working at Gyeongsang National University as an assistant professor; and Mee-Jung Han, an assistant professor in the Environmental Engineering and Energy Department at Dongyang University. Jin Young Kim, a senior researcher at the Korea Basic Science Institute, also participated in the research.
This research was supported by the Technology Development Program to Solve Climate Change’s research project entitled “Systems Metabolic Engineering for Biorefineries” from the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea (NRF-2012M1A2A2026556).
Schematic Diagram of C. tyrobutyricum’s Genome Sequence and Its Proteome Profiles
The picture below shows the complete genome sequence, global protein expression profiles, and the genome-based metabolic characteristics during batch fermentation of C. tyrobutyricum.
2016.06.20 View 11684 -
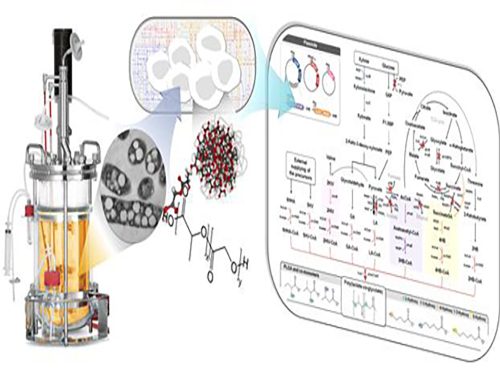 Non-Natural Biomedical Polymers Produced from Microorganisms
KAIST researchers have developed metabolically engineered Escherichia coli strains to synthesize non-natural, biomedically important polymers including poly(lactate-co-glycolate) (PLGA), previously considered impossible to obtain from biobased materials.
Renewable non-food biomass could potentially replace petrochemical raw materials to produce energy sources, useful chemicals, or a vast array of petroleum-based end products such as plastics, lubricants, paints, fertilizers, and vitamin capsules. In recent years, biorefineries which transform non-edible biomass into fuel, heat, power, chemicals, and materials have received a great deal of attention as a sustainable alternative to decreasing the reliance on fossil fuels.
A research team headed by Distinguished Professor Sang Yup Lee of the Chemical and Biomolecular Engineering Department at KAIST has established a biorefinery system to create non-natural polymers from natural sources, allowing various plastics to be made in an environmentally-friendly and sustainable manner. The research results were published online in Nature Biotechnology on March 7, 2016. The print version will be issued in April 2016.
The research team adopted a systems metabolic engineering approach to develop a microorganism that can produce diverse non-natural, biomedically important polymers and succeeded in synthesizing poly(lactate-co-glycolate) (PLGA), a copolymer of two different polymer monomers, lactic and glycolic acid. PLGA is biodegradable, biocompatible, and non-toxic, and has been widely used in biomedical and therapeutic applications such as surgical sutures, prosthetic devices, drug delivery, and tissue engineering.
Inspired by the biosynthesis process for polyhydroxyalkanoates (PHAs), biologically-derived polyesters produced in nature by the bacterial fermentation of sugar or lipids, the research team designed a metabolic pathway for the biosynthesis of PLGA through microbial fermentation directly from carbohydrates in Escherichia coli (E. coli) strains.
The team had previously reported a recombinant E. coli producing PLGA by using the glyoxylate shunt pathway for the generation of glycolate from glucose, which was disclosed in their patents KR10-1575585-0000 (filing date of March 11, 2011), US08883463 and JP5820363. However, they discovered that the polymer content and glycolate fraction of PLGA could not be significantly enhanced via further engineering techniques. Thus, in this research, the team introduced a heterologous pathway to produce glycolate from xylose and succeeded in developing the recombinant E. coli producing PLGA and various novel copolymers much more efficiently.
In order to produce PLGA by microbial fermentation directly from carbohydrates, the team incorporated external and engineered enzymes as catalysts to co-polymerize PLGA while establishing a few additional metabolic pathways for the biosynthesis to produce a range of different non-natural polymers, some for the first time. This bio-based synthetic process for PLGA and other polymers could substitute for the existing complicated chemical production that involves the preparation and purification of precursors, chemical polymerization processes, and the elimination of metal catalysts.
Professor Lee and his team performed in silico genome-scale metabolic simulations of the E. coli cell to predict and analyze changes in the metabolic fluxes of cells which were caused by the introduction of external metabolic pathways. Based on these results, genes are manipulated to optimize metabolic fluxes by eliminating the genes responsible for byproducts formation and enhancing the expression levels of certain genes, thereby achieving the effective production of target polymers as well as stimulating cell growth.
The team utilized the structural basis of broad substrate specificity of the key synthesizing enzyme, PHA synthase, to incorporate various co-monomers with main and side chains of different lengths. These monomers were produced inside the cell by metabolic engineering, and then copolymerized to improve the material properties of PLGA. As a result, a variety of PLGA copolymers with different monomer compositions such as the US Food and Drug Administration (FDA)-approved monomers, 3-hydroxyburate, 4-hydroxyburate, and 6-hydroxyhexanoate, were produced. Newly applied bioplastics such as 5-hydroxyvalerate and 2-hydroxyisovalerate were also made.
The team employed a systems metabolic engineering application which, according to the researchers, is the first successful example of biological production of PGLA and several novel copolymers from renewable biomass by one-step direct fermentation of metabolically engineered E.coli.
Professor Lee said, “We presented important findings that non-natural polymers, such as PLGA which is commonly used for drug delivery or biomedical devices, were produced by a metabolically engineered gut bacterium. Our research is meaningful in that it proposes a platform strategy in metabolic engineering, which can be further utilized in the development of numerous non-natural, useful polymers.”
Director Ilsub Baek at the Platform Technology Division of the Ministry of Science, ICT and Future Planning of Korea, who oversees the Technology Development Program to Solve Climate Change, said, “Professor Lee has led one of our research projects, the Systems Metabolic Engineering for Biorefineries, which began as part of the Ministry’s Technology Development Program to Solve Climate Change. He and his team have continuously achieved promising results and been attracting greater interest from the global scientific community. As climate change technology grows more important, this research on the biological production of non-natural, high value polymers will have a great impact on science and industry.”
The title of the research paper is “One-step Fermentative Production of Poly(lactate-co-glycolate) from Carbohydrates in Escherichia coli (DOI: 10.1038/nbt.3485).” The lead authors are So Young Choi, a Ph.D. candidate in the Department of Chemical and Biomolecular Engineering at KAIST, and Si Jae Park, Assistant Professor of the Environmental Engineering and Energy Department at Myongji University. Won Jun Kim and Jung Eun Yang, both doctoral students in the Department of Chemical and Biomolecular Engineering at KAIST, also participated in the research.
This research was supported by the Technology Development Program to Solve Climate Change’s research project titled “Systems Metabolic Engineering for Biorefineries” from the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea (NRF-2012M1A2A2026556).
Figure: Production of PLGA and Other Non-Natural Copolymers
This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced a variety of PLGAs with different monomer compositions, proposing the chemosynthetic process of non-natural polymers from biomass. The non-natural polymer PLGA and its other copolymers, which were produced by engineered bacteria developed by taking a systems metabolic engineering approach, accumulate in granule forms within a cell.
