Department+of+Bio
-
 Two Dimensions of Value: Dopamine Neurons Represent Reward but not Aversiveness
Professor Christopher D. Fiorillo of the Bio & Brain Engineering (http://ineuron.kaist.ac.kr/web/home.html) at KAIST published a research paper in the August 2 issue of Science. The title of the paper is “Two Dimensions of Value: Dopamine Neurons Represent Reward but not Aversiveness.” The following is an introduction of his research work:
To make decisions, we need to estimate the value of sensory stimuli and motor actions, their “goodness” and “badness.” We can imagine that good and bad are two ends of a single continuum, or dimension, of value. This would be analogous to the single dimension of light intensity, which ranges from dark on one end to bright light on the other, with many shades of gray in between. Past models of behavior and learning have been based on a single continuum of value, and it has been proposed that a particular group of neurons (brain cells) that use dopamine as a neurotransmitter (chemical messenger) represent the single dimension of value, signaling both good and bad.
The experiments reported here show that dopamine neurons are sensitive to the value of reward but not punishment (like the aversiveness of a bitter taste). This demonstrates that reward and aversiveness are represented as two discrete dimensions (or categories) in the brain. “Reward” refers to the category of good things (food, water, sex, money, etc.), and “punishment” to the category of bad things (stimuli associated with harm to the body and that cause pain or other unpleasant sensations or emotions).
Rather than having one neurotransmitter (dopamine) to represent a single dimension of value, the present results imply the existence of four neurotransmitters to represent two dimensions of value. Dopamine signals evidence for reward (“gains”) and some other neurotransmitter presumably signals evidence against reward (“losses”). Likewise, there should be a neurotransmitter for evidence of danger and another for evidence of safety. It is interesting that there are three other neurotransmitters that are analogous to dopamine in many respects (serotonin, norepinephrine, and acetylcholine), and it is possible that they could represent the other three value signals.
For the research article, please visit: http://www.sciencemag.org/content/341/6145/546.abstract
For the Science 2nd issue, please visit: http://www.sciencemag.org/content/current#ResearchArticles
Illustration of Value Dimension
2013.08.08 View 9153
Two Dimensions of Value: Dopamine Neurons Represent Reward but not Aversiveness
Professor Christopher D. Fiorillo of the Bio & Brain Engineering (http://ineuron.kaist.ac.kr/web/home.html) at KAIST published a research paper in the August 2 issue of Science. The title of the paper is “Two Dimensions of Value: Dopamine Neurons Represent Reward but not Aversiveness.” The following is an introduction of his research work:
To make decisions, we need to estimate the value of sensory stimuli and motor actions, their “goodness” and “badness.” We can imagine that good and bad are two ends of a single continuum, or dimension, of value. This would be analogous to the single dimension of light intensity, which ranges from dark on one end to bright light on the other, with many shades of gray in between. Past models of behavior and learning have been based on a single continuum of value, and it has been proposed that a particular group of neurons (brain cells) that use dopamine as a neurotransmitter (chemical messenger) represent the single dimension of value, signaling both good and bad.
The experiments reported here show that dopamine neurons are sensitive to the value of reward but not punishment (like the aversiveness of a bitter taste). This demonstrates that reward and aversiveness are represented as two discrete dimensions (or categories) in the brain. “Reward” refers to the category of good things (food, water, sex, money, etc.), and “punishment” to the category of bad things (stimuli associated with harm to the body and that cause pain or other unpleasant sensations or emotions).
Rather than having one neurotransmitter (dopamine) to represent a single dimension of value, the present results imply the existence of four neurotransmitters to represent two dimensions of value. Dopamine signals evidence for reward (“gains”) and some other neurotransmitter presumably signals evidence against reward (“losses”). Likewise, there should be a neurotransmitter for evidence of danger and another for evidence of safety. It is interesting that there are three other neurotransmitters that are analogous to dopamine in many respects (serotonin, norepinephrine, and acetylcholine), and it is possible that they could represent the other three value signals.
