National+Research+Foundation
-
 Play Games With No Latency
One of the most challenging issues for game players looks to be resolved soon with the introduction of a zero-latency gaming environment. A KAIST team developed a technology that helps game players maintain zero-latency performance. The new technology transforms the shapes of game design according to the amount of latency.
Latency in human-computer interactions is often caused by various factors related to the environment and performance of the devices, networks, and data processing. The term ‘lag’ is used to refer to any latency during gaming which impacts the user’s performance.
Professor Byungjoo Lee at the Graduate School of Culture Technology in collaboration with Aalto University in Finland presented a mathematical model for predicting players' behavior by understanding the effects of latency on players. This cognitive model is capable of predicting the success rate of a user when there is latency in a 'moving target selection' task which requires button input in a time constrained situation.
The model predicts the players’ task success rate when latency is added to the gaming environment. Using these predicted success rates, the design elements of the game are geometrically modified to help players maintain similar success rates as they would achieve in a zero-latency environment. In fact, this research succeeded in modifying the pillar heights of the Flappy Bird game, allowing the players to maintain their gaming performance regardless of the added latency.
Professor Lee said, "This technique is unique in the sense that it does not interfere with a player's gaming flow, unlike traditional methods which manipulate the game clock by the amount of latency. This study can be extended to various games such as reducing the size of obstacles in the latent computing environment.”
This research, in collaboration with Dr. Sunjun Kim from Aalto University and led by PhD candidate Injung Lee, was presented during the 2019 CHI Conference on Human Factors in Computing Systems last month in Glasgow in the UK.
This research was supported by the National Research Foundation of Korea (NRF) (2017R1C1B2002101, 2018R1A5A7025409), and the Aalto University Seed Funding Granted to the GamerLab respectively.
Figure 1. Overview of Geometric Compensation
Publication:
Injung Lee, Sunjun Kim, and Byungjoo Lee. 2019. Geometrically Compensating Effect of End-to-End Latency in Moving-Target Selection Games. In Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems (CHI’19) . ACM, New York, NY, USA, Article 560, 12 pages. https://doi.org/10.1145/3290605.3300790
Video Material:
https://youtu.be/TTi7dipAKJs
Profile: Prof. Byungjoo Lee, MD, PhD
byungjoo.lee@kaist.ac.kr
http://kiml.org/
Assistant Professor
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Injung Lee, PhD Candidate
edndn@kaist.ac.kr
PhD Candidate
Interactive Media Lab
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon 34141, Korea
Profile: Postdoc. Sunjun Kim, MD, PhD
kuaa.net@gmail.com
Postdoctoral Researcher
User Interfaces Group
Aalto University
https://www.aalto.fi Espoo 02150, Finland
(END)
2019.06.11 View 48252
Play Games With No Latency
One of the most challenging issues for game players looks to be resolved soon with the introduction of a zero-latency gaming environment. A KAIST team developed a technology that helps game players maintain zero-latency performance. The new technology transforms the shapes of game design according to the amount of latency.
Latency in human-computer interactions is often caused by various factors related to the environment and performance of the devices, networks, and data processing. The term ‘lag’ is used to refer to any latency during gaming which impacts the user’s performance.
Professor Byungjoo Lee at the Graduate School of Culture Technology in collaboration with Aalto University in Finland presented a mathematical model for predicting players' behavior by understanding the effects of latency on players. This cognitive model is capable of predicting the success rate of a user when there is latency in a 'moving target selection' task which requires button input in a time constrained situation.
The model predicts the players’ task success rate when latency is added to the gaming environment. Using these predicted success rates, the design elements of the game are geometrically modified to help players maintain similar success rates as they would achieve in a zero-latency environment. In fact, this research succeeded in modifying the pillar heights of the Flappy Bird game, allowing the players to maintain their gaming performance regardless of the added latency.
Professor Lee said, "This technique is unique in the sense that it does not interfere with a player's gaming flow, unlike traditional methods which manipulate the game clock by the amount of latency. This study can be extended to various games such as reducing the size of obstacles in the latent computing environment.”
This research, in collaboration with Dr. Sunjun Kim from Aalto University and led by PhD candidate Injung Lee, was presented during the 2019 CHI Conference on Human Factors in Computing Systems last month in Glasgow in the UK.
This research was supported by the National Research Foundation of Korea (NRF) (2017R1C1B2002101, 2018R1A5A7025409), and the Aalto University Seed Funding Granted to the GamerLab respectively.
Figure 1. Overview of Geometric Compensation
Publication:
Injung Lee, Sunjun Kim, and Byungjoo Lee. 2019. Geometrically Compensating Effect of End-to-End Latency in Moving-Target Selection Games. In Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems (CHI’19) . ACM, New York, NY, USA, Article 560, 12 pages. https://doi.org/10.1145/3290605.3300790
Video Material:
https://youtu.be/TTi7dipAKJs
Profile: Prof. Byungjoo Lee, MD, PhD
byungjoo.lee@kaist.ac.kr
http://kiml.org/
Assistant Professor
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Injung Lee, PhD Candidate
edndn@kaist.ac.kr
PhD Candidate
Interactive Media Lab
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon 34141, Korea
Profile: Postdoc. Sunjun Kim, MD, PhD
kuaa.net@gmail.com
Postdoctoral Researcher
User Interfaces Group
Aalto University
https://www.aalto.fi Espoo 02150, Finland
(END)
2019.06.11 View 48252 -
 Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke
(Professor Young Seok Ju at the Graduate School of Medical Science and Engineering)
Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancers develop.
Researchers at KAIST, Seoul National University and their collaborators confirmed that gene fusions in non-smokers mostly occur early on, sometimes as early as childhood or adolescence, and on average about three decades before cancer is diagnosed. The study showed that these mutant lung cells, harboring oncogenic seeds, remain dormant for several decades until a number of further mutations accumulate sufficiently for progression into cancer. This is the first study to reveal the landscape of genome structural variations in lung adenocarcinoma.
Lung cancer is the leading cause of cancer-related deaths worldwide, and lung adenocarcinoma is its most common type. Most lung adenocarcinomas are associated with chronic smoking, but about a fourth develop in non-smokers. Precisely what happens in non-smokers for this cancer to develop is not clearly understood.
Researchers analyzed the genomes of 138 lung adenocarcinoma patients, including smokers and non-smokers, with whole-genome sequencing technologies. They explored DNA damage that induced neoplastic transformation.
Lung adenocarcinomas that originated from chronic smoking, referred to as signature 4-high (S4-high) cancers in the study, showed several distinguishing features compared to smoking-unrelated cancers (S4-low).
People in the S4-high group were largely older, men and had more frequent mutations in a cancer-related gene called KRAS. Cancer genomes in the S4-high group were hypermutated with simple mutational classes, such as the substitution, insertion, or deletion of a single base, the building block of DNA.