2016.03.08 View 12887
Non-Natural Biomedical Polymers Produced from Microorganisms
KAIST researchers have developed metabolically engineered Escherichia coli strains to synthesize non-natural, biomedically important polymers including poly(lactate-co-glycolate) (PLGA), previously considered impossible to obtain from biobased materials.
Renewable non-food biomass could potentially replace petrochemical raw materials to produce energy sources, useful chemicals, or a vast array of petroleum-based end products such as plastics, lubricants, paints, fertilizers, and vitamin capsules. In recent years, biorefineries which transform non-edible biomass into fuel, heat, power, chemicals, and materials have received a great deal of attention as a sustainable alternative to decreasing the reliance on fossil fuels.
A research team headed by Distinguished Professor Sang Yup Lee of the Chemical and Biomolecular Engineering Department at KAIST has established a biorefinery system to create non-natural polymers from natural sources, allowing various plastics to be made in an environmentally-friendly and sustainable manner. The research results were published online in Nature Biotechnology on March 7, 2016. The print version will be issued in April 2016.
The research team adopted a systems metabolic engineering approach to develop a microorganism that can produce diverse non-natural, biomedically important polymers and succeeded in synthesizing poly(lactate-co-glycolate) (PLGA), a copolymer of two different polymer monomers, lactic and glycolic acid. PLGA is biodegradable, biocompatible, and non-toxic, and has been widely used in biomedical and therapeutic applications such as surgical sutures, prosthetic devices, drug delivery, and tissue engineering.
Inspired by the biosynthesis process for polyhydroxyalkanoates (PHAs), biologically-derived polyesters produced in nature by the bacterial fermentation of sugar or lipids, the research team designed a metabolic pathway for the biosynthesis of PLGA through microbial fermentation directly from carbohydrates in Escherichia coli (E. coli) strains.
The team had previously reported a recombinant E. coli producing PLGA by using the glyoxylate shunt pathway for the generation of glycolate from glucose, which was disclosed in their patents KR10-1575585-0000 (filing date of March 11, 2011), US08883463 and JP5820363. However, they discovered that the polymer content and glycolate fraction of PLGA could not be significantly enhanced via further engineering techniques. Thus, in this research, the team introduced a heterologous pathway to produce glycolate from xylose and succeeded in developing the recombinant E. coli producing PLGA and various novel copolymers much more efficiently.
In order to produce PLGA by microbial fermentation directly from carbohydrates, the team incorporated external and engineered enzymes as catalysts to co-polymerize PLGA while establishing a few additional metabolic pathways for the biosynthesis to produce a range of different non-natural polymers, some for the first time. This bio-based synthetic process for PLGA and other polymers could substitute for the existing complicated chemical production that involves the preparation and purification of precursors, chemical polymerization processes, and the elimination of metal catalysts.
Professor Lee and his team performed in silico genome-scale metabolic simulations of the E. coli cell to predict and analyze changes in the metabolic fluxes of cells which were caused by the introduction of external metabolic pathways. Based on these results, genes are manipulated to optimize metabolic fluxes by eliminating the genes responsible for byproducts formation and enhancing the expression levels of certain genes, thereby achieving the effective production of target polymers as well as stimulating cell growth.
The team utilized the structural basis of broad substrate specificity of the key synthesizing enzyme, PHA synthase, to incorporate various co-monomers with main and side chains of different lengths. These monomers were produced inside the cell by metabolic engineering, and then copolymerized to improve the material properties of PLGA. As a result, a variety of PLGA copolymers with different monomer compositions such as the US Food and Drug Administration (FDA)-approved monomers, 3-hydroxyburate, 4-hydroxyburate, and 6-hydroxyhexanoate, were produced. Newly applied bioplastics such as 5-hydroxyvalerate and 2-hydroxyisovalerate were also made.
The team employed a systems metabolic engineering application which, according to the researchers, is the first successful example of biological production of PGLA and several novel copolymers from renewable biomass by one-step direct fermentation of metabolically engineered E.coli.
Professor Lee said, “We presented important findings that non-natural polymers, such as PLGA which is commonly used for drug delivery or biomedical devices, were produced by a metabolically engineered gut bacterium. Our research is meaningful in that it proposes a platform strategy in metabolic engineering, which can be further utilized in the development of numerous non-natural, useful polymers.”
Director Ilsub Baek at the Platform Technology Division of the Ministry of Science, ICT and Future Planning of Korea, who oversees the Technology Development Program to Solve Climate Change, said, “Professor Lee has led one of our research projects, the Systems Metabolic Engineering for Biorefineries, which began as part of the Ministry’s Technology Development Program to Solve Climate Change. He and his team have continuously achieved promising results and been attracting greater interest from the global scientific community. As climate change technology grows more important, this research on the biological production of non-natural, high value polymers will have a great impact on science and industry.”
The title of the research paper is “One-step Fermentative Production of Poly(lactate-co-glycolate) from Carbohydrates in Escherichia coli (DOI: 10.1038/nbt.3485).” The lead authors are So Young Choi, a Ph.D. candidate in the Department of Chemical and Biomolecular Engineering at KAIST, and Si Jae Park, Assistant Professor of the Environmental Engineering and Energy Department at Myongji University. Won Jun Kim and Jung Eun Yang, both doctoral students in the Department of Chemical and Biomolecular Engineering at KAIST, also participated in the research.
This research was supported by the Technology Development Program to Solve Climate Change’s research project titled “Systems Metabolic Engineering for Biorefineries” from the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea (NRF-2012M1A2A2026556).
Figure: Production of PLGA and Other Non-Natural Copolymers
This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced a variety of PLGAs with different monomer compositions, proposing the chemosynthetic process of non-natural polymers from biomass. The non-natural polymer PLGA and its other copolymers, which were produced by engineered bacteria developed by taking a systems metabolic engineering approach, accumulate in granule forms within a cell.
2016.03.08 View 12887 -
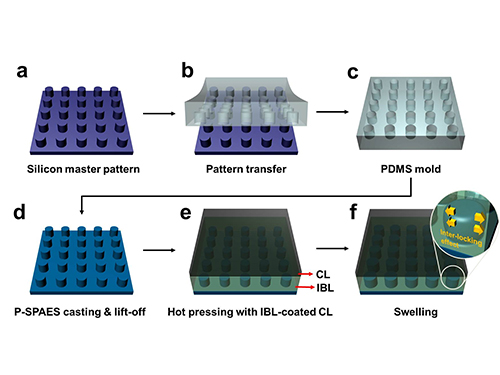 Membrane
Scientists at KAIST have developed a new way of making fuel cell membranes using nanoscale fasteners, paving the way for lower-cost, higher-efficiency and more easily manufactured fuel cells.
The internal workings of fuel cells vary, but basically all types mix hydrogen and oxygen to produce a chemical reaction that delivers usable electricity and exhausts ordinary water as a by-product. One of the most efficient types is the proton exchange membrane (PEM) fuel cell, which operates at low enough temperatures to be used in homes and vehicles.
To generate electricity, PEM fuel cells rely on two chemical compartments separated by a permeable catalyst membrane. This membrane acts as an electrolyte; a negative electrode is bonded to one side of the membrane and a positive electrode is bonded to the other. The electrolyte membrane is often based on a polymer of perfluorosulfonic acid. Due to its high cost, however, a less expensive hydrocarbon-based electrolyte membrane has attracted interest in this technology sector.