For the research article, please visit: http://www.sciencemag.org/content/341/6145/546.abstract
For the Science 2nd issue, please visit: http://www.sciencemag.org/content/current#ResearchArticles
Illustration of Value Dimension
2013.08.08 View 9153 -
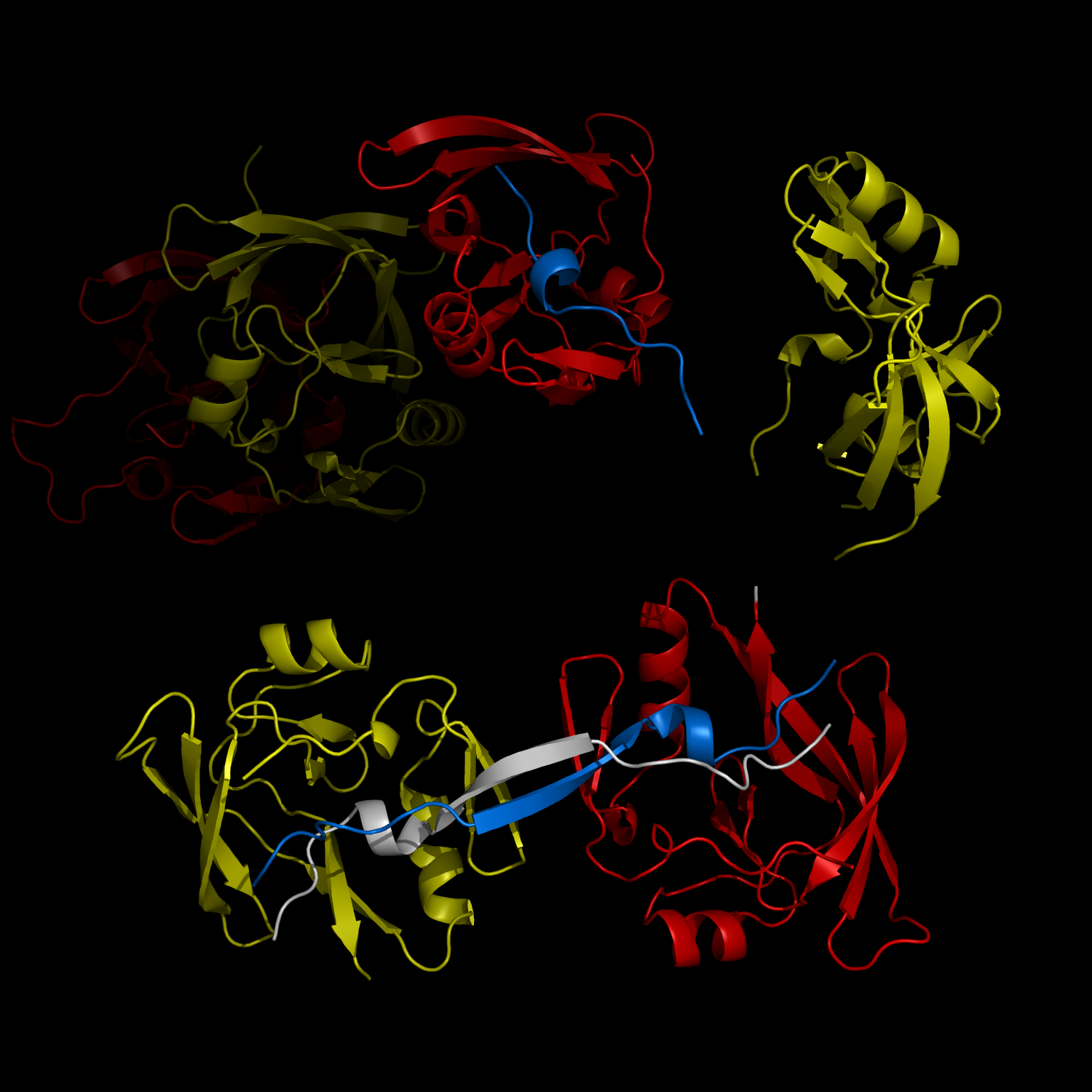 New Structural Insight into Neurodegenerative Disease
A research team from the Korea Advanced Institute of Science and Technology (KAIST) released their results on the structure and molecular details of the neurodegenerative disease-associated protein Ataxin-1. Mutations in Ataxin-1 cause the neurological disease, Spinocerebella Ataxia Type 1 (SCA1), which is characterized by a loss of muscular coordination and balance (ataxia), as is seen in Parkinson’s, Alzheimer’s, and Huntington’s diseases.
SCA1-causing mutations in the ATAXIN1 gene alter the length of a glutamine stretch in the Ataxin-1 protein. The research team provides the first structural insight into the complex formation of ATAXIN-1 with its binding partner, Capicua (CIC). The team, led by Professor Ji-Joon Song from the Department of Biological Sciences at KAIST, solved the structure of Ataxin-1 and CIC complex in atomic level revealing molecular details of the interaction between Ataxin-1 and CIC.
Professor Song explained his recent research work,
“We are able to see the intricate process of complex formation and reconfiguration of the two proteins when they interact with each other. Our work, we expect, will provide a new therapeutic target to modulate SCA1 neurodegenerative disease.”
Understanding structural and molecular details of proteins at the atomic level will help researchers to track the molecular pathogenesis of the disease and, ultimately, design targeted therapies or treatments for patients, rather than just relieving the symptoms of diseases.
Professor Song’s research paper, entitled “Structural Basis of Protein Complex Formation and Reconfiguration by Polyglutamine Disease Protein ATAXIN-1 and Capicua,” will be published in the March 15th issue of Genes & Development (www.genesdev.org).
Complex Formation and Reconfiguration of ATAXIN-1 and Capicua
The complex formation between a polyglutamine disease protein, ATXIN-1 and the transcriptional repressor Capicua (CIC) plays a critical role in SCA 1 pathogenesis. The image shows that the homodimerization of ATXIN-1 (yellow and red) is disrupted upon binding of CIC (blue). Furthermore, the binding of CIC to the ATXIN-1 induces a new form of ATXIN-1 dimerization mediated by CICs (ATXIN-1 AXH domains are shown in yellow and red, and CIC peptides shown in blue and white).
2013.04.02 View 10292
New Structural Insight into Neurodegenerative Disease
A research team from the Korea Advanced Institute of Science and Technology (KAIST) released their results on the structure and molecular details of the neurodegenerative disease-associated protein Ataxin-1. Mutations in Ataxin-1 cause the neurological disease, Spinocerebella Ataxia Type 1 (SCA1), which is characterized by a loss of muscular coordination and balance (ataxia), as is seen in Parkinson’s, Alzheimer’s, and Huntington’s diseases.
SCA1-causing mutations in the ATAXIN1 gene alter the length of a glutamine stretch in the Ataxin-1 protein. The research team provides the first structural insight into the complex formation of ATAXIN-1 with its binding partner, Capicua (CIC). The team, led by Professor Ji-Joon Song from the Department of Biological Sciences at KAIST, solved the structure of Ataxin-1 and CIC complex in atomic level revealing molecular details of the interaction between Ataxin-1 and CIC.
Professor Song explained his recent research work,
“We are able to see the intricate process of complex formation and reconfiguration of the two proteins when they interact with each other. Our work, we expect, will provide a new therapeutic target to modulate SCA1 neurodegenerative disease.”
Understanding structural and molecular details of proteins at the atomic level will help researchers to track the molecular pathogenesis of the disease and, ultimately, design targeted therapies or treatments for patients, rather than just relieving the symptoms of diseases.
Professor Song’s research paper, entitled “Structural Basis of Protein Complex Formation and Reconfiguration by Polyglutamine Disease Protein ATAXIN-1 and Capicua,” will be published in the March 15th issue of Genes & Development (www.genesdev.org).