But the story was very different in the S4-low group. Generally, mutational profiles in this group were much more silent than the S4-high group. However, all cancer-related gene fusions, which are abnormally activated from the merging of two originally separate genes, were exclusively observed in the S4-low group.
The patterns of genomic structural changes underlying gene fusions suggest that about three in four cases of gene fusions emerged from a single cellular crisis causing massive genomic fragmentation and subsequent imprecise repair in normal lung epithelium.
Most strikingly, these major genomic rearrangements, which led to the development of lung adenocarcinoma, are very likely to be acquired decades before cancer diagnosis. The researchers used genomic archaeology techniques to trace the timing of when the catastrophes took place.
Researchers started this study seven years ago when they discovered the expression of the KIF5B-RET gene fusion in lung adenocarcinoma for the first time. Professor Young-Seok Ju, co-lead author from the Graduate School of Medical Science and Engineering at KAIST says, “It is remarkable that oncogenesis can begin by a massive shattering of chromosomes early in life. Our study immediately raises a new question: What induces the mutational catastrophe in our normal lung epithelium.”
Professor Young Tae Kim, co-lead author from Seoul National University says, “We hope this work will help us get one step closer to precision medicine for lung cancer patients.”
The research team plans to further focus on the molecular mechanisms that stimulate complex rearrangements in the body, through screening the genomic structures of fusion genes in other cancer types.
This study was supported by the National Research Foundation of Korea (NRF), Korea Health Industry Development Institute (KHIDI), Suh Kyungbae Foundation, the College of Medicine Research Foundations at Seoul National University and others.
Figure.
(Smoking-unrelated oncogenesis of lung cancers by gene fusions)
Publication.
Jake June-Koo Lee, Seongyeol Park et al., Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma
Cell 177, June 13 2019, online publication ahead of print at May 30, 2019
https://doi.org/10.1016/j.cell.2019.05.013
Profile: Prof Young Seok Ju, MD, PhD
ysju@kaist.ac.kr
http://julab.kaist.ac.kr
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon 34141, Korea
Profile: Prof Young Tae Kim, MD, PhD
ytkim@snu.ac.kr
Professor
Seoul National University Cancer Research Institute
Department of Thoracic and Cardiovascular Surgery
Seoul National University Hospital Seoul 03080, Korea
2019.05.31 View 58361
Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke
(Professor Young Seok Ju at the Graduate School of Medical Science and Engineering)
Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancers develop.
Researchers at KAIST, Seoul National University and their collaborators confirmed that gene fusions in non-smokers mostly occur early on, sometimes as early as childhood or adolescence, and on average about three decades before cancer is diagnosed. The study showed that these mutant lung cells, harboring oncogenic seeds, remain dormant for several decades until a number of further mutations accumulate sufficiently for progression into cancer. This is the first study to reveal the landscape of genome structural variations in lung adenocarcinoma.
Lung cancer is the leading cause of cancer-related deaths worldwide, and lung adenocarcinoma is its most common type. Most lung adenocarcinomas are associated with chronic smoking, but about a fourth develop in non-smokers. Precisely what happens in non-smokers for this cancer to develop is not clearly understood.
Researchers analyzed the genomes of 138 lung adenocarcinoma patients, including smokers and non-smokers, with whole-genome sequencing technologies. They explored DNA damage that induced neoplastic transformation.
Lung adenocarcinomas that originated from chronic smoking, referred to as signature 4-high (S4-high) cancers in the study, showed several distinguishing features compared to smoking-unrelated cancers (S4-low).
People in the S4-high group were largely older, men and had more frequent mutations in a cancer-related gene called KRAS. Cancer genomes in the S4-high group were hypermutated with simple mutational classes, such as the substitution, insertion, or deletion of a single base, the building block of DNA.
But the story was very different in the S4-low group. Generally, mutational profiles in this group were much more silent than the S4-high group. However, all cancer-related gene fusions, which are abnormally activated from the merging of two originally separate genes, were exclusively observed in the S4-low group.
The patterns of genomic structural changes underlying gene fusions suggest that about three in four cases of gene fusions emerged from a single cellular crisis causing massive genomic fragmentation and subsequent imprecise repair in normal lung epithelium.
Most strikingly, these major genomic rearrangements, which led to the development of lung adenocarcinoma, are very likely to be acquired decades before cancer diagnosis. The researchers used genomic archaeology techniques to trace the timing of when the catastrophes took place.
Researchers started this study seven years ago when they discovered the expression of the KIF5B-RET gene fusion in lung adenocarcinoma for the first time. Professor Young-Seok Ju, co-lead author from the Graduate School of Medical Science and Engineering at KAIST says, “It is remarkable that oncogenesis can begin by a massive shattering of chromosomes early in life. Our study immediately raises a new question: What induces the mutational catastrophe in our normal lung epithelium.”
Professor Young Tae Kim, co-lead author from Seoul National University says, “We hope this work will help us get one step closer to precision medicine for lung cancer patients.”
The research team plans to further focus on the molecular mechanisms that stimulate complex rearrangements in the body, through screening the genomic structures of fusion genes in other cancer types.
This study was supported by the National Research Foundation of Korea (NRF), Korea Health Industry Development Institute (KHIDI), Suh Kyungbae Foundation, the College of Medicine Research Foundations at Seoul National University and others.
Figure.
(Smoking-unrelated oncogenesis of lung cancers by gene fusions)
Publication.
Jake June-Koo Lee, Seongyeol Park et al., Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma
Cell 177, June 13 2019, online publication ahead of print at May 30, 2019
https://doi.org/10.1016/j.cell.2019.05.013
Profile: Prof Young Seok Ju, MD, PhD
ysju@kaist.ac.kr
http://julab.kaist.ac.kr
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon 34141, Korea
Profile: Prof Young Tae Kim, MD, PhD
ytkim@snu.ac.kr
Professor
Seoul National University Cancer Research Institute
Department of Thoracic and Cardiovascular Surgery
Seoul National University Hospital Seoul 03080, Korea
2019.05.31 View 58361 -
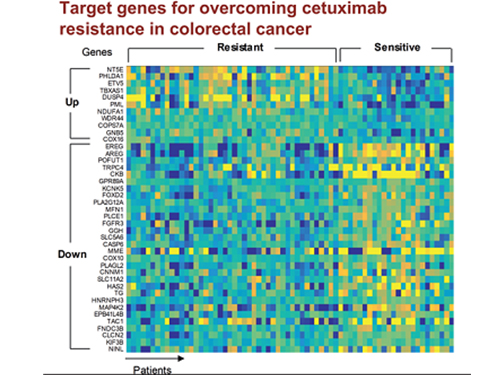 5 Biomarkers for Overcoming Colorectal Cancer Drug Resistance Identified
< Professor Kwang-Hyun Cho's Team >
KAIST researchers have identified five biomarkers that will help them address resistance to cancer-targeting therapeutics. This new treatment strategy will bring us one step closer to precision medicine for patients who showed resistance.