Until now, the challenge in adopting such a hydrocarbon membrane has been that the interface between the electrode and hydrocarbon membrane is weak. This causes the membrane to delaminate relatively easily, falling apart and losing efficiency with use.
Professor Hee-Tak Kim of the Department of Chemical and Biomolecular Engineering at the Korea Advanced Institute of Science and Technology (KAIST) and his research team have developed a new fastening system that bonds the two materials mechanically rather than chemically. This opens the way to the development of fuel cell membranes that are less expensive, easier to manufacture, stronger and more efficient.
The researchers achieved this by moulding a pattern of tiny cylindrical pillars on the face of the hydrocarbon membrane. The pillars protrude into a softened skin of the electrode with heat. The mechanical bond sets and strengthens as the material cools and absorbs water. The pillar-patterned hydrocarbon membrane is cast using silicone moulds.
Professor Kim said, “This physically fastened bond is almost five times stronger and harder to separate than current bonds between the same layers.”
The new interlocking method also appears to offer a way to bond many types of hydrocarbon membranes that, until now, have been rejected because they couldn’t be fastened robustly. This would make hydrocarbon membranes practical for a number of applications beyond fuel cells such as rechargeable “redox flow” batteries.
The research team is now developing a stronger and more scalable interlocking interface for their nanoscale fasteners.
Picture: Schematic Diagram of the Fabrication of the Pillar P-SPAES Membrane and Its Working Principle of Interlocking Effects
2015.11.06 View 10685
Membrane
Scientists at KAIST have developed a new way of making fuel cell membranes using nanoscale fasteners, paving the way for lower-cost, higher-efficiency and more easily manufactured fuel cells.
The internal workings of fuel cells vary, but basically all types mix hydrogen and oxygen to produce a chemical reaction that delivers usable electricity and exhausts ordinary water as a by-product. One of the most efficient types is the proton exchange membrane (PEM) fuel cell, which operates at low enough temperatures to be used in homes and vehicles.
To generate electricity, PEM fuel cells rely on two chemical compartments separated by a permeable catalyst membrane. This membrane acts as an electrolyte; a negative electrode is bonded to one side of the membrane and a positive electrode is bonded to the other. The electrolyte membrane is often based on a polymer of perfluorosulfonic acid. Due to its high cost, however, a less expensive hydrocarbon-based electrolyte membrane has attracted interest in this technology sector.
Until now, the challenge in adopting such a hydrocarbon membrane has been that the interface between the electrode and hydrocarbon membrane is weak. This causes the membrane to delaminate relatively easily, falling apart and losing efficiency with use.
Professor Hee-Tak Kim of the Department of Chemical and Biomolecular Engineering at the Korea Advanced Institute of Science and Technology (KAIST) and his research team have developed a new fastening system that bonds the two materials mechanically rather than chemically. This opens the way to the development of fuel cell membranes that are less expensive, easier to manufacture, stronger and more efficient.
The researchers achieved this by moulding a pattern of tiny cylindrical pillars on the face of the hydrocarbon membrane. The pillars protrude into a softened skin of the electrode with heat. The mechanical bond sets and strengthens as the material cools and absorbs water. The pillar-patterned hydrocarbon membrane is cast using silicone moulds.
Professor Kim said, “This physically fastened bond is almost five times stronger and harder to separate than current bonds between the same layers.”
The new interlocking method also appears to offer a way to bond many types of hydrocarbon membranes that, until now, have been rejected because they couldn’t be fastened robustly. This would make hydrocarbon membranes practical for a number of applications beyond fuel cells such as rechargeable “redox flow” batteries.
The research team is now developing a stronger and more scalable interlocking interface for their nanoscale fasteners.
Picture: Schematic Diagram of the Fabrication of the Pillar P-SPAES Membrane and Its Working Principle of Interlocking Effects
2015.11.06 View 10685 -
 KAIST and Hanwha Chemical Agree on Research Collaboration
KAIST signed a memorandum of understanding (MOU) with Hanwha Chemical Co., Ltd., a Korean chemical and auto manufacturer, on November 2, 2015 to establish a research center on campus.
The research center, which will be named “KAIST-Hanwha Chemical Future Technology Research Center,” will implement joint research projects for five years beginning from 2016 to develop innovative, green technologies that will help the Korean chemical industry boost its global competitiveness and to nurture top researchers and engineers in chemical engineering.
The research center will lead the development of next-generation petrochemical materials and manufacturing technology and the establishment of pure high-refining processes which are more energy-efficient and environmentally friendly. KAIST and Hanwha will strive to secure new technologies that have the greatest commercialization potential in the global market. They will also establish a scholarship fund for 15 KAIST doctoral students in the Department of Chemical and Biomolecular Engineering.
Many professors from the Chemical and Biomolecular Engineering Department including Distinguished Professor Sang Yup Lee, who was listed in the Top 20 Translational Researchers of 2014 by Nature Biotechnology this year, and Professor Hyunjoo Lee who received the Woman Scholar award at the 2015 World Chemistry Conference, will work at the research center.
Professor Lee, the head of the research center, said, “Collaborating with Hanwha will give us a strong basis for our efforts to carry out original research and train the best researchers in the field.”
Chang-Bum Kim, the Chief Executive Officer (CEO) of Hanwha Chemical, said,
“We hope our collaborations with KAIST will go beyond the typical industry and university cooperation. The two organizations will indeed jointly operate the research center, and this will become a new model for industry and university cooperation. We expect that the research center will play a crucial role in the development of new products and technologies to grow the Korean chemical industry.”
In the photo, President Steve Kang of KAIST (fourth from left) and CEO Chang-Bum Kim of Hanwha Chemical (fifth from left) hold the MOU together.
2015.11.01 View 13355
KAIST and Hanwha Chemical Agree on Research Collaboration
KAIST signed a memorandum of understanding (MOU) with Hanwha Chemical Co., Ltd., a Korean chemical and auto manufacturer, on November 2, 2015 to establish a research center on campus.
The research center, which will be named “KAIST-Hanwha Chemical Future Technology Research Center,” will implement joint research projects for five years beginning from 2016 to develop innovative, green technologies that will help the Korean chemical industry boost its global competitiveness and to nurture top researchers and engineers in chemical engineering.
The research center will lead the development of next-generation petrochemical materials and manufacturing technology and the establishment of pure high-refining processes which are more energy-efficient and environmentally friendly. KAIST and Hanwha will strive to secure new technologies that have the greatest commercialization potential in the global market. They will also establish a scholarship fund for 15 KAIST doctoral students in the Department of Chemical and Biomolecular Engineering.
Many professors from the Chemical and Biomolecular Engineering Department including Distinguished Professor Sang Yup Lee, who was listed in the Top 20 Translational Researchers of 2014 by Nature Biotechnology this year, and Professor Hyunjoo Lee who received the Woman Scholar award at the 2015 World Chemistry Conference, will work at the research center.
Professor Lee, the head of the research center, said, “Collaborating with Hanwha will give us a strong basis for our efforts to carry out original research and train the best researchers in the field.”
Chang-Bum Kim, the Chief Executive Officer (CEO) of Hanwha Chemical, said,
“We hope our collaborations with KAIST will go beyond the typical industry and university cooperation. The two organizations will indeed jointly operate the research center, and this will become a new model for industry and university cooperation. We expect that the research center will play a crucial role in the development of new products and technologies to grow the Korean chemical industry.”