Complex Formation and Reconfiguration of ATAXIN-1 and Capicua
The complex formation between a polyglutamine disease protein, ATXIN-1 and the transcriptional repressor Capicua (CIC) plays a critical role in SCA 1 pathogenesis. The image shows that the homodimerization of ATXIN-1 (yellow and red) is disrupted upon binding of CIC (blue). Furthermore, the binding of CIC to the ATXIN-1 induces a new form of ATXIN-1 dimerization mediated by CICs (ATXIN-1 AXH domains are shown in yellow and red, and CIC peptides shown in blue and white).
2013.04.02 View 10292 -
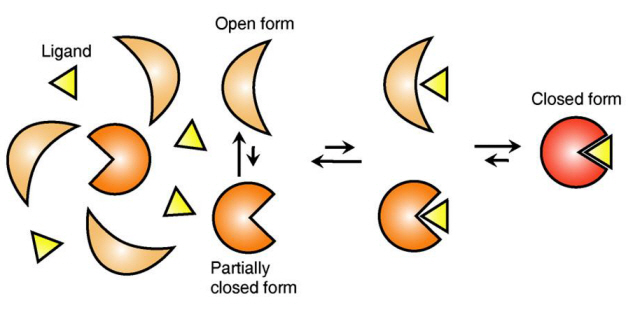 Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 13426
Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 13426 -
 Novel material that prevents health decline with age found
Professor Kim Dae Soo (Department of Biological Science), his research team, the Choong Nam University Medicine School, and various companies conducted collaborative research succeeded in developing a novel material that prevents health decline with age. The result was published in PLoS One Journal with the title “Beta-lapachone, a modulator of NAD metabolism, prevents health declines in aged mice”.
Longevity and health can be obtained with reducing consumption of food and aerobic exercise.
Professor Kim’s team focused on the fact that reduced consumption of food and aerobic exercise increase the coenzyme (NAD+) which suppresses the aging of cells. The research team discovered that by activating NQO1 enzyme with Beta-lapachone, the amount of NAD+ in the body increases even without reduction of food consumption or aerobic exercise.
Even consumption of Beta-lapachone by aging mice caused an improved on the brain and exercise ability of the mice. It is expected that commercialization of Beta-lapachone will be possible as it is a chemical that is commonly found in herbs used in both the orient and the oxidant.
2012.12.21 View 8387
Novel material that prevents health decline with age found
Professor Kim Dae Soo (Department of Biological Science), his research team, the Choong Nam University Medicine School, and various companies conducted collaborative research succeeded in developing a novel material that prevents health decline with age. The result was published in PLoS One Journal with the title “Beta-lapachone, a modulator of NAD metabolism, prevents health declines in aged mice”.
Longevity and health can be obtained with reducing consumption of food and aerobic exercise.
Professor Kim’s team focused on the fact that reduced consumption of food and aerobic exercise increase the coenzyme (NAD+) which suppresses the aging of cells. The research team discovered that by activating NQO1 enzyme with Beta-lapachone, the amount of NAD+ in the body increases even without reduction of food consumption or aerobic exercise.
Even consumption of Beta-lapachone by aging mice caused an improved on the brain and exercise ability of the mice. It is expected that commercialization of Beta-lapachone will be possible as it is a chemical that is commonly found in herbs used in both the orient and the oxidant.
2012.12.21 View 8387 -
 Firefly inspired high efficiency LED technology developed
A firefly inspired, high efficiency self-illuminating LED has been developed.
Professor Jeong Gi Hoon (Department of Bio and Brain Engineering) mimicked the nanostructure of the external layer of the illumination organ of a firefly and succeeded in fabricating high illumination efficiency LED lenses.
Conventional lenses required expensive anti-reflection coating. The developed lenses utilize the bio-inspired nanostructure on the surface of the lenses themselves to reduce the reflectivity of the lenses thereby decreasing production costs.
The developed antireflection nanostructure is expected to be applied to various digital devices and lighting fixtures.
Antireflective structures have been applied in various fields in order to enhance light efficiency However these structures have been limited to flat surfaces and therefore was difficult to implement to curved surfaces like LED lenses.
Professor Jeong’s team solved this problem by using three dimensional micro molding processes.
The team fabricated the nanostructure by forming a single nanoparticle layer on the silicon oxide and performing dry etching. On this nanostructure PDMS was poured and manipulated to fabricate a lens structure similar to that of a firefly.
The fabricated lens showed similar efficiency as conventional antireflection coating.
2012.11.29 View 9540
Firefly inspired high efficiency LED technology developed
A firefly inspired, high efficiency self-illuminating LED has been developed.
Professor Jeong Gi Hoon (Department of Bio and Brain Engineering) mimicked the nanostructure of the external layer of the illumination organ of a firefly and succeeded in fabricating high illumination efficiency LED lenses.
Conventional lenses required expensive anti-reflection coating. The developed lenses utilize the bio-inspired nanostructure on the surface of the lenses themselves to reduce the reflectivity of the lenses thereby decreasing production costs.
The developed antireflection nanostructure is expected to be applied to various digital devices and lighting fixtures.
Antireflective structures have been applied in various fields in order to enhance light efficiency However these structures have been limited to flat surfaces and therefore was difficult to implement to curved surfaces like LED lenses.
Professor Jeong’s team solved this problem by using three dimensional micro molding processes.
The team fabricated the nanostructure by forming a single nanoparticle layer on the silicon oxide and performing dry etching. On this nanostructure PDMS was poured and manipulated to fabricate a lens structure similar to that of a firefly.
The fabricated lens showed similar efficiency as conventional antireflection coating.
2012.11.29 View 9540 -
 Liver Damage Mechanism of Hepatitis C Proven
KAIST researchers found mechanics behind a Hepatitis C virus, thereby taking a step closer to the development of a cure for Hepatitis C.