Colorectal cancer is one of the most common types of cancer worldwide. The number of patients has surpassed 1 million, and its five-year survival rate significantly drops to about 20 percent when metastasized. In Korea, the surge of colorectal cancer has been the highest in the last 10 years due to increasing Westernized dietary patterns and obesity. It is expected that the number and mortality rates of colorectal cancer patients will increase sharply as the nation is rapidly facing an increase in its aging population.
Recently, anticancer agents targeting only specific molecules of colon cancer cells have been developed. Unlike conventional anticancer medications, these selectively treat only specific target factors, so they can significantly reduce some of the side-effects of anticancer therapy while enhancing drug efficacy.
Cetuximab is the most well-known FDA approved anticancer medication. It is a biomarker that predicts drug reactivity and utilizes the presence of the ‘KRAS’ gene mutation. Cetuximab is prescribed to patients who don’t carry the KRAS gene mutation.
However, even in patients without the KRAS gene mutation, the response rate of Cetuximab is only about fifty percent, and there is also resistance to drugs after targeted chemotherapy. Compared with conventional chemotherapy alone, the life expectancy only lasts five months on average.
In research featured in the FEBS Journal as the cover paper for the April 7 edition, the KAIST research team led by Professor Kwang-Hyun Cho at the Department of Bio and Brain Engineering presented five additional biomarkers that could increase Cetuximab responsiveness using systems biology approach that combines genomic data analysis, mathematical modeling, and cell experiments. The experimental inhibition of newly discovered biomarkers DUSP4, ETV5, GNB5, NT5E, and PHLDA1 in colorectal cancer cells has been shown to overcome Cetuximab resistance in KRAS-normal genes. The research team confirmed that when suppressing GNB5, one of the new biomarkers, it was shown to overcome resistance to Cetuximab regardless of having a mutation in the KRAS gene.
Professor Cho said, “There has not been an example of colorectal cancer treatment involving regulation of the GNB5 gene.” He continued, “Identifying the principle of drug resistance in cancer cells through systems biology and discovering new biomarkers that could be a new molecular target to overcome drug resistance suggest real potential to actualize precision medicine.”
This study was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT (2017R1A2A1A17069642 and 2015M3A9A7067220).
Image 1. The cover of FEBS Journal for April 2019
2019.05.27 View 60914
5 Biomarkers for Overcoming Colorectal Cancer Drug Resistance Identified
< Professor Kwang-Hyun Cho's Team >
KAIST researchers have identified five biomarkers that will help them address resistance to cancer-targeting therapeutics. This new treatment strategy will bring us one step closer to precision medicine for patients who showed resistance.
Colorectal cancer is one of the most common types of cancer worldwide. The number of patients has surpassed 1 million, and its five-year survival rate significantly drops to about 20 percent when metastasized. In Korea, the surge of colorectal cancer has been the highest in the last 10 years due to increasing Westernized dietary patterns and obesity. It is expected that the number and mortality rates of colorectal cancer patients will increase sharply as the nation is rapidly facing an increase in its aging population.
Recently, anticancer agents targeting only specific molecules of colon cancer cells have been developed. Unlike conventional anticancer medications, these selectively treat only specific target factors, so they can significantly reduce some of the side-effects of anticancer therapy while enhancing drug efficacy.
Cetuximab is the most well-known FDA approved anticancer medication. It is a biomarker that predicts drug reactivity and utilizes the presence of the ‘KRAS’ gene mutation. Cetuximab is prescribed to patients who don’t carry the KRAS gene mutation.
However, even in patients without the KRAS gene mutation, the response rate of Cetuximab is only about fifty percent, and there is also resistance to drugs after targeted chemotherapy. Compared with conventional chemotherapy alone, the life expectancy only lasts five months on average.
In research featured in the FEBS Journal as the cover paper for the April 7 edition, the KAIST research team led by Professor Kwang-Hyun Cho at the Department of Bio and Brain Engineering presented five additional biomarkers that could increase Cetuximab responsiveness using systems biology approach that combines genomic data analysis, mathematical modeling, and cell experiments. The experimental inhibition of newly discovered biomarkers DUSP4, ETV5, GNB5, NT5E, and PHLDA1 in colorectal cancer cells has been shown to overcome Cetuximab resistance in KRAS-normal genes. The research team confirmed that when suppressing GNB5, one of the new biomarkers, it was shown to overcome resistance to Cetuximab regardless of having a mutation in the KRAS gene.
Professor Cho said, “There has not been an example of colorectal cancer treatment involving regulation of the GNB5 gene.” He continued, “Identifying the principle of drug resistance in cancer cells through systems biology and discovering new biomarkers that could be a new molecular target to overcome drug resistance suggest real potential to actualize precision medicine.”
This study was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT (2017R1A2A1A17069642 and 2015M3A9A7067220).
Image 1. The cover of FEBS Journal for April 2019
2019.05.27 View 60914 -
 Scientist of October, Professor Haeshin Lee
(Professor Haeshin Lee from the Department of Chemistry)
Professor Haeshin Lee from the Department of Chemistry received the ‘Science and Technology Award of October’ from the Ministry of Science and ICT and the National Research Foundation of Korea for his contribution to developing an antibleeding injection needle. This novel outcome will fundamentally prevent the problem of secondary infections of AIDS, Ebola and Hepatitis viruses transmitting from patients to medical teams.
This needle’s surface is coated with hemostatic materials. Its concept is simple and the key to this technology is to make materials that are firmly coated on the needle so that they can endure frictional force when being injected into skin and blood vessels. Moreover, the materials should be adhesive to skin and the interior of blood vessels, but harmless to humans.
Professor Lee found a solution from natural polymer ingredients. Catecholamine can be found in mussels. Professor Lee conjugated catechol groups on the chitosan backbone. He applied this mussel-inspired adhesive polymer Chitosan-catechol, which immediately forms an adhesive layer with blood, as a bioadhesion for the antibleeding injection needle.
Professor Lee said, “Chitosan-catechol, which copies the adhesive mechanism of mussels, shows high solubility in physiological saline as well as great mucoadhesion. Hence, it is perfectly suitable for coating the injection needle. Combining it with proteins allows for efficient drug delivery to the heart, which is a challenging injection location, so it will be also useful for treating incurable heart disease.”
2018.10.05 View 11961
Scientist of October, Professor Haeshin Lee
(Professor Haeshin Lee from the Department of Chemistry)
Professor Haeshin Lee from the Department of Chemistry received the ‘Science and Technology Award of October’ from the Ministry of Science and ICT and the National Research Foundation of Korea for his contribution to developing an antibleeding injection needle. This novel outcome will fundamentally prevent the problem of secondary infections of AIDS, Ebola and Hepatitis viruses transmitting from patients to medical teams.