In the photo, President Steve Kang of KAIST (fourth from left) and CEO Chang-Bum Kim of Hanwha Chemical (fifth from left) hold the MOU together.
2015.11.01 View 13355 -
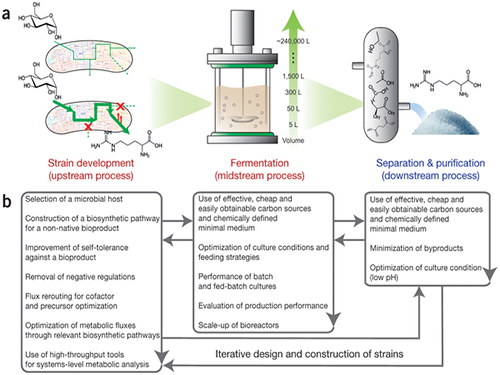 Establishment of System Metabolic Engineering Strategies
Although conventional petrochemical processes have generated chemicals and materials which have been useful to mankind, they have also triggered a variety of environmental problems including climate change and relied too much on nonrenewable natural resources. To ameliorate this, researchers have actively pursued the development of industrial microbial strains around the globe in order to overproduce industrially useful chemicals and materials from microbes using renewable biomass. This discipline is called metabolic engineering.
Thanks to advances in genetic engineering and our knowledge of cellular metabolism, conventional metabolic engineering efforts have succeeded to a certain extent in developing microbial strains that overproduce bioproducts at an industrial level. However, many metabolic engineering projects launched in academic labs do not reach commercial markets due to a failure to fully integrate industrial bioprocesses.
In response to this, Distinguished Professor Sang Yup Lee and Dr. Hyun Uk Kim, both from the Department of Chemical and Biomolecular Engineering at KAIST, have recently suggested ten general strategies of systems metabolic engineering to successfully develop industrial microbial strains. Systems metabolic engineering differs from conventional metabolic engineering by incorporating traditional metabolic engineering approaches along with tools of other fields, such as systems biology, synthetic biology, and molecular evolution.
The ten strategies of systems metabolic engineering have been featured in Nature Biotechnology released online in October 2015, which is entitled "Systems strategies for developing industrial microbial strains."
The strategies cover economic, state-of-the-art biological techniques and traditional bioprocess aspects. Specifically, they consist of: 1) project design including economic evaluation of a target bioproduct; 2) selection of host strains to be used for overproduction of a bioproduct; 3) metabolic pathway reconstruction for bioproducts that are not naturally produced in the selected host strains; 4) increasing tolerance of a host strain against the bioproduct; 5) removing negative regulatory circuits in the microbial host limiting overproduction of a bioproduct; 6) rerouting intracellular fluxes to optimize cofactor and precursor availability necessary for the bioproduct formation; 7) diagnosing and optimizing metabolic fluxes towards product formation; 8) diagnosis and optimization of microbial culture conditions including carbon sources; 9) system-wide gene manipulation to further increase the host strain's production performance using high-throughput genome-scale engineering and computational tools; and 10) scale-up fermentation of the developed strain and diagnosis for the reproducibility of the strain's production performance.
These ten strategies were articulated with successful examples of the production of L-arginine using Corynebacterium glutamicum, 1,4-butanediol using Escherichia coli, and L-lysine and bio-nylon using C. glutamicum.
Professor Sang Yup Lee said, "At the moment, the chance of commercializing microbial strains developed in academic labs is very low. The strategies of systems metabolic engineering outlined in this analysis can serve as guidelines when developing industrial microbial strains. We hope that these strategies contribute to improving opportunities to commercialize microbial strains developed in academic labs with drastically reduced costs and efforts, and that a large fraction of petroleum-based processes will be replaced with sustainable bioprocesses."
Lee S. Y. & Kim, H. U. Systems Strategies for Developing Industrial Microbial Strains. Nature Biotechnology (2015).
This work was supported by the Technology Development Program to Solve Climate Change on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556) and by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) from the Ministry of Science, ICT and Future Planning (MSIP), Korea, and through the National Research Foundation (NRF) of Korea. This work was also supported by the Novo Nordisk Foundation.
Picture: Concept of the Systems Metabolic Engineering Framework
(a) Three major bioprocess stages (b) Considerations in systems metabolic engineering to optimize the whole bioprocess. List of considerations for the strain development and fermentation contribute to improving microbial strain's production performance (red), whereas those for the separation and purification help in reducing overall operation costs by facilitating the downstream process (blue). Some of the considerations can be repeated in the course of systems metabolic engineering.
2015.10.19 View 12003
Establishment of System Metabolic Engineering Strategies
Although conventional petrochemical processes have generated chemicals and materials which have been useful to mankind, they have also triggered a variety of environmental problems including climate change and relied too much on nonrenewable natural resources. To ameliorate this, researchers have actively pursued the development of industrial microbial strains around the globe in order to overproduce industrially useful chemicals and materials from microbes using renewable biomass. This discipline is called metabolic engineering.
Thanks to advances in genetic engineering and our knowledge of cellular metabolism, conventional metabolic engineering efforts have succeeded to a certain extent in developing microbial strains that overproduce bioproducts at an industrial level. However, many metabolic engineering projects launched in academic labs do not reach commercial markets due to a failure to fully integrate industrial bioprocesses.
In response to this, Distinguished Professor Sang Yup Lee and Dr. Hyun Uk Kim, both from the Department of Chemical and Biomolecular Engineering at KAIST, have recently suggested ten general strategies of systems metabolic engineering to successfully develop industrial microbial strains. Systems metabolic engineering differs from conventional metabolic engineering by incorporating traditional metabolic engineering approaches along with tools of other fields, such as systems biology, synthetic biology, and molecular evolution.
The ten strategies of systems metabolic engineering have been featured in Nature Biotechnology released online in October 2015, which is entitled "Systems strategies for developing industrial microbial strains."
The strategies cover economic, state-of-the-art biological techniques and traditional bioprocess aspects. Specifically, they consist of: 1) project design including economic evaluation of a target bioproduct; 2) selection of host strains to be used for overproduction of a bioproduct; 3) metabolic pathway reconstruction for bioproducts that are not naturally produced in the selected host strains; 4) increasing tolerance of a host strain against the bioproduct; 5) removing negative regulatory circuits in the microbial host limiting overproduction of a bioproduct; 6) rerouting intracellular fluxes to optimize cofactor and precursor availability necessary for the bioproduct formation; 7) diagnosing and optimizing metabolic fluxes towards product formation; 8) diagnosis and optimization of microbial culture conditions including carbon sources; 9) system-wide gene manipulation to further increase the host strain's production performance using high-throughput genome-scale engineering and computational tools; and 10) scale-up fermentation of the developed strain and diagnosis for the reproducibility of the strain's production performance.
These ten strategies were articulated with successful examples of the production of L-arginine using Corynebacterium glutamicum, 1,4-butanediol using Escherichia coli, and L-lysine and bio-nylon using C. glutamicum.
Professor Sang Yup Lee said, "At the moment, the chance of commercializing microbial strains developed in academic labs is very low. The strategies of systems metabolic engineering outlined in this analysis can serve as guidelines when developing industrial microbial strains. We hope that these strategies contribute to improving opportunities to commercialize microbial strains developed in academic labs with drastically reduced costs and efforts, and that a large fraction of petroleum-based processes will be replaced with sustainable bioprocesses."