Professor Choi Chul Hui (Department of Biological and Brain Engineering) and Professor Shin Eui Chul (Graduate School of Medical Sciences) proved, for the first time in the world, the mechanism behind liver damage of a patient with Hepatitis C.
It is anticipated that this discovery will allow for the development of a Hepatitis C cure that has no side effects and little Liver damage.
Hepatitis C is an immune response of the body to the Hepatitis C virus and causes liver irritation.
Around 170million people are infected with Hepatitis C worldwide including 1% of the Korean population. Once infected, most cases turn into chronic cases and may lead to liver cancer.
However it was impossible to infect Hepatitis C within a test tube cell environment until 2005 and up till then Chimpanzees were used to study the virus which proved to be a huge barrier to research.
The research team used cells infected with Hepatitis C virus and found out that the virus works by increasing the destruction of cells by the TNF-a protein responsible for the cell’s immune response.
In addition the protein structure of the virus that causes this reaction was successfully found.
Conventionally the Hepatitis C medication focused on the suppressing the growth of the virus and therefore had many side effects.
The experimental results allow new medication aimed at suppressing the actual mechanism of liver damage to be discovered.
The result was selected as the cover dissertation of the September Edition of the Hepatolog magazine.
2012.09.11 View 14433
Liver Damage Mechanism of Hepatitis C Proven
KAIST researchers found mechanics behind a Hepatitis C virus, thereby taking a step closer to the development of a cure for Hepatitis C.
Professor Choi Chul Hui (Department of Biological and Brain Engineering) and Professor Shin Eui Chul (Graduate School of Medical Sciences) proved, for the first time in the world, the mechanism behind liver damage of a patient with Hepatitis C.
It is anticipated that this discovery will allow for the development of a Hepatitis C cure that has no side effects and little Liver damage.
Hepatitis C is an immune response of the body to the Hepatitis C virus and causes liver irritation.
Around 170million people are infected with Hepatitis C worldwide including 1% of the Korean population. Once infected, most cases turn into chronic cases and may lead to liver cancer.
However it was impossible to infect Hepatitis C within a test tube cell environment until 2005 and up till then Chimpanzees were used to study the virus which proved to be a huge barrier to research.
The research team used cells infected with Hepatitis C virus and found out that the virus works by increasing the destruction of cells by the TNF-a protein responsible for the cell’s immune response.
In addition the protein structure of the virus that causes this reaction was successfully found.
Conventionally the Hepatitis C medication focused on the suppressing the growth of the virus and therefore had many side effects.
The experimental results allow new medication aimed at suppressing the actual mechanism of liver damage to be discovered.
The result was selected as the cover dissertation of the September Edition of the Hepatolog magazine.
2012.09.11 View 14433 -
 Systems biology demystifies the resistance mechanism of targeted cancer medication
Korean researchers have found the fundamental resistance mechanism of the MEK inhibitor, a recently highlighted chemotherapy method, laying the foundation for future research on overcoming cancer drug resistance and improving cancer survival rates. This research is meaningful because it was conducted through systems biology, a fusion of IT and biotechnology.
The research was conducted by Professor Gwang hyun Cho’s team from the Department of Biology at KAIST and was supported by the Ministry of Education, Science and Technology and the National Research Foundation of Korea.
The research was published as the cover paper for the June edition of the Journal of Molecular Cell Biology (Title: The cross regulation between ERK and PI3K signaling pathways determines the tumoricidal efficacy of MEK inhibitor).
Targeted anticancer medication targets certain molecules in the signaling pathway of the tumor cell and not only has fewer side effects than pre-existing anticancer medication, but also has high clinical efficacy. The technology also allows the creation of personalized medication and has been widely praised by scientists worldwide.
However, resistances to the targeted medication have often been found before or during the clinical stage, eventually causing the medications to fail to reach the drug development stage. Moreover, even if the drug is effective, the survival rate is low and the redevelopment rate is high.
An active pathway in most tumor cells is the ERK (Extracellular signal-regulated kinases) signaling pathway. This pathway is especially important in the development of skin cancer or thyroid cancer, which are developed by the mutation of the BRAF gene inside the path. In these cases, the MEK (Extracellular signal-regulated kinases) inhibitor is an effective treatment because it targets the pathway itself. However, the built-up resistance to the inhibitor commonly leads to the redevelopment of cancer.
Professor Cho’s research team used large scale computer simulations to analyze the fundamental resistance mechanism of the MEK inhibitor and used molecular cell biological experiments as well as bio-imaging* techniques to verify the results.
* Bio-imaging: Checking biological phenomena at the cellular and molecular levels using imagery
The research team used different mutational variables, which revealed that the use of the MEK inhibitor reduced the transmission of the ERK signal but led to the activation of another signaling pathway (the PI3K signaling pathway), reducing the effectiveness of the medication. Professor Cho’s team also found that this response originated from the complex interaction between the signaling matter as well as the feedback network structure, suggesting that the mix of the MEK inhibitor with other drugs could improve the effects of the targeted anticancer medication.
Professor Cho stated that this research was the first of its kind to examine the drug resistivity against the MEK inhibitor at the systematic dimension and showed how the effects of drugs on the signaling pathways of cells could be predicted using computer simulation. It also showed how basic research on signaling networks can be applied to clinical drug use, successfully suggesting a new research platform on overcoming resistance to targeting medication using its fundamental mechanism.
2012.07.06 View 13422
Systems biology demystifies the resistance mechanism of targeted cancer medication
Korean researchers have found the fundamental resistance mechanism of the MEK inhibitor, a recently highlighted chemotherapy method, laying the foundation for future research on overcoming cancer drug resistance and improving cancer survival rates. This research is meaningful because it was conducted through systems biology, a fusion of IT and biotechnology.
The research was conducted by Professor Gwang hyun Cho’s team from the Department of Biology at KAIST and was supported by the Ministry of Education, Science and Technology and the National Research Foundation of Korea.