This needle’s surface is coated with hemostatic materials. Its concept is simple and the key to this technology is to make materials that are firmly coated on the needle so that they can endure frictional force when being injected into skin and blood vessels. Moreover, the materials should be adhesive to skin and the interior of blood vessels, but harmless to humans.
Professor Lee found a solution from natural polymer ingredients. Catecholamine can be found in mussels. Professor Lee conjugated catechol groups on the chitosan backbone. He applied this mussel-inspired adhesive polymer Chitosan-catechol, which immediately forms an adhesive layer with blood, as a bioadhesion for the antibleeding injection needle.
Professor Lee said, “Chitosan-catechol, which copies the adhesive mechanism of mussels, shows high solubility in physiological saline as well as great mucoadhesion. Hence, it is perfectly suitable for coating the injection needle. Combining it with proteins allows for efficient drug delivery to the heart, which is a challenging injection location, so it will be also useful for treating incurable heart disease.”
2018.10.05 View 11961 -
 Center for Industrial Future Strategy Takes Off at KAIST
(Professor Wonjoon Kim from the School of Business and Technology Management)
Professors from KAIST and major international universities launched a mega-scale research center focusing on the Fourth Industrial Revolution, named the Center for Industrial Future Strategy (CIFS).
This center is funded by the National Research Foundation Korea and will receive 2.25 billion KRW over four years.
Directed by Professor Wonjoon Kim from the School of Business and Technology Management, the center is comprised of ten top-tier researchers and four research associates, including Professor Hawoon Jeong (KAIST), Professor Scott Stern (MIT), Professor Aaron Chatterji (Duke University), Dr. Yong Suk Lee (Stanford University) and Professor Hyejin Youn (Northwestern University).
The center will conduct research on technical, social, and economic changes derived by a new paradigm of technological innovation.
Moreover, they will study policies and strategies in relation to innovation in the corporate and government sectors to achieve economic growth in a sustainable manner. The center will also propose policies and strategies in a variety of economic and industrial settings to establish a sustainable and global innovation ecosystem.
To carry out these studies successfully, CIFS will further expand the AIEA-NBER Conference with the Asia Innovation and Entrepreneurship Association (AIEA) and the National Bureau of Economic Research (NBER) in which numerous Nobel Laureates in Economics are affiliated. They will also comprise thematic research teams with co-founding universities to build stronger cooperation with one another.
Besides the academic cooperation, the center will also build partnerships with international organizations, including the Asian Development Bank and the Inter-American Development Bank to carry out their missions at multilateral levels.
Their research topics include changes to value chains in a new paradigm of technological innovation, labor market changes in the Fourth Industrial Revolution, sharing economies and social interests, big data, artificial intelligence & privacy policy, and innovation & ethical and institutional countermeasures to AI technology.
Professor Kim said, “The new paradigm of technological innovation is evolving social, economic, and industrial structures, such as R&D, industry, technology, labor, finance, and institutions. The Center will contribute to proposing policies and strategies so that Korea, as well as the international community, can take appropriate measures to these big changes.”
2018.09.11 View 11981
Center for Industrial Future Strategy Takes Off at KAIST
(Professor Wonjoon Kim from the School of Business and Technology Management)
Professors from KAIST and major international universities launched a mega-scale research center focusing on the Fourth Industrial Revolution, named the Center for Industrial Future Strategy (CIFS).
This center is funded by the National Research Foundation Korea and will receive 2.25 billion KRW over four years.
Directed by Professor Wonjoon Kim from the School of Business and Technology Management, the center is comprised of ten top-tier researchers and four research associates, including Professor Hawoon Jeong (KAIST), Professor Scott Stern (MIT), Professor Aaron Chatterji (Duke University), Dr. Yong Suk Lee (Stanford University) and Professor Hyejin Youn (Northwestern University).
The center will conduct research on technical, social, and economic changes derived by a new paradigm of technological innovation.
Moreover, they will study policies and strategies in relation to innovation in the corporate and government sectors to achieve economic growth in a sustainable manner. The center will also propose policies and strategies in a variety of economic and industrial settings to establish a sustainable and global innovation ecosystem.
To carry out these studies successfully, CIFS will further expand the AIEA-NBER Conference with the Asia Innovation and Entrepreneurship Association (AIEA) and the National Bureau of Economic Research (NBER) in which numerous Nobel Laureates in Economics are affiliated. They will also comprise thematic research teams with co-founding universities to build stronger cooperation with one another.
Besides the academic cooperation, the center will also build partnerships with international organizations, including the Asian Development Bank and the Inter-American Development Bank to carry out their missions at multilateral levels.
Their research topics include changes to value chains in a new paradigm of technological innovation, labor market changes in the Fourth Industrial Revolution, sharing economies and social interests, big data, artificial intelligence & privacy policy, and innovation & ethical and institutional countermeasures to AI technology.
Professor Kim said, “The new paradigm of technological innovation is evolving social, economic, and industrial structures, such as R&D, industry, technology, labor, finance, and institutions. The Center will contribute to proposing policies and strategies so that Korea, as well as the international community, can take appropriate measures to these big changes.”
2018.09.11 View 11981 -
 Professor Hee-Sung Park Named Scientist of May
(Professor Hee-Sung Park)
Professor Hee-Sung Park from the Department of Chemistry was named ‘Scientist of May’ sponsored by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Park was honored in recognition of his developing a tool to engineer designer proteins via diverse chemical modifications. This approach provides a novel platform for investigating numerous diseases such as cancer and dementia.
His research focuses on the production of synthetic proteins and the generation of diverse protein functions as well as the designing and engineering of new translation machinery for genetic code expansion, and the application of synthetic biology techniques for basic cell biology and applied medical science.
Post-translational modifications (PTMs) are constantly taking place during or after protein biosynthesis. PTMs play a vital role in expanding protein functional diversity and, as a result, critically affect numerous biological processes. Abnormal PTMs have been known to trigger various diseases including cancer and dementia. Therefore, this technology enables proteins to reproduce with specific modifications at selected residues and will significantly help establish experimental strategies to investigate fundamental biological mechanisms including the development of targeted cancer therapies.
Professor Park also received 10 million KRW in prize money.
2018.05.04 View 11885
Professor Hee-Sung Park Named Scientist of May
(Professor Hee-Sung Park)
Professor Hee-Sung Park from the Department of Chemistry was named ‘Scientist of May’ sponsored by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Park was honored in recognition of his developing a tool to engineer designer proteins via diverse chemical modifications. This approach provides a novel platform for investigating numerous diseases such as cancer and dementia.