Lee S. Y. & Kim, H. U. Systems Strategies for Developing Industrial Microbial Strains. Nature Biotechnology (2015).
This work was supported by the Technology Development Program to Solve Climate Change on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556) and by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) from the Ministry of Science, ICT and Future Planning (MSIP), Korea, and through the National Research Foundation (NRF) of Korea. This work was also supported by the Novo Nordisk Foundation.
Picture: Concept of the Systems Metabolic Engineering Framework
(a) Three major bioprocess stages (b) Considerations in systems metabolic engineering to optimize the whole bioprocess. List of considerations for the strain development and fermentation contribute to improving microbial strain's production performance (red), whereas those for the separation and purification help in reducing overall operation costs by facilitating the downstream process (blue). Some of the considerations can be repeated in the course of systems metabolic engineering.
2015.10.19 View 12003 -
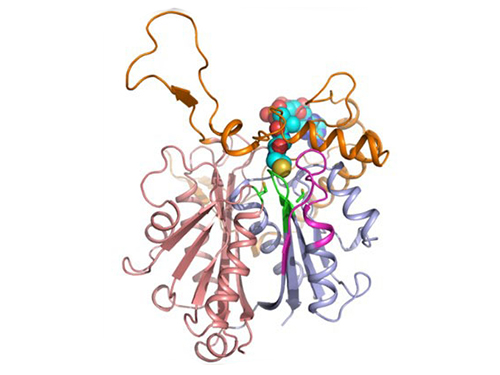 Discovery of Redox-Switch of KEenzyme Involved in N-Butanol Biosynthesis
Research teams at KAIST and Kyungpook National University (KNU) have succeeded in uncovering the redox-switch of thiolase, a key enzyme for n-butanol production in Clostridium acetobutylicum, one of the best known butanol-producing bacteria.
Biological n-butanol production was first reported by Louis Pasteur in 1861, and the bioprocess was industrialized usingClostridium acetobutylicum. The fermentation process by Clostridium strains has been known to be the most efficient one for n-butanol production. Due to growing world-wide issues such as energy security and climate change, the biological production of n-butanol has been receiving much renewed interest. This is because n-butanol possesses much better fuel characteristics compared to ethanol, such as higher energy content (29.2 MJ/L vs 19.6 MJ/L), less corrosiveness, less hygroscopy, and the ease with which it can be blended with gasoline and diesel.
In the paper published in Nature Communications, a broad-scope, online-only, and open access journal issued by the Nature Publishing Group (NPG), on September 22, 2015, Professor Kyung-Jin Kim at the School of Life Sciences, KNU, and Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, KAIST, have proved that the redox-switch of thiolase plays a role in a regulation of metabolic flux in C. acetobutylicum by using in silico modeling and simulation tools.
The research team has redesigned thiolase with enhanced activity on the basis of the 3D structure of the wild-type enzyme. To reinforce a metabolic flux toward butanol production, the metabolic network of C. acetobutylicum strain was engineered with the redesigned enzyme. The combination of the discovery of 3D enzyme structure and systems metabolic engineering approaches resulted in increased n-butanol production in C. acetobutylicum, which allows the production of this important industrial chemical to be cost competitive.
Professors Kim and Lee said, "We have reported the 3D structure of C. acetobutylicum thiolase-a key enzyme involved in n-butanol biosynthesis, for the first time. Further study will be done to produce butanol more economically on the basis of the 3D structure of C. acetobutylicum thiolase."
This work was published online in Nature Communications on September 22, 2015.
Reference: Kim et al. "Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum," Nature Communications
This research was supported by the Technology Development Program to Solve Climate Changes from the Ministry of Education, Science and Technology (MEST), Korea, the National Research Foundation of Korea, and the Advanced Biomass Center through the Global Frontier Research Program of the MEST, Korea.
For further information, contact Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930); and Dr. Kyung-Jin Kim, Professor, KNU, Daegu, Korea (kkim@knu.ac.kr, +82-53-950-6088).
Figure 1: A redox-switch of thiolase involves in butanol biosynthesis in Clostridium acetobutylicum. Thiolase condenses two acetyl-CoA molecules for initiating four carbon flux towards butanol.
Figure 2: Thiolase catalyzes the condensation reaction of acetyl-CoA to acetoacetyl-CoA. Two catalytic cysteine residues at 88th and 378th are oxidized and formed an intermolecular disulfide bond in an oxidized status, which results in inactivation of the enzyme for n-butanol biosynthesis. The intermolecular disulfide bond is broken enabling the n-butanol biosynthesis, when the environment status is reduced.
2015.09.23 View 11795
Discovery of Redox-Switch of KEenzyme Involved in N-Butanol Biosynthesis
Research teams at KAIST and Kyungpook National University (KNU) have succeeded in uncovering the redox-switch of thiolase, a key enzyme for n-butanol production in Clostridium acetobutylicum, one of the best known butanol-producing bacteria.
Biological n-butanol production was first reported by Louis Pasteur in 1861, and the bioprocess was industrialized usingClostridium acetobutylicum. The fermentation process by Clostridium strains has been known to be the most efficient one for n-butanol production. Due to growing world-wide issues such as energy security and climate change, the biological production of n-butanol has been receiving much renewed interest. This is because n-butanol possesses much better fuel characteristics compared to ethanol, such as higher energy content (29.2 MJ/L vs 19.6 MJ/L), less corrosiveness, less hygroscopy, and the ease with which it can be blended with gasoline and diesel.
In the paper published in Nature Communications, a broad-scope, online-only, and open access journal issued by the Nature Publishing Group (NPG), on September 22, 2015, Professor Kyung-Jin Kim at the School of Life Sciences, KNU, and Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, KAIST, have proved that the redox-switch of thiolase plays a role in a regulation of metabolic flux in C. acetobutylicum by using in silico modeling and simulation tools.
The research team has redesigned thiolase with enhanced activity on the basis of the 3D structure of the wild-type enzyme. To reinforce a metabolic flux toward butanol production, the metabolic network of C. acetobutylicum strain was engineered with the redesigned enzyme. The combination of the discovery of 3D enzyme structure and systems metabolic engineering approaches resulted in increased n-butanol production in C. acetobutylicum, which allows the production of this important industrial chemical to be cost competitive.
Professors Kim and Lee said, "We have reported the 3D structure of C. acetobutylicum thiolase-a key enzyme involved in n-butanol biosynthesis, for the first time. Further study will be done to produce butanol more economically on the basis of the 3D structure of C. acetobutylicum thiolase."
This work was published online in Nature Communications on September 22, 2015.
Reference: Kim et al. "Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum," Nature Communications
This research was supported by the Technology Development Program to Solve Climate Changes from the Ministry of Education, Science and Technology (MEST), Korea, the National Research Foundation of Korea, and the Advanced Biomass Center through the Global Frontier Research Program of the MEST, Korea.
For further information, contact Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930); and Dr. Kyung-Jin Kim, Professor, KNU, Daegu, Korea (kkim@knu.ac.kr, +82-53-950-6088).
Figure 1: A redox-switch of thiolase involves in butanol biosynthesis in Clostridium acetobutylicum. Thiolase condenses two acetyl-CoA molecules for initiating four carbon flux towards butanol.