The research was published as the cover paper for the June edition of the Journal of Molecular Cell Biology (Title: The cross regulation between ERK and PI3K signaling pathways determines the tumoricidal efficacy of MEK inhibitor).
Targeted anticancer medication targets certain molecules in the signaling pathway of the tumor cell and not only has fewer side effects than pre-existing anticancer medication, but also has high clinical efficacy. The technology also allows the creation of personalized medication and has been widely praised by scientists worldwide.
However, resistances to the targeted medication have often been found before or during the clinical stage, eventually causing the medications to fail to reach the drug development stage. Moreover, even if the drug is effective, the survival rate is low and the redevelopment rate is high.
An active pathway in most tumor cells is the ERK (Extracellular signal-regulated kinases) signaling pathway. This pathway is especially important in the development of skin cancer or thyroid cancer, which are developed by the mutation of the BRAF gene inside the path. In these cases, the MEK (Extracellular signal-regulated kinases) inhibitor is an effective treatment because it targets the pathway itself. However, the built-up resistance to the inhibitor commonly leads to the redevelopment of cancer.
Professor Cho’s research team used large scale computer simulations to analyze the fundamental resistance mechanism of the MEK inhibitor and used molecular cell biological experiments as well as bio-imaging* techniques to verify the results.
* Bio-imaging: Checking biological phenomena at the cellular and molecular levels using imagery
The research team used different mutational variables, which revealed that the use of the MEK inhibitor reduced the transmission of the ERK signal but led to the activation of another signaling pathway (the PI3K signaling pathway), reducing the effectiveness of the medication. Professor Cho’s team also found that this response originated from the complex interaction between the signaling matter as well as the feedback network structure, suggesting that the mix of the MEK inhibitor with other drugs could improve the effects of the targeted anticancer medication.
Professor Cho stated that this research was the first of its kind to examine the drug resistivity against the MEK inhibitor at the systematic dimension and showed how the effects of drugs on the signaling pathways of cells could be predicted using computer simulation. It also showed how basic research on signaling networks can be applied to clinical drug use, successfully suggesting a new research platform on overcoming resistance to targeting medication using its fundamental mechanism.
2012.07.06 View 13422 -
 The hereditary factor of autism revealed
Korean researchers have successfully investigated the causes and hereditary factors for autistic behavior and proposed a new treatment method with fewer side effects.
This research was jointly supported by the Ministry of Education, Science and Technology and the National Research Foundation as part of the Leading Researcher and Science Research Center Program
The research findings were publishing in the June edition of Nature magazine and will also be introduced in the July edition of Nature Reviews Drug Discovery, under the title ‘Autistic-like social behavior in Shank2-mutant mice improved by restoring NMDA receptor function’.
The research team found that lack of Shank2 genes in mice, which are responsible for the production of synapse proteins, caused autistic-like behavior. The results strongly suggested that the Shank2 gene was linked to autistic behavior and that Shank2 deficiency induced autistic behaviors.
Autism is a neural development disorder characterized by impaired social interaction, repetitive behavior, mental retardation, anxiety and hyperactivity. Around 100 million people worldwide display symptoms of autistic behavior. Recent studies conducted by the University of Washington revealed that 1 out of 3 young adults who display autistic behavior do not fit into the workplace or get accepted to college, a much higher rate than any other disorder. However, an effective cure has not yet been developed and current treatments are limited to reducing repetitive behavior.
The research team confirmed autistic-like social behavior in mice without the Shank2 genes and that the mice had decreased levels of neurotransmission in the NMDA receptor. The mice also showed damaged synaptic plasticity* in the hippocampus**.
* Plasticity: ability of the connectionbetween two neurons to change in strength in response to transmission of information
**Hippocampus: part of the brain responsible for short-term and long-term memory as well as spatial navigation.
The research team also found out that, to restore the function of the NMDA receptor, the passive stimulation of certain receptors, such as the mGLuR5, yielded better treatment results than the direct stimulation of the NMDA. This greatly reduces the side effects associated with the direct stimulation of receptors, resulting in a more effective treatment method.
This research successfully investigated the function of the Shank2 gene in the nerve tissue and showed how the reduced function of the NMDA receptor, due to the lack of the gene, resulted in autistic behavior. It also provided new possibilities for the treatment of autistic behavior and impaired social interaction
2012.06.24 View 14280
The hereditary factor of autism revealed
Korean researchers have successfully investigated the causes and hereditary factors for autistic behavior and proposed a new treatment method with fewer side effects.
This research was jointly supported by the Ministry of Education, Science and Technology and the National Research Foundation as part of the Leading Researcher and Science Research Center Program
The research findings were publishing in the June edition of Nature magazine and will also be introduced in the July edition of Nature Reviews Drug Discovery, under the title ‘Autistic-like social behavior in Shank2-mutant mice improved by restoring NMDA receptor function’.
The research team found that lack of Shank2 genes in mice, which are responsible for the production of synapse proteins, caused autistic-like behavior. The results strongly suggested that the Shank2 gene was linked to autistic behavior and that Shank2 deficiency induced autistic behaviors.
Autism is a neural development disorder characterized by impaired social interaction, repetitive behavior, mental retardation, anxiety and hyperactivity. Around 100 million people worldwide display symptoms of autistic behavior. Recent studies conducted by the University of Washington revealed that 1 out of 3 young adults who display autistic behavior do not fit into the workplace or get accepted to college, a much higher rate than any other disorder. However, an effective cure has not yet been developed and current treatments are limited to reducing repetitive behavior.
The research team confirmed autistic-like social behavior in mice without the Shank2 genes and that the mice had decreased levels of neurotransmission in the NMDA receptor. The mice also showed damaged synaptic plasticity* in the hippocampus**.