His research focuses on the production of synthetic proteins and the generation of diverse protein functions as well as the designing and engineering of new translation machinery for genetic code expansion, and the application of synthetic biology techniques for basic cell biology and applied medical science.
Post-translational modifications (PTMs) are constantly taking place during or after protein biosynthesis. PTMs play a vital role in expanding protein functional diversity and, as a result, critically affect numerous biological processes. Abnormal PTMs have been known to trigger various diseases including cancer and dementia. Therefore, this technology enables proteins to reproduce with specific modifications at selected residues and will significantly help establish experimental strategies to investigate fundamental biological mechanisms including the development of targeted cancer therapies.
Professor Park also received 10 million KRW in prize money.
2018.05.04 View 11885 -
 Scientist of March, Professor Hee-Seung Lee
(Professor Hee-Seung Lee)
Professor Hee-Seung Lee from the Department of Chemistry at KAIST received the ‘Science and Technology Award of the Month’ awarded by the Ministry of ICT and Science, and the National Research Foundation of Korea for March 2018.
Professor Lee has been recognized for successfully producing peptide-based molecular machines, which used to be made of metals.
The methodology can be translated into magnetotactic behavior at the macroscopic scale, which is reminiscent of magnetosomes in magnetotactic bacteria.
The team employed foldectures, self-assembled molecular architectures of β-peptide foldamers, to develop the peptide-based molecular machines that uniformly align with respect to an applied static magnetic field.
Professor Lee said, “Molecular machines are widely used in the field of medical engineering or material science; however, there were limitations for developing the machines using magnetic fields. By developing peptide-based molecular machines, we were able to develop body-friendly molecular machines.”
Every month, the Ministry of ICT and Science and the National Research Foundation of Korea award a cash prize worth 10,000,000 KRW to a scientist who has contributed to science and technology with outstanding research and development performance.
2018.03.15 View 10749
Scientist of March, Professor Hee-Seung Lee
(Professor Hee-Seung Lee)
Professor Hee-Seung Lee from the Department of Chemistry at KAIST received the ‘Science and Technology Award of the Month’ awarded by the Ministry of ICT and Science, and the National Research Foundation of Korea for March 2018.
Professor Lee has been recognized for successfully producing peptide-based molecular machines, which used to be made of metals.
The methodology can be translated into magnetotactic behavior at the macroscopic scale, which is reminiscent of magnetosomes in magnetotactic bacteria.
The team employed foldectures, self-assembled molecular architectures of β-peptide foldamers, to develop the peptide-based molecular machines that uniformly align with respect to an applied static magnetic field.
Professor Lee said, “Molecular machines are widely used in the field of medical engineering or material science; however, there were limitations for developing the machines using magnetic fields. By developing peptide-based molecular machines, we were able to develop body-friendly molecular machines.”
Every month, the Ministry of ICT and Science and the National Research Foundation of Korea award a cash prize worth 10,000,000 KRW to a scientist who has contributed to science and technology with outstanding research and development performance.
2018.03.15 View 10749 -
 Scientist of November, Professor Hyung Jin Sung
Professor Hyung Jin Sung from the Department of Mechanical Engineering at KAIST received a ‘Science and Technology Award of the Month’ given by the Ministry of ICT and Science and the National Research Foundation of Korea for November 2017. He developed technology that can exquisitely control a micrometer-scaled liquid drop on a dime-sized lab-on-a-chip. With his work, he was recognized for reinforcing research capability on microfluidics.
Lab-on-a-chip is an emerging experiment and diagnostic technology in the form of a bio-microchip that facilitates complex and various experiments with only a minimal sample size required. This technology draws a lot of attention not only from medical and pharmaceutical areas, but also the health and environmental field. The biggest problem was that technology for the temperature control of a fluid sample, which is one of the core technologies in microfluidics, has low accuracy. This limit had to be overcome in order to use the lab-on-a-chip more widely.
Professor Sung developed an acoustic and thermal method which controls the temperature of a droplet quickly and meticulously by using sound and energy. This is a thermal method that uses heat generated during the absorption of an acoustic wave into viscoelastic substances. It facilitates a rapid heating rate and spatial-temporal temperature control, allowing heating in desired areas. In addition, Professor Sung applied his technology to polymerase chain reactions, which are used to amplify DNA.
Through this experiment, he successfully shortened the reaction time from 1-2 hours to only three minutes, making this a groundbreaking achievement.
Professor Sung said, “My research is significant for enhancing the applicability of microfluidics. I expect that it will lead to technological innovations in healthcare fields including biochemistry, medical checkups, and new medicine development.”
2017.11.03 View 11480
Scientist of November, Professor Hyung Jin Sung
Professor Hyung Jin Sung from the Department of Mechanical Engineering at KAIST received a ‘Science and Technology Award of the Month’ given by the Ministry of ICT and Science and the National Research Foundation of Korea for November 2017. He developed technology that can exquisitely control a micrometer-scaled liquid drop on a dime-sized lab-on-a-chip. With his work, he was recognized for reinforcing research capability on microfluidics.
Lab-on-a-chip is an emerging experiment and diagnostic technology in the form of a bio-microchip that facilitates complex and various experiments with only a minimal sample size required. This technology draws a lot of attention not only from medical and pharmaceutical areas, but also the health and environmental field. The biggest problem was that technology for the temperature control of a fluid sample, which is one of the core technologies in microfluidics, has low accuracy. This limit had to be overcome in order to use the lab-on-a-chip more widely.
Professor Sung developed an acoustic and thermal method which controls the temperature of a droplet quickly and meticulously by using sound and energy. This is a thermal method that uses heat generated during the absorption of an acoustic wave into viscoelastic substances. It facilitates a rapid heating rate and spatial-temporal temperature control, allowing heating in desired areas. In addition, Professor Sung applied his technology to polymerase chain reactions, which are used to amplify DNA.
Through this experiment, he successfully shortened the reaction time from 1-2 hours to only three minutes, making this a groundbreaking achievement.
Professor Sung said, “My research is significant for enhancing the applicability of microfluidics. I expect that it will lead to technological innovations in healthcare fields including biochemistry, medical checkups, and new medicine development.”
2017.11.03 View 11480 -
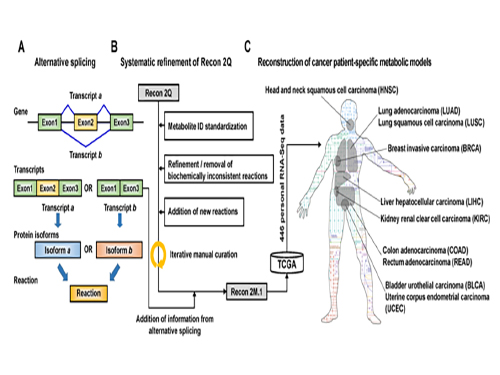 Development of a Highly-Accurate Computational Model of Human Metabolism
A research team from KAIST developed a computational framework that enables the reconstruction of a comprehensive computational model of human metabolism, which allows for an accurate prediction of personal metabolic features (or phenotypes).