Figure 2: Thiolase catalyzes the condensation reaction of acetyl-CoA to acetoacetyl-CoA. Two catalytic cysteine residues at 88th and 378th are oxidized and formed an intermolecular disulfide bond in an oxidized status, which results in inactivation of the enzyme for n-butanol biosynthesis. The intermolecular disulfide bond is broken enabling the n-butanol biosynthesis, when the environment status is reduced.
2015.09.23 View 11795 -
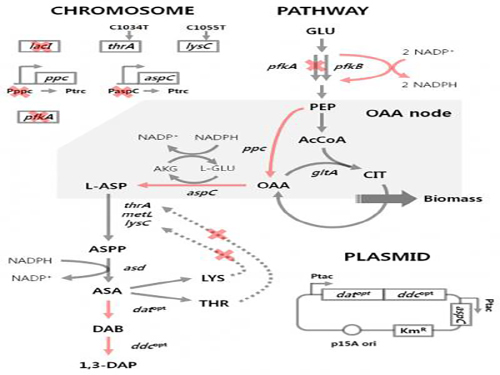 'Engineered Bacterium Produces 1,3-Diaminopropane'
A research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported, for the first time, the production of 1,3-diaminopropane via fermentation of an engineered bacterium.
1,3-Diaminopropane is a three carbon diamine, which has a wide range of industrial applications including epoxy resin and cross-linking agents, as well as precursors for pharmaceuticals, agrochemicals, and organic chemicals. It can also be polymerized with dicarboxylic acids to make polyamides (nylons) for use as engineering plastics, medical materials, and adhesives.
Traditionally, 1,3-diaminopropane is derived from petroleum-based processes. In effort to address critical problems such as the depletion of petroleum and environmental issues inherent to the petroleum-based processes, the research team has developed an Escherichia coli (E. coli) strain capable of producing 1,3-diaminopropane. Using this technology, 1,3-diaminopropane can now be produced from renewable biomass instead of petroleum.
E. coli as found in nature is unable to produce 1,3-diaminopropane. Metabolic engineering, a technology to transform microorganisms into highly efficient microbial cell factories capable of producing chemical compounds of interest, was utilized to engineer the E. coli strain. First, naturally existing metabolic pathways for the biosynthesis of 1,3-diaminopropane were introduced into a virtual cell in silico to determine the most efficient metabolic pathway for the 1,3-diaminopropane production. The metabolic pathway selected was then introduced into an E. coli strain and successfully produced 1,3-diaminopropane for the first time in the world.
The research team applied metabolic engineering additionally, and the production titer of 1,3-diaminopropane increased about 21 fold. The Fed-batch fermentation of the engineered E. coli strain produced 13 grams per liter of 1,3-diaminoproapne. With this technology, 1,3-diaminopropane can be produced using renewable biomass, and it will be the starting point for replacing the current petroleum-based processes with bio-based processes.
Professor Lee said, “Our study suggested a possibility to produce 1,3-diaminopropane based on biorefinery. Further study will be done to increase the titer and productivity of 1,3-diaminopropane.”
This work was published online in Scientific Reports on August 11, 2015.
Reference: Chae, T.U. et al. "Metabolic engineering of Escherichia coli for the production of 1,3-diaminopropane, a three carbon diamine," Scientific Reports:
http://www.nature.com/articles/srep13040
This research was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from Ministry of Science, ICT and Future Planning (MSIP) through the National Research Foundation (NRF) of Korea.
Figure 1: Metabolic engineering strategies for 1,3-diaminopropane production using C4 pathway
Figure 2: Fed-batch fermentation profiles of two final engineered E. coli strains
2015.08.12 View 11969
'Engineered Bacterium Produces 1,3-Diaminopropane'
A research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported, for the first time, the production of 1,3-diaminopropane via fermentation of an engineered bacterium.
1,3-Diaminopropane is a three carbon diamine, which has a wide range of industrial applications including epoxy resin and cross-linking agents, as well as precursors for pharmaceuticals, agrochemicals, and organic chemicals. It can also be polymerized with dicarboxylic acids to make polyamides (nylons) for use as engineering plastics, medical materials, and adhesives.
Traditionally, 1,3-diaminopropane is derived from petroleum-based processes. In effort to address critical problems such as the depletion of petroleum and environmental issues inherent to the petroleum-based processes, the research team has developed an Escherichia coli (E. coli) strain capable of producing 1,3-diaminopropane. Using this technology, 1,3-diaminopropane can now be produced from renewable biomass instead of petroleum.
E. coli as found in nature is unable to produce 1,3-diaminopropane. Metabolic engineering, a technology to transform microorganisms into highly efficient microbial cell factories capable of producing chemical compounds of interest, was utilized to engineer the E. coli strain. First, naturally existing metabolic pathways for the biosynthesis of 1,3-diaminopropane were introduced into a virtual cell in silico to determine the most efficient metabolic pathway for the 1,3-diaminopropane production. The metabolic pathway selected was then introduced into an E. coli strain and successfully produced 1,3-diaminopropane for the first time in the world.
The research team applied metabolic engineering additionally, and the production titer of 1,3-diaminopropane increased about 21 fold. The Fed-batch fermentation of the engineered E. coli strain produced 13 grams per liter of 1,3-diaminoproapne. With this technology, 1,3-diaminopropane can be produced using renewable biomass, and it will be the starting point for replacing the current petroleum-based processes with bio-based processes.
Professor Lee said, “Our study suggested a possibility to produce 1,3-diaminopropane based on biorefinery. Further study will be done to increase the titer and productivity of 1,3-diaminopropane.”
This work was published online in Scientific Reports on August 11, 2015.
Reference: Chae, T.U. et al. "Metabolic engineering of Escherichia coli for the production of 1,3-diaminopropane, a three carbon diamine," Scientific Reports:
http://www.nature.com/articles/srep13040
This research was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from Ministry of Science, ICT and Future Planning (MSIP) through the National Research Foundation (NRF) of Korea.
Figure 1: Metabolic engineering strategies for 1,3-diaminopropane production using C4 pathway
Figure 2: Fed-batch fermentation profiles of two final engineered E. coli strains
2015.08.12 View 11969 -
 Affordable Genetic Diagnostic Technique for Target DNA Analysis Developed
Professor Hyun-Gyu Park of the Department of Chemical and Biomolecular Engineering at KAIST has developed a technique to analyze various target DNAs using an aptamer, a DNA fragment that can recognize and bind to a specific protein or enzyme. This technique will allow the development of affordable genetic diagnoses for new bacteria or virus, such as Middle Ease Respiratory Syndrome (MERS). The research findings were published in the June issue of Chemical Communications, issued by the Royal Society of Chemistry in the United Kingdom. The paper was selected as a lead article of the journal.
The existing genetic diagnosis technique, based on molecular beacon probes, requires a new beacon probe whenever a target DNA mutates. As a result, it was costly to analyze various target DNA fragments. To address this problem, Professor Park’s team designed an aptamer that binds and deactivates DNA polymerase. The technique was used in reverse, so that the aptemer did not bind to the polymerase, maintaining its activated state, only if the target DNA was present. These probes are called TagMan probes.