* Plasticity: ability of the connectionbetween two neurons to change in strength in response to transmission of information
**Hippocampus: part of the brain responsible for short-term and long-term memory as well as spatial navigation.
The research team also found out that, to restore the function of the NMDA receptor, the passive stimulation of certain receptors, such as the mGLuR5, yielded better treatment results than the direct stimulation of the NMDA. This greatly reduces the side effects associated with the direct stimulation of receptors, resulting in a more effective treatment method.
This research successfully investigated the function of the Shank2 gene in the nerve tissue and showed how the reduced function of the NMDA receptor, due to the lack of the gene, resulted in autistic behavior. It also provided new possibilities for the treatment of autistic behavior and impaired social interaction
2012.06.24 View 14280 -
 Successful Development of Excavation System of Biomarkers containing Protein Decomposition Control Enzyme Information
A Korean team of researchers successfully developed a biomarker excavation system named E3Net that excavates biomarkers containing information of the enzymes that control the decomposition of proteins. The development of the system paved the possibility of development of new high quality biomarkers.
*Biomarker: Molecular information of unique patterns derived from genes and proteins that allow the monitoring of physical changes from genetic or environmental causes.
Professor Lee Kwan Soo’s team (Department of Biological Sciences) composed of Doctorate candidate Han Young Woong, Lee Ho Dong Ph.D. and Professor Park Jong Chul published a dissertation in the April edition of Molecular and Cellular Proteomics. (Dissertation Title: A system for exploring E3-mediated regulatory networks of cellular functions).
Professor Lee’s team compiled all available information of the enzyme that controls protein decomposition (E3 enzyme) and successfully compiled the inter-substrate network by extracting information from 20,000 biology related data base dissertations. The result was the development of the E3Net system that analyzes the related cell function and disease.
Cells have a system that produces, destroys, and recycles proteins in response to the ever changing environmental conditions. Error in these processes leads to disease.
Therefore finding the relationship between E3 enzymes that control the decomposition of proteins and the substrates will allow disease curing and prevention to become much easier. E3 enzyme is responsible for 80% of the protein decomposition and is therefore predicted to be related to various diseases.
However the information on E3 enzyme and inter-substrate behavior are spread out among numerous dissertations and data bases which prevented methodological analysis of the role of the related cells and characteristics of the disease itself.
Professor Lee’s team was successful in creating the E3Net that compiled 2,201 pieces of E3 substrate information, 4,896 pieces of substrate information, and 1,671 pieces of inter-substrate relationship information. This compilation allows for the systematic analysis of cells and diseases.
The newly created network is 10 times larger than the existing network and is the first case where it is possible to accurately find the cell function and related diseases.
It is anticipated that the use of the E3Net will allow the excavation of new biomarkers for the development of personalized drug systems.
The research team applied the E3Net to find tens of new candidate biomarkers related to the major modern diseases like diabetes and cancer.
2012.05.30 View 14645
Successful Development of Excavation System of Biomarkers containing Protein Decomposition Control Enzyme Information
A Korean team of researchers successfully developed a biomarker excavation system named E3Net that excavates biomarkers containing information of the enzymes that control the decomposition of proteins. The development of the system paved the possibility of development of new high quality biomarkers.
*Biomarker: Molecular information of unique patterns derived from genes and proteins that allow the monitoring of physical changes from genetic or environmental causes.
Professor Lee Kwan Soo’s team (Department of Biological Sciences) composed of Doctorate candidate Han Young Woong, Lee Ho Dong Ph.D. and Professor Park Jong Chul published a dissertation in the April edition of Molecular and Cellular Proteomics. (Dissertation Title: A system for exploring E3-mediated regulatory networks of cellular functions).
Professor Lee’s team compiled all available information of the enzyme that controls protein decomposition (E3 enzyme) and successfully compiled the inter-substrate network by extracting information from 20,000 biology related data base dissertations. The result was the development of the E3Net system that analyzes the related cell function and disease.
Cells have a system that produces, destroys, and recycles proteins in response to the ever changing environmental conditions. Error in these processes leads to disease.
Therefore finding the relationship between E3 enzymes that control the decomposition of proteins and the substrates will allow disease curing and prevention to become much easier. E3 enzyme is responsible for 80% of the protein decomposition and is therefore predicted to be related to various diseases.
However the information on E3 enzyme and inter-substrate behavior are spread out among numerous dissertations and data bases which prevented methodological analysis of the role of the related cells and characteristics of the disease itself.
Professor Lee’s team was successful in creating the E3Net that compiled 2,201 pieces of E3 substrate information, 4,896 pieces of substrate information, and 1,671 pieces of inter-substrate relationship information. This compilation allows for the systematic analysis of cells and diseases.
The newly created network is 10 times larger than the existing network and is the first case where it is possible to accurately find the cell function and related diseases.
It is anticipated that the use of the E3Net will allow the excavation of new biomarkers for the development of personalized drug systems.
The research team applied the E3Net to find tens of new candidate biomarkers related to the major modern diseases like diabetes and cancer.
2012.05.30 View 14645 -
 The output of terahertz waves enhanced by KAIST team
KAIST researchers have greatly improved the output of terahertz waves, the blue ocean of the optics world. This technology is expected to be applied to portable X-ray cameras, small bio-diagnostic systems, and in many other devices.
Professor Ki-Hun Jeong"s research team from the Department of Bio and Brain Engineering used optical nano-antenna technology to increase the output of terahertz waves by three times.
Terahertz waves are electromagnetic waves with frequencies between 100GHz to 30THz. They are produced when a femtosecond (10^-15 s) pulse laser is shone on a semiconductor substrate with photoconduction antennas, causing a photocurrent pulse of one picosecond (10^-12 s). Their long wavelengths, in comparison to visible light and infrared rays, give terahertz waves a high penetration power with less energy than X-rays, making them less harmful to humans.
These qualities allow us to see through objects, just as X-rays do, but because terahertz waves absorb certain frequencies, we can detect hidden explosives or drugs, which was not possible with X-rays. We can even identify fake drugs. Furthermore, using the spectral information, we can analyze a material"s innate qualities without chemical processing, making it possible to identify skin diseases without harming the body. However, the output was not sufficient to be used in biosensors and other applications.