Understanding personal metabolic phenotypes allows us to design effective therapeutic strategies for various chronic and infectious diseases. A human computational model called the genome-scale metabolic model (GEM) contains information on thousands of metabolic genes and their corresponding reactions and metabolites, and has played an important role in predicting metabolic phenotypes. Although several versions of human GEMs have been released, they had room for further development, especially as to incorporating biological information coming from a human genetics mechanism called “alternative splicing.” Alternative splicing is a genetic mechanism that allows a gene to give rise to multiple reactions, and is strongly associated with pathology.
To tackle this problem, Jae Yong Ryu (a Ph.D. student), Dr. Hyun Uk Kim (Research Fellow), and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at KAIST, developed a computational framework that systematically generates metabolic reactions, and adds them to the human GEM. The resulting human GEM was demonstrated to accurately predict metabolic phenotypes under varied environmental conditions. The research results were published online in Proceedings of the National Academy of Sciences (PNAS) on October 24, 2017, under the title “Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism.”
The research team first updated the biological contents of a previous version of the human GEM. The updated biological contents include metabolic genes and their corresponding metabolites and reactions. In particular, metabolic reactions catalyzed by already-known protein isoforms were additionally incorporated into the human GEM; protein isoforms are multiple variants of proteins generated from individual genes through the alternative splicing process. Each protein isoform is often responsible for the operation of a metabolic reaction. Although multiple protein isoforms generated from one gene can play different functions by having different sets of protein domains and/or subcellular localizations, such information was not properly considered in previous versions of human GEMs.
Upon the initial update of the human GEM, named Recon 2M.1, the research team subsequently implemented a computational framework that systematically generates information on Gene-Transcript-Protein-Reaction Associations (GeTPRA) in order to identify protein isoforms that were previously not identified. This framework was developed in this study. As a result of the implementation of the framework for GeTPRA, more than 11,000 GeTPRA were automatically predicted, and thoroughly validated. Additional metabolic reactions were then added to Recon 2M.1 based on the predicted GeTPRA for the previously uncharacterized protein isoforms; Recon 2M.1 was renamed Recon 2M.2 from this upgrade.
Finally, Recon 2M.2 was integrated with 446 sets of personal biological data (RNA-Seq data) in order to build patient-specific cancer models. These patient-specific cancer models were used to predict cancer metabolism activities and anticancer targets.
The development of a new version of human GEMs along with the computational framework for GeTPRA is expected to boost studies in fundamental human genetics and medicine. Model files of the human GEMs Recon 2M.1 and 2M.2, a full list of the GeTPRA and the source code for the computational framework to predict the GeTPRA are all available as part of the publication of this study.
Distinguished Professor Lee said, “The predicted GeTPRA from the computational framework is expected to serve as a guideline for future experiments on human genetics and biochemistry, whereas the resulting Recon 2M.2 can be used to predict drug targets for various human diseases.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea.
(Figure 1:A scheme of Recon 2M.1 development and its use in reconstructing personal genome-scale metabolic models (GEMs). (A) A concept of alternative splicing of human genes and its use in Gene-Transcript-Protein-Reaction Associations (GeTPRA) of Recon 2M.1. (B) A procedure of systematic refinement of the Recon 2Q. Recon 2Q is one of the previously released human GEMs. Biochemically inconsistent reactions include unbalanced, artificial, blocked, and/or redundant reactions. Iterative manual curation was conducted while validating the Recon 2M.1. (C) Reconstruction of cancer patient-specific GEMs using Recon 2M.1 for further simulation studies. In this study, personal biological data (RNA-Seq data) were obtained from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/ ) across the ten cancer types.
(Figure 2: Computational framework for the systematic generation of Gene-Transcript-Protein-Reaction Associations (GeTPRA; red box in the flowchart). Peptide sequences of metabolic genes defined in Recon 2M.1 were retrieved from a database called Ensembl. EC numbers and subcellular localizations of all the protein isoforms of metabolic genes in Recon 2M.1 were predicted using software programs EFICAz2.5 and Wolf PSort, respectively. Information on the newly predicted GeTPRA was systematically incorporated into the Recon 2M.1, thereby resulting in Recon 2M.2.)
2017.10.25 View 10907
Development of a Highly-Accurate Computational Model of Human Metabolism
A research team from KAIST developed a computational framework that enables the reconstruction of a comprehensive computational model of human metabolism, which allows for an accurate prediction of personal metabolic features (or phenotypes).
Understanding personal metabolic phenotypes allows us to design effective therapeutic strategies for various chronic and infectious diseases. A human computational model called the genome-scale metabolic model (GEM) contains information on thousands of metabolic genes and their corresponding reactions and metabolites, and has played an important role in predicting metabolic phenotypes. Although several versions of human GEMs have been released, they had room for further development, especially as to incorporating biological information coming from a human genetics mechanism called “alternative splicing.” Alternative splicing is a genetic mechanism that allows a gene to give rise to multiple reactions, and is strongly associated with pathology.
To tackle this problem, Jae Yong Ryu (a Ph.D. student), Dr. Hyun Uk Kim (Research Fellow), and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at KAIST, developed a computational framework that systematically generates metabolic reactions, and adds them to the human GEM. The resulting human GEM was demonstrated to accurately predict metabolic phenotypes under varied environmental conditions. The research results were published online in Proceedings of the National Academy of Sciences (PNAS) on October 24, 2017, under the title “Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism.”
The research team first updated the biological contents of a previous version of the human GEM. The updated biological contents include metabolic genes and their corresponding metabolites and reactions. In particular, metabolic reactions catalyzed by already-known protein isoforms were additionally incorporated into the human GEM; protein isoforms are multiple variants of proteins generated from individual genes through the alternative splicing process. Each protein isoform is often responsible for the operation of a metabolic reaction. Although multiple protein isoforms generated from one gene can play different functions by having different sets of protein domains and/or subcellular localizations, such information was not properly considered in previous versions of human GEMs.
Upon the initial update of the human GEM, named Recon 2M.1, the research team subsequently implemented a computational framework that systematically generates information on Gene-Transcript-Protein-Reaction Associations (GeTPRA) in order to identify protein isoforms that were previously not identified. This framework was developed in this study. As a result of the implementation of the framework for GeTPRA, more than 11,000 GeTPRA were automatically predicted, and thoroughly validated. Additional metabolic reactions were then added to Recon 2M.1 based on the predicted GeTPRA for the previously uncharacterized protein isoforms; Recon 2M.1 was renamed Recon 2M.2 from this upgrade.