The controlled activation and deactivation of DNA polymerase enables nucleic acid to elongate or dwindle, making it possible to measure fluorescence signals coming from TaqMan probes. This same probe can be used to detect various target DNAs, leading to the development of a new and sensitive genetic diagnostic technique.
Unlike the existing molecular beacon probe technique which requires a new probe for every target DNA, this new technique uses the same fluorescent TaqMan probe, which is cheaper and easier to detect a number of different target nucleic acid fragments. The application of this technique will make the process of identifying and detecting foreign DNAs from pathogens such as virus and bacteria more affordable and simple.
Professor Park said, “This technique will enable us to develop simpler diagnostic kits for new pathogens, such as MERS, allowing a faster response to various diseases. Our technology can also be applied widely in the field of genetic diagnostics.”
Picture: A schematic image of target nucleic acid extracted through the activation and deactivation of DNA polymerase
2015.07.31 View 11103
Affordable Genetic Diagnostic Technique for Target DNA Analysis Developed
Professor Hyun-Gyu Park of the Department of Chemical and Biomolecular Engineering at KAIST has developed a technique to analyze various target DNAs using an aptamer, a DNA fragment that can recognize and bind to a specific protein or enzyme. This technique will allow the development of affordable genetic diagnoses for new bacteria or virus, such as Middle Ease Respiratory Syndrome (MERS). The research findings were published in the June issue of Chemical Communications, issued by the Royal Society of Chemistry in the United Kingdom. The paper was selected as a lead article of the journal.
The existing genetic diagnosis technique, based on molecular beacon probes, requires a new beacon probe whenever a target DNA mutates. As a result, it was costly to analyze various target DNA fragments. To address this problem, Professor Park’s team designed an aptamer that binds and deactivates DNA polymerase. The technique was used in reverse, so that the aptemer did not bind to the polymerase, maintaining its activated state, only if the target DNA was present. These probes are called TagMan probes.
The controlled activation and deactivation of DNA polymerase enables nucleic acid to elongate or dwindle, making it possible to measure fluorescence signals coming from TaqMan probes. This same probe can be used to detect various target DNAs, leading to the development of a new and sensitive genetic diagnostic technique.
Unlike the existing molecular beacon probe technique which requires a new probe for every target DNA, this new technique uses the same fluorescent TaqMan probe, which is cheaper and easier to detect a number of different target nucleic acid fragments. The application of this technique will make the process of identifying and detecting foreign DNAs from pathogens such as virus and bacteria more affordable and simple.
Professor Park said, “This technique will enable us to develop simpler diagnostic kits for new pathogens, such as MERS, allowing a faster response to various diseases. Our technology can also be applied widely in the field of genetic diagnostics.”
Picture: A schematic image of target nucleic acid extracted through the activation and deactivation of DNA polymerase
2015.07.31 View 11103 -
 2015 QS World University Rankings by Subject: KAIST's Chemical Engineering ranks 17th and 19th for Materials Science in the World
Chemical Engineering (1st in Korea)
1
MIT (US)
2
UC Berkeley (US)
3
Stanford University (US)
4
University of Cambridge (UK)
5
National University of Singapore (Singapore)
17
KAIST (Korea)
Materials Science and Engineering (1st in Korea)
1
MIT (US)
2
Stanford University (US)
3
UC Berkeley (US)
4
University of Cambridge (UK)
5
North Western University (US)
19
KAIST (Korea)
Electrical and Electronic Engineering (1st in Korea)
1
MIT (US)
2
Stanford University (US)
3
UC Berkeley (US)
4
Harvard University (US)
5
ETH Zurich – Swiss Federal Institute of Technology (Switzerland)
22
KAIST (Korea)
Civil and Structural Engineering (1st in Korea)
1
MIT (US)
2
Delft University of Technology (The Netherlands)
3
National University of Singapore (Singapore)
4
Imperial College London (UK)
5
University of Cambridge (UK)
22
KAIST (Korea)
Mechanical, Aeronautical and Manufacturing Engineering (1st in Korea)
1
MIT (US)
2
Stanford University (US)
3
University of Cambridge (UK)
4
UC Berkeley (US)
5
Michigan University (US)
26
KAIST (Korea)
Chemistry (2nd in Korea)
1
MIT (US)
2
UC Berkeley (US)
3
University of Cambridge (UK)
4
Harvard University (US)
5
University of Oxford (UK)
26
KAIST (Korea)
Computer Science and Information Systems (1st in Korea)
1
MIT (US)
2
Stanford University (US)
3
University of Oxford (UK)
4
Carnegie Mellon University (US)
Harvard University (US)
39
KAIST (Korea)
The QS World University Rankings released its 2015 rankings by subject on April 29, 2015.
According to the rankings, KAIST’s Chemical and Biomolecular Engineering and Materials Science Engineering were listed in the top 20 global universities, 17th and 19th, respectively.
KAIST took first place in six subjects among Korean universities, including electrical and electronic engineering; civil and structural engineering; mechanical, aeronautical and manufacturing engineering; and computer science and information systems.
The QS World University Rankings by Subject highlights the world’s top universities in a range of popular subject areas, covering 36 subjects as of this year. Published annually since 2011, the rankings are based on academic reputation, employer reputation, citation count, and research impact.
For a full list of the rankings: http://www.topuniversities.com/subject-rankings/2015
2015.04.29 View 6240
2015 QS World University Rankings by Subject: KAIST's Chemical Engineering ranks 17th and 19th for Materials Science in the World
Chemical Engineering (1st in Korea)
1
MIT (US)
2
UC Berkeley (US)
3
Stanford University (US)
4
University of Cambridge (UK)
5
National University of Singapore (Singapore)
17
KAIST (Korea)
Materials Science and Engineering (1st in Korea)
1
MIT (US)
2
Stanford University (US)
3
UC Berkeley (US)
4
University of Cambridge (UK)
5
North Western University (US)
19
KAIST (Korea)
Electrical and Electronic Engineering (1st in Korea)
1
MIT (US)
2
Stanford University (US)
3
UC Berkeley (US)
4
Harvard University (US)
5
ETH Zurich – Swiss Federal Institute of Technology (Switzerland)
22
KAIST (Korea)
Civil and Structural Engineering (1st in Korea)
1
MIT (US)
2
Delft University of Technology (The Netherlands)
3
National University of Singapore (Singapore)
4
Imperial College London (UK)
5
University of Cambridge (UK)
22
KAIST (Korea)
Mechanical, Aeronautical and Manufacturing Engineering (1st in Korea)
1
MIT (US)
2
Stanford University (US)
3
University of Cambridge (UK)
4
UC Berkeley (US)
5
Michigan University (US)
26
KAIST (Korea)
Chemistry (2nd in Korea)
1
MIT (US)
2
UC Berkeley (US)
3
University of Cambridge (UK)
4
Harvard University (US)
5
University of Oxford (UK)
26
KAIST (Korea)
Computer Science and Information Systems (1st in Korea)
1
MIT (US)
2
Stanford University (US)
3
University of Oxford (UK)
4
Carnegie Mellon University (US)
Harvard University (US)
39
KAIST (Korea)
The QS World University Rankings released its 2015 rankings by subject on April 29, 2015.
According to the rankings, KAIST’s Chemical and Biomolecular Engineering and Materials Science Engineering were listed in the top 20 global universities, 17th and 19th, respectively.