Prof. Jeong"s team added optical nano-antennas, made from gold nano-rods, in between the photoconduction antennas and optimized the structure. This resulted in nanoplasmonic resonance in the photoconduction substrate, increasing the degree of integration of the photocurrent pulse and resulting in a three times larger output.
Hence, it is not only possible to see through objects more clearly, but it is also possible to analyze components without a biopsy.
Professor Jeong explained, "This technology, coupled with the miniaturization of terahertz devices, can be applied to endoscopes to detect early epithelial cancer" and that he will focus on creating and commercializing these biosensor systems.
This research was published in the March issue of the international nanotechnology journal ACS Nano and was funded by the Korea Evaluation Institute of Industrial Technology and the National Research Foundation of Korea.
Figure: Mimetic diagram of a THz generator with nano-antennas
2012.04.29 View 14327
The output of terahertz waves enhanced by KAIST team
KAIST researchers have greatly improved the output of terahertz waves, the blue ocean of the optics world. This technology is expected to be applied to portable X-ray cameras, small bio-diagnostic systems, and in many other devices.
Professor Ki-Hun Jeong"s research team from the Department of Bio and Brain Engineering used optical nano-antenna technology to increase the output of terahertz waves by three times.
Terahertz waves are electromagnetic waves with frequencies between 100GHz to 30THz. They are produced when a femtosecond (10^-15 s) pulse laser is shone on a semiconductor substrate with photoconduction antennas, causing a photocurrent pulse of one picosecond (10^-12 s). Their long wavelengths, in comparison to visible light and infrared rays, give terahertz waves a high penetration power with less energy than X-rays, making them less harmful to humans.
These qualities allow us to see through objects, just as X-rays do, but because terahertz waves absorb certain frequencies, we can detect hidden explosives or drugs, which was not possible with X-rays. We can even identify fake drugs. Furthermore, using the spectral information, we can analyze a material"s innate qualities without chemical processing, making it possible to identify skin diseases without harming the body. However, the output was not sufficient to be used in biosensors and other applications.
Prof. Jeong"s team added optical nano-antennas, made from gold nano-rods, in between the photoconduction antennas and optimized the structure. This resulted in nanoplasmonic resonance in the photoconduction substrate, increasing the degree of integration of the photocurrent pulse and resulting in a three times larger output.
Hence, it is not only possible to see through objects more clearly, but it is also possible to analyze components without a biopsy.
Professor Jeong explained, "This technology, coupled with the miniaturization of terahertz devices, can be applied to endoscopes to detect early epithelial cancer" and that he will focus on creating and commercializing these biosensor systems.
This research was published in the March issue of the international nanotechnology journal ACS Nano and was funded by the Korea Evaluation Institute of Industrial Technology and the National Research Foundation of Korea.
Figure: Mimetic diagram of a THz generator with nano-antennas
2012.04.29 View 14327 -
 Creation of Synthetic Antibodies: Professor Hak Seong Kim
Synthetics antibodies which can replace antibodies from humans used as ingredients of medicines have been developed. It can increase the costs to 1/100 of the current costs and is much easier to develop. It is expected that the development period will be shortened from 10 years to 5.
Prof. Hak Seong Kim from the Biology department of KAIST conducted a joint research with Prof. Dong Seob Kim to reconstruct proteins and has succeeded.
The synthetic antibody displays much strength in terms of its productivity, structural formation, and bonding capability, and is thus regarded as an ideal protein. It can replace the antigens that are currently in use. It is expected that Korea will therefore be able to lead the world market for protein medicines which is a 192trillion won industry.
The original antibody has been used for not only treating diseases, but also for various other applications in the fields of medical sciences and biology. However, it is produced through a very complex process involving the incubation of animal cells, and is therefore very expensive. Also, most antibodies are already patented by more developed countries, so a high royalty fee must be paid.
Because of this, many countries including Korea has been concentrating on developing biosimilars copying the antibody medicines for which the patents have already expired. This causes Korea to be behind in the development of antibody protein pharmaceuticals.
Prof. Kim’s research team has focused on the face that the protein existing in some eels are not antibodies but functions as one, and has been successful in developing a synthetic antibody.
The synthetic antibody can be mass produced from the colon bacillus, which allows it to be produced at 1/100 the original cost. It is in a module structure which allows the structuring of the antibody into the desired structure, enabling it to be developed into a protein-based medicine within 5 years.
Together with this, the coherence with the important antigens can be easily controlled, thus allowing for highly effective treatments, less side-effects, high security regarding heat and pH, and the immunogen levels being negligeable. This suggests a very high rate of the antibody being converted into a protein based medication.
The synthetic antibody technology has been tested as a sample for the cure for lung diseases and rheumatism and has been proven to be appropriate. Animal testing will be conducted soon.
Prof Kim said “The original antibodies had a small area allowing the bonding with antibodies, creating barriers for raising bonding strength and structuring. The newly created antibody carries only the strengths and will become a new protein based medicine purely created by Korean technology to replace the antibodies currently used in medications.”
Furthermore, he added that, “The synthesized antibody structuring and designing technology will be widely used in the areas of detecting, diagnosing, and analyzing diseases.”
At the same time, this research result has been published in the Feb 10th issue of the PNAS, and has been supported by the future promising pioneer business program held by the Ministry of Education and Technology.
2012.04.04 View 12592
Creation of Synthetic Antibodies: Professor Hak Seong Kim
Synthetics antibodies which can replace antibodies from humans used as ingredients of medicines have been developed. It can increase the costs to 1/100 of the current costs and is much easier to develop. It is expected that the development period will be shortened from 10 years to 5.
Prof. Hak Seong Kim from the Biology department of KAIST conducted a joint research with Prof. Dong Seob Kim to reconstruct proteins and has succeeded.