Finally, Recon 2M.2 was integrated with 446 sets of personal biological data (RNA-Seq data) in order to build patient-specific cancer models. These patient-specific cancer models were used to predict cancer metabolism activities and anticancer targets.
The development of a new version of human GEMs along with the computational framework for GeTPRA is expected to boost studies in fundamental human genetics and medicine. Model files of the human GEMs Recon 2M.1 and 2M.2, a full list of the GeTPRA and the source code for the computational framework to predict the GeTPRA are all available as part of the publication of this study.
Distinguished Professor Lee said, “The predicted GeTPRA from the computational framework is expected to serve as a guideline for future experiments on human genetics and biochemistry, whereas the resulting Recon 2M.2 can be used to predict drug targets for various human diseases.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea.
(Figure 1:A scheme of Recon 2M.1 development and its use in reconstructing personal genome-scale metabolic models (GEMs). (A) A concept of alternative splicing of human genes and its use in Gene-Transcript-Protein-Reaction Associations (GeTPRA) of Recon 2M.1. (B) A procedure of systematic refinement of the Recon 2Q. Recon 2Q is one of the previously released human GEMs. Biochemically inconsistent reactions include unbalanced, artificial, blocked, and/or redundant reactions. Iterative manual curation was conducted while validating the Recon 2M.1. (C) Reconstruction of cancer patient-specific GEMs using Recon 2M.1 for further simulation studies. In this study, personal biological data (RNA-Seq data) were obtained from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/ ) across the ten cancer types.
(Figure 2: Computational framework for the systematic generation of Gene-Transcript-Protein-Reaction Associations (GeTPRA; red box in the flowchart). Peptide sequences of metabolic genes defined in Recon 2M.1 were retrieved from a database called Ensembl. EC numbers and subcellular localizations of all the protein isoforms of metabolic genes in Recon 2M.1 were predicted using software programs EFICAz2.5 and Wolf PSort, respectively. Information on the newly predicted GeTPRA was systematically incorporated into the Recon 2M.1, thereby resulting in Recon 2M.2.)
2017.10.25 View 10907 -
 Photoacoustic Imaging and Photothermal Cancer Therapy Using BR Nanoparticles
(Professor Sangyong Jon and PhD Candidate Dong Yun Lee)
Sangyong Jon, a professor in the Department of Biological Sciences at KAIST, and his team developed combined photoacoustic imaging and photothermal therapy for cancer by using Bilirubin (BR) nanoparticles.
The research team applied the properties of a bile pigment called BR, which exerts potent antioxidant and anti-inflammatory effects, to this research.
The team expects this research, which shows high biocompatibility as well as outstanding photoacoustic imaging and photothermal therapy, to be an appropriate system in the field of treatment for cancer.
In the past, the research team developed a PEGylated bilirubin-based nanoparticle system by combining water-insoluble BR with water-soluble Polyethylene Glycol (PEG).
This technology facilitated BR exerting antioxidants yet prevented them from being accumulated in the body. Its efficiency and safety was identified in an animal disease model, for conditions such as inflammatory bowel disease, islet cell transportation, and asthma.
Differing from previous research methods, this research applied the different physicochemical properties of BR to cancer treatment.
When the causative agent of jaundice, yellow BR, is exposed to a certain wavelength of blue light, the agent becomes a photonic nanomaterial as it responses to the light. This light-responsive nanomaterial can be used to cure jaundice because it allows for active excretion in infants.
Secondly, the team identified that BR is a major component of black pigment gallstones which can be often found in gall bladders or bile ducts under certain pathological conditions. The findings show that BR forms black pigment gallstones without the role of an intermediate or cation, such as calcium and copper. The research team combined cisplatin, a platinum metal-based anticancer drug, with BR so that BR nanoparticles changed the solution color from yellow to purple.
The team also examined the possibility of cisplatin-chelated BR nanoparticles as a probe for photoacoustic images. They found that considerable photoacoustic activity was shown when it was exposed to near infrared light. In fact, the photoacoustic signal was increased significantly in tumors of animals with colorectal cancer when the nanoparticles were administered to it intravenously. The team expects a more accurate diagnosis of tumors through this technology.
Moreover, the team assessed the photothermal effects of cisplatin-chelated BR nanoparticles. The research showed that the temperature of tumors increased by 25 degrees Celsius within five minutes when they were exposed to near infrared light, due to the photothermal effect. After two weeks, their size was reduced compared to that of other groups, and sometimes the tumors were even necrotized.
Professor Jon said, “Existing substances have a low biocompatibility and limitation for clinical therapy because they are artificially oriented; therefore, they might have toxicity. I am hoping that these cisplatin-chelated BR-based nanoparticles will provide a new platform for preclinical, translational research and clinical adaptation of the photoacoustic imaging and photothermal therapy.”
The paper (Dong Yun Lee as a first author) was published online in the renowned journal in the field of applied chemistry, Angewandte Chemi International Edition, on September 4. This research was sponsored by the National Research Foundation of Korea.
(Schematic diagram of the research)
(From left: Bilirubin nanoparticles, cisplatin-chelated Bilirubin nanoparticles)
2017.09.26 View 9671
Photoacoustic Imaging and Photothermal Cancer Therapy Using BR Nanoparticles
(Professor Sangyong Jon and PhD Candidate Dong Yun Lee)
Sangyong Jon, a professor in the Department of Biological Sciences at KAIST, and his team developed combined photoacoustic imaging and photothermal therapy for cancer by using Bilirubin (BR) nanoparticles.
The research team applied the properties of a bile pigment called BR, which exerts potent antioxidant and anti-inflammatory effects, to this research.
The team expects this research, which shows high biocompatibility as well as outstanding photoacoustic imaging and photothermal therapy, to be an appropriate system in the field of treatment for cancer.
In the past, the research team developed a PEGylated bilirubin-based nanoparticle system by combining water-insoluble BR with water-soluble Polyethylene Glycol (PEG).
This technology facilitated BR exerting antioxidants yet prevented them from being accumulated in the body. Its efficiency and safety was identified in an animal disease model, for conditions such as inflammatory bowel disease, islet cell transportation, and asthma.
Differing from previous research methods, this research applied the different physicochemical properties of BR to cancer treatment.
When the causative agent of jaundice, yellow BR, is exposed to a certain wavelength of blue light, the agent becomes a photonic nanomaterial as it responses to the light. This light-responsive nanomaterial can be used to cure jaundice because it allows for active excretion in infants.
Secondly, the team identified that BR is a major component of black pigment gallstones which can be often found in gall bladders or bile ducts under certain pathological conditions. The findings show that BR forms black pigment gallstones without the role of an intermediate or cation, such as calcium and copper. The research team combined cisplatin, a platinum metal-based anticancer drug, with BR so that BR nanoparticles changed the solution color from yellow to purple.