KAIST took first place in six subjects among Korean universities, including electrical and electronic engineering; civil and structural engineering; mechanical, aeronautical and manufacturing engineering; and computer science and information systems.
The QS World University Rankings by Subject highlights the world’s top universities in a range of popular subject areas, covering 36 subjects as of this year. Published annually since 2011, the rankings are based on academic reputation, employer reputation, citation count, and research impact.
For a full list of the rankings: http://www.topuniversities.com/subject-rankings/2015
2015.04.29 View 6240 -
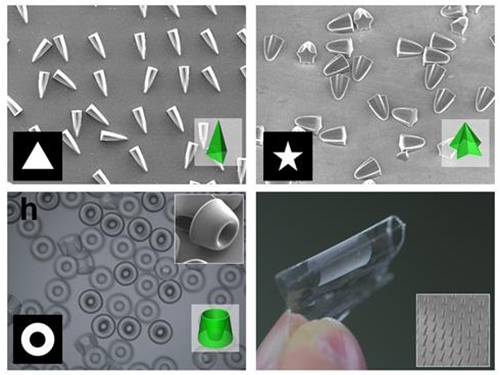 Novel Photolithographic Technology Enabling 3D Control over Functional Shapes of Microstructures
Professor Shin-Hyun Kim and his research team in the Department of Chemical and Biomolecular Engineering at KAIST have developed a novel photolithographic technology enabling control over the functional shapes of micropatterns using oxygen diffusion.
The research was published online in the March 13th issue of Nature Communications and was selected as a featured image for the journal.
Photolithography is a standard optical process for transferring micropatterns on to a substrate by exposing specific regions of the photoresist layer to ultraviolet (UV) light. It is used widely throughout industries that require micropatterns, especially in the semiconductor manufacturing industry.
Conventional photolithography relied on photomasks which protected certain regions of the substrate from the input UV light. Areas covered by the photomasks remain intact with the base layer while the areas exposed to the UV light are washed away, thus creating a micropattern. This technology was limited to a two-dimensional, disc-shaped design as the boundaries between the exposed and roofed regions are always in a parallel arrangement with the direction of the light.
Professor Kim’s research team discovered that: 1) the areas exposed to UV light lowered the concentration of oxygen and thus resulted in oxygen diffusion; and 2) manipulation of the diffusion speed and direction allowed control of the growth, shape and size of the polymers. Based on these findings, the team developed a new photolithographic technology that enabled the production of micropatterns with three-dimensional structures in various shapes and sizes.
Oxygen was considered an inhibitor during photopolymerization. Photoresist under UV light creates radicals which initialize a chemical reaction. These radicals are eliminated with the presence of oxygen and thus prevents the reaction. This suggests that the photoresist must be exposed to UV light for an extended time to completely remove oxygen for a chemical reaction to begin.
The research team, however, exploited the presence of oxygen. While the region affected by the UV light lowered oxygen concentration, the concentration in the untouched region remained unchanged. This difference in the concentrations caused a diffusion of oxygen to the region under UV light.
When the speed of the oxygen flow is slow, the diffusion occurs in parallel with the direction of the UV light. When fast, the diffusion process develops horizontally, outward from the area affected by the UV light. Professor Kim and his team proved this phenomenon both empirically and theoretically. Furthermore, by injecting an external oxygen source, the team was able to manipulate diffusion strength and direction, and thus control the shape and size of the polymer. The use of the polymerization inhibitors enabled and facilitated the fabrication of complex, three-dimensional micropatterns.
Professor Kim said, “While 3D printing is considered an innovative manufacturing technology, it cannot be used for mass-production of microscopic products. The new photolithographic technology will have a broad impact on both the academia and industry especially because existing, conventional photolithographic equipment can be used for the development of more complex micropatterns.” His newest technology will enhance the manufacturing process of three-dimensional polymers which were considered difficult to be commercialized.
The research was also dedicated to the late Professor Seung-Man Yang of the Department of Chemical and Biomolecular Engineering at KAIST. He was considered one of the greatest scholars in Korea in the field of hydrodynamics and colloids.
Picture 1: Featured Image of Nature Communications, March 2015
Picture 2: Polymers with various shapes and sizes produced with the new photolithographic technology developed by Professor Kim
2015.04.06 View 11675
Novel Photolithographic Technology Enabling 3D Control over Functional Shapes of Microstructures
Professor Shin-Hyun Kim and his research team in the Department of Chemical and Biomolecular Engineering at KAIST have developed a novel photolithographic technology enabling control over the functional shapes of micropatterns using oxygen diffusion.
The research was published online in the March 13th issue of Nature Communications and was selected as a featured image for the journal.
Photolithography is a standard optical process for transferring micropatterns on to a substrate by exposing specific regions of the photoresist layer to ultraviolet (UV) light. It is used widely throughout industries that require micropatterns, especially in the semiconductor manufacturing industry.
Conventional photolithography relied on photomasks which protected certain regions of the substrate from the input UV light. Areas covered by the photomasks remain intact with the base layer while the areas exposed to the UV light are washed away, thus creating a micropattern. This technology was limited to a two-dimensional, disc-shaped design as the boundaries between the exposed and roofed regions are always in a parallel arrangement with the direction of the light.
Professor Kim’s research team discovered that: 1) the areas exposed to UV light lowered the concentration of oxygen and thus resulted in oxygen diffusion; and 2) manipulation of the diffusion speed and direction allowed control of the growth, shape and size of the polymers. Based on these findings, the team developed a new photolithographic technology that enabled the production of micropatterns with three-dimensional structures in various shapes and sizes.
Oxygen was considered an inhibitor during photopolymerization. Photoresist under UV light creates radicals which initialize a chemical reaction. These radicals are eliminated with the presence of oxygen and thus prevents the reaction. This suggests that the photoresist must be exposed to UV light for an extended time to completely remove oxygen for a chemical reaction to begin.
The research team, however, exploited the presence of oxygen. While the region affected by the UV light lowered oxygen concentration, the concentration in the untouched region remained unchanged. This difference in the concentrations caused a diffusion of oxygen to the region under UV light.
When the speed of the oxygen flow is slow, the diffusion occurs in parallel with the direction of the UV light. When fast, the diffusion process develops horizontally, outward from the area affected by the UV light. Professor Kim and his team proved this phenomenon both empirically and theoretically. Furthermore, by injecting an external oxygen source, the team was able to manipulate diffusion strength and direction, and thus control the shape and size of the polymer. The use of the polymerization inhibitors enabled and facilitated the fabrication of complex, three-dimensional micropatterns.
Professor Kim said, “While 3D printing is considered an innovative manufacturing technology, it cannot be used for mass-production of microscopic products. The new photolithographic technology will have a broad impact on both the academia and industry especially because existing, conventional photolithographic equipment can be used for the development of more complex micropatterns.” His newest technology will enhance the manufacturing process of three-dimensional polymers which were considered difficult to be commercialized.
The research was also dedicated to the late Professor Seung-Man Yang of the Department of Chemical and Biomolecular Engineering at KAIST. He was considered one of the greatest scholars in Korea in the field of hydrodynamics and colloids.
Picture 1: Featured Image of Nature Communications, March 2015
Picture 2: Polymers with various shapes and sizes produced with the new photolithographic technology developed by Professor Kim
2015.04.06 View 11675