The synthetic antibody displays much strength in terms of its productivity, structural formation, and bonding capability, and is thus regarded as an ideal protein. It can replace the antigens that are currently in use. It is expected that Korea will therefore be able to lead the world market for protein medicines which is a 192trillion won industry.
The original antibody has been used for not only treating diseases, but also for various other applications in the fields of medical sciences and biology. However, it is produced through a very complex process involving the incubation of animal cells, and is therefore very expensive. Also, most antibodies are already patented by more developed countries, so a high royalty fee must be paid.
Because of this, many countries including Korea has been concentrating on developing biosimilars copying the antibody medicines for which the patents have already expired. This causes Korea to be behind in the development of antibody protein pharmaceuticals.
Prof. Kim’s research team has focused on the face that the protein existing in some eels are not antibodies but functions as one, and has been successful in developing a synthetic antibody.
The synthetic antibody can be mass produced from the colon bacillus, which allows it to be produced at 1/100 the original cost. It is in a module structure which allows the structuring of the antibody into the desired structure, enabling it to be developed into a protein-based medicine within 5 years.
Together with this, the coherence with the important antigens can be easily controlled, thus allowing for highly effective treatments, less side-effects, high security regarding heat and pH, and the immunogen levels being negligeable. This suggests a very high rate of the antibody being converted into a protein based medication.
The synthetic antibody technology has been tested as a sample for the cure for lung diseases and rheumatism and has been proven to be appropriate. Animal testing will be conducted soon.
Prof Kim said “The original antibodies had a small area allowing the bonding with antibodies, creating barriers for raising bonding strength and structuring. The newly created antibody carries only the strengths and will become a new protein based medicine purely created by Korean technology to replace the antibodies currently used in medications.”
Furthermore, he added that, “The synthesized antibody structuring and designing technology will be widely used in the areas of detecting, diagnosing, and analyzing diseases.”
At the same time, this research result has been published in the Feb 10th issue of the PNAS, and has been supported by the future promising pioneer business program held by the Ministry of Education and Technology.
2012.04.04 View 12592 -
 Seeing Inside Cells with Fiber Optics
Professor Jiho Park’s research team was successful in receiving minute optical signals from inside the cell using optical nano fibers.
Through the invention of this technology, we can now look inside cells in high resolution without the use of equipment such as endoscopes that damage cells. We will be able to study the biological phenomena within cells, and thus cure diseases more effectively.
Recently, ultra high resolution microscopes have been used to analyze incubated cells. However, because of the need for a very complex and large system, it had been impossible to monitor cells in the less transparent areas of the body in real time.
The research team created the wire with a semiconductor created with tin oxides to be only 100 nanometers in diameter (1nanometer= 1/1billion meters).
The nanowire is connected to the end of the optical fiber, and the light that comes through the optical fiber is transmitted to particular spots in the cell, and the optical signals from the cell are retrieved back from the cell as well
Together with this, based on the fact that nanowires do not damage cells, the research team covered the end of the wire with a photo reactive material and entered this into the cell. They were able to check that the material reacted to light and entered the cell when they transmitted light
Accordingly, this showed the possibilities of the use of this technology as a method of treatment to effectively transfer the medication into the cells.
Prof. Jiho Park stated that “in this research, we only used cells incubated outside the human body, but soon we will use this technology to stimulate and control cells within the body in a minute scale” as well as that “soon, we will be able to study the biological phenomena inside a cell to study diseases and apply this to cure them more effectively”.
This research result has been published in the online publication of ‘Nature Nanotechnology’ on December 18.
This study was done through the cooperation of various schools. Besides Prof. Jiho Park, Prof. Seungman Yang from the Biochemistry department, and Doctor Chuljoon Huh from KAIST, Prof. Yeonho Choi from Biomedical Science department of Korea University, Professor Peidon Yang and Doctor Ruoxue Yan from UC Berkeley’s chemistry department, and Luke Lee from UC Berkeley’s bioengineering department participated in the project.
2012.01.31 View 10876
Seeing Inside Cells with Fiber Optics
Professor Jiho Park’s research team was successful in receiving minute optical signals from inside the cell using optical nano fibers.
Through the invention of this technology, we can now look inside cells in high resolution without the use of equipment such as endoscopes that damage cells. We will be able to study the biological phenomena within cells, and thus cure diseases more effectively.
Recently, ultra high resolution microscopes have been used to analyze incubated cells. However, because of the need for a very complex and large system, it had been impossible to monitor cells in the less transparent areas of the body in real time.
The research team created the wire with a semiconductor created with tin oxides to be only 100 nanometers in diameter (1nanometer= 1/1billion meters).
The nanowire is connected to the end of the optical fiber, and the light that comes through the optical fiber is transmitted to particular spots in the cell, and the optical signals from the cell are retrieved back from the cell as well
Together with this, based on the fact that nanowires do not damage cells, the research team covered the end of the wire with a photo reactive material and entered this into the cell. They were able to check that the material reacted to light and entered the cell when they transmitted light
Accordingly, this showed the possibilities of the use of this technology as a method of treatment to effectively transfer the medication into the cells.
Prof. Jiho Park stated that “in this research, we only used cells incubated outside the human body, but soon we will use this technology to stimulate and control cells within the body in a minute scale” as well as that “soon, we will be able to study the biological phenomena inside a cell to study diseases and apply this to cure them more effectively”.
This research result has been published in the online publication of ‘Nature Nanotechnology’ on December 18.
This study was done through the cooperation of various schools. Besides Prof. Jiho Park, Prof. Seungman Yang from the Biochemistry department, and Doctor Chuljoon Huh from KAIST, Prof. Yeonho Choi from Biomedical Science department of Korea University, Professor Peidon Yang and Doctor Ruoxue Yan from UC Berkeley’s chemistry department, and Luke Lee from UC Berkeley’s bioengineering department participated in the project.
2012.01.31 View 10876