The team also examined the possibility of cisplatin-chelated BR nanoparticles as a probe for photoacoustic images. They found that considerable photoacoustic activity was shown when it was exposed to near infrared light. In fact, the photoacoustic signal was increased significantly in tumors of animals with colorectal cancer when the nanoparticles were administered to it intravenously. The team expects a more accurate diagnosis of tumors through this technology.
Moreover, the team assessed the photothermal effects of cisplatin-chelated BR nanoparticles. The research showed that the temperature of tumors increased by 25 degrees Celsius within five minutes when they were exposed to near infrared light, due to the photothermal effect. After two weeks, their size was reduced compared to that of other groups, and sometimes the tumors were even necrotized.
Professor Jon said, “Existing substances have a low biocompatibility and limitation for clinical therapy because they are artificially oriented; therefore, they might have toxicity. I am hoping that these cisplatin-chelated BR-based nanoparticles will provide a new platform for preclinical, translational research and clinical adaptation of the photoacoustic imaging and photothermal therapy.”
The paper (Dong Yun Lee as a first author) was published online in the renowned journal in the field of applied chemistry, Angewandte Chemi International Edition, on September 4. This research was sponsored by the National Research Foundation of Korea.
(Schematic diagram of the research)
(From left: Bilirubin nanoparticles, cisplatin-chelated Bilirubin nanoparticles)
2017.09.26 View 9671 -
 Research Center for Smart Submerged Floating Tunnel Systems Opens
(Distinguished guests including President Shin (fourth from the right) and Director Lee (third from left) at the opening ceremony)
The Research Center for a Smart Submerged Floating Tunnel Systems was recently established at KAIST with the purpose of taking the lead in developing fundamental and applicable technology for submerged floating tunnels as well as fostering creative and talented people. Haeng-Ki Lee, a professor in the Department of Civil & Environmental Engineering at KAIST is heading the center.
KAIST held its opening ceremony on September 7, 2017 in the Applied Engineering Building located on the main campus.
Distinguished guests, including KAIST president Sung-Chul Shin, the President of the Korea Institute of Ocean Science and Technology Gi-Hoon Hong, the President of the Korean Society of Civil Engineering Young-Seok Park, and the Director in the Division of Engineering at the National Research Foundation of Korea Joong-Kon Park attended the ceremony.
The National Research Foundation of Korea provides Engineering Research Center (ERC) projects which find and foster groups with outstanding research performance in a field of engineering. The projects support these groups so that they can strengthen their global competitiveness while enhancing national competence in basic research.
The ‘Research Center for Smart Submerged Floating Tunnel Systems’ was selected as one of the ERC projects in 2017. For the next seven years, the research center will work to develop a submerged floating tunnel system resistant depths greater than 100 meters.
To achieve its goal, the center has defined crucial research topics including: i) a structural analysis program and integrated design technology specific for submerged floating tunnel systems, ii) high-durability marine construction materials and submerged construction integrated systems, and iii) safety and maintenance integrated technology for smart submerged floating tunnel systems.
The ‘Research Center for Smart Submerged Floating Tunnel Systems’ will devote itself to developing a variety of fundamental and applicable technology that will be leading global maritime construction. Moreover, it will concentrate on fostering professional research manpower in related areas.
The Director of the Center Lee said, “The center will cooperate with KAIST researchers who are experts in various fields, including structures, materials, construction, and maritime research. Based on this collaboration, the center will contribute to achieving autonomous technologies by developing fundamental and applicable technology related with submerged floating tunnel systems. It will also take the role of a leading global research hub in the field of submerged floating tunnels as well as construction technologies.”
2017.09.07 View 10956
Research Center for Smart Submerged Floating Tunnel Systems Opens
(Distinguished guests including President Shin (fourth from the right) and Director Lee (third from left) at the opening ceremony)
The Research Center for a Smart Submerged Floating Tunnel Systems was recently established at KAIST with the purpose of taking the lead in developing fundamental and applicable technology for submerged floating tunnels as well as fostering creative and talented people. Haeng-Ki Lee, a professor in the Department of Civil & Environmental Engineering at KAIST is heading the center.
KAIST held its opening ceremony on September 7, 2017 in the Applied Engineering Building located on the main campus.
Distinguished guests, including KAIST president Sung-Chul Shin, the President of the Korea Institute of Ocean Science and Technology Gi-Hoon Hong, the President of the Korean Society of Civil Engineering Young-Seok Park, and the Director in the Division of Engineering at the National Research Foundation of Korea Joong-Kon Park attended the ceremony.
The National Research Foundation of Korea provides Engineering Research Center (ERC) projects which find and foster groups with outstanding research performance in a field of engineering. The projects support these groups so that they can strengthen their global competitiveness while enhancing national competence in basic research.
The ‘Research Center for Smart Submerged Floating Tunnel Systems’ was selected as one of the ERC projects in 2017. For the next seven years, the research center will work to develop a submerged floating tunnel system resistant depths greater than 100 meters.
To achieve its goal, the center has defined crucial research topics including: i) a structural analysis program and integrated design technology specific for submerged floating tunnel systems, ii) high-durability marine construction materials and submerged construction integrated systems, and iii) safety and maintenance integrated technology for smart submerged floating tunnel systems.
The ‘Research Center for Smart Submerged Floating Tunnel Systems’ will devote itself to developing a variety of fundamental and applicable technology that will be leading global maritime construction. Moreover, it will concentrate on fostering professional research manpower in related areas.
The Director of the Center Lee said, “The center will cooperate with KAIST researchers who are experts in various fields, including structures, materials, construction, and maritime research. Based on this collaboration, the center will contribute to achieving autonomous technologies by developing fundamental and applicable technology related with submerged floating tunnel systems. It will also take the role of a leading global research hub in the field of submerged floating tunnels as well as construction technologies.”
2017.09.07 View 10956 -
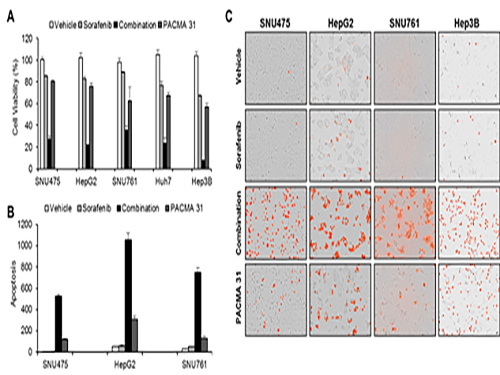 Discovery of an Optimal Drug Combination: Overcoming Resistance to Targeted Drugs for Liver Cancer
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences. Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)
(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)
(Figure 3. ER stress network model)
2017.08.30 View 13013
Discovery of an Optimal Drug Combination: Overcoming Resistance to Targeted Drugs for Liver Cancer
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences. Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)
(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)
(Figure 3. ER stress network model)
2017.08.30 View 13013