ACT
-
 Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 11941
Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 11941 -
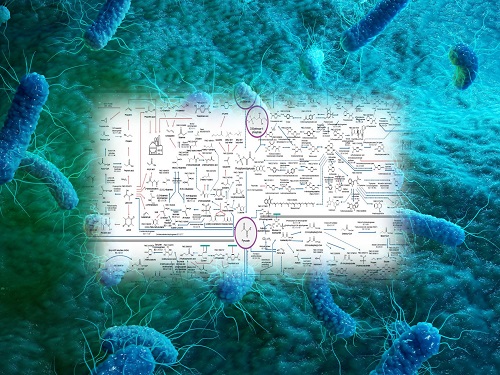 Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 13026
Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 13026 -
 Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 9220
Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 9220 -
 Professor Juho Kim’s Team Wins Best Paper Award at ACM CHI 2022
The research team led by Professor Juho Kim from the KAIST School of Computing won a Best Paper Award and an Honorable Mention Award at the Association for Computing Machinery Conference on Human Factors in Computing Systems (ACM CHI) held between April 30 and May 6.
ACM CHI is the world’s most recognized conference in the field of human computer interactions (HCI), and is ranked number one out of all HCI-related journals and conferences based on Google Scholar’s h-5 index. Best paper awards are given to works that rank in the top one percent, and honorable mention awards are given to the top five percent of the papers accepted by the conference.
Professor Juho Kim presented a total of seven papers at ACM CHI 2022, and tied for the largest number of papers. A total of 19 papers were affiliated with KAIST, putting it fifth out of all participating institutes and thereby proving KAIST’s competence in research.
One of Professor Kim’s research teams composed of Jeongyeon Kim (first author, MS graduate) from the School of Computing, MS candidate Yubin Choi from the School of Electrical Engineering, and Dr. Meng Xia (post-doctoral associate in the School of Computing, currently a post-doctoral associate at Carnegie Mellon University) received a best paper award for their paper, “Mobile-Friendly Content Design for MOOCs: Challenges, Requirements, and Design Opportunities”.
The study analyzed the difficulties experienced by learners watching video-based educational content in a mobile environment and suggests guidelines for solutions. The research team analyzed 134 survey responses and 21 interviews, and revealed that texts that are too small or overcrowded are what mainly brings down the legibility of video contents. Additionally, lighting, noise, and surrounding environments that change frequently are also important factors that may disturb a learning experience.
Based on these findings, the team analyzed the aptness of 41,722 frames from 101 video lectures for mobile environments, and confirmed that they generally show low levels of adequacy. For instance, in the case of text sizes, only 24.5% of the frames were shown to be adequate for learning in mobile environments. To overcome this issue, the research team suggested a guideline that may improve the legibility of video contents and help overcome the difficulties arising from mobile learning environments.
The importance of and dependency on video-based learning continue to rise, especially in the wake of the pandemic, and it is meaningful that this research suggested a means to analyze and tackle the difficulties of users that learn from the small screens of mobile devices. Furthermore, the paper also suggested technology that can solve problems related to video-based learning through human-AI collaborations, enhancing existing video lectures and improving learning experiences. This technology can be applied to various video-based platforms and content creation.
Meanwhile, a research team composed of Ph.D. candidate Tae Soo Kim (first author), MS candidate DaEun Choi, and Ph.D. candidate Yoonseo Choi from the School of Computing received an honorable mention award for their paper, “Stylette: styling the Web with Natural Language”.
The research team developed a novel interface technology that allows nonexperts who are unfamiliar with technical jargon to edit website features through speech. People often find it difficult to use or find the information they need from various websites due to accessibility issues, device-related constraints, inconvenient design, style preferences, etc. However, it is not easy for laymen to edit website features without expertise in programming or design, and most end up just putting up with the inconveniences. But what if the system could read the intentions of its users from their everyday language like “emphasize this part a little more”, or “I want a more modern design”, and edit the features automatically?
Based on this question, Professor Kim’s research team developed ‘Stylette’, a system in which AI analyses its users’ speech expressed in their natural language and automatically recommends a new style that best fits their intentions. The research team created a new system by putting together language AI, visual AI, and user interface technologies. On the linguistic side, a large-scale language model AI converts the intentions of the users expressed through their everyday language into adequate style elements. On the visual side, computer vision AI compares 1.7 million existing web design features and recommends a style adequate for the current website. In an experiment where 40 nonexperts were asked to edit a website design, the subjects that used this system showed double the success rate in a time span that was 35% shorter compared to the control group.
It is meaningful that this research proposed a practical case in which AI technology constructs intuitive interactions with users. The developed technology can be applied to existing design applications and web browsers in a plug-in format, and can be utilized to improve websites or for advertisements by collecting the natural intention data of users on a large scale.
2022.06.13 View 7897
Professor Juho Kim’s Team Wins Best Paper Award at ACM CHI 2022
The research team led by Professor Juho Kim from the KAIST School of Computing won a Best Paper Award and an Honorable Mention Award at the Association for Computing Machinery Conference on Human Factors in Computing Systems (ACM CHI) held between April 30 and May 6.
ACM CHI is the world’s most recognized conference in the field of human computer interactions (HCI), and is ranked number one out of all HCI-related journals and conferences based on Google Scholar’s h-5 index. Best paper awards are given to works that rank in the top one percent, and honorable mention awards are given to the top five percent of the papers accepted by the conference.
Professor Juho Kim presented a total of seven papers at ACM CHI 2022, and tied for the largest number of papers. A total of 19 papers were affiliated with KAIST, putting it fifth out of all participating institutes and thereby proving KAIST’s competence in research.
One of Professor Kim’s research teams composed of Jeongyeon Kim (first author, MS graduate) from the School of Computing, MS candidate Yubin Choi from the School of Electrical Engineering, and Dr. Meng Xia (post-doctoral associate in the School of Computing, currently a post-doctoral associate at Carnegie Mellon University) received a best paper award for their paper, “Mobile-Friendly Content Design for MOOCs: Challenges, Requirements, and Design Opportunities”.
The study analyzed the difficulties experienced by learners watching video-based educational content in a mobile environment and suggests guidelines for solutions. The research team analyzed 134 survey responses and 21 interviews, and revealed that texts that are too small or overcrowded are what mainly brings down the legibility of video contents. Additionally, lighting, noise, and surrounding environments that change frequently are also important factors that may disturb a learning experience.
Based on these findings, the team analyzed the aptness of 41,722 frames from 101 video lectures for mobile environments, and confirmed that they generally show low levels of adequacy. For instance, in the case of text sizes, only 24.5% of the frames were shown to be adequate for learning in mobile environments. To overcome this issue, the research team suggested a guideline that may improve the legibility of video contents and help overcome the difficulties arising from mobile learning environments.
The importance of and dependency on video-based learning continue to rise, especially in the wake of the pandemic, and it is meaningful that this research suggested a means to analyze and tackle the difficulties of users that learn from the small screens of mobile devices. Furthermore, the paper also suggested technology that can solve problems related to video-based learning through human-AI collaborations, enhancing existing video lectures and improving learning experiences. This technology can be applied to various video-based platforms and content creation.
Meanwhile, a research team composed of Ph.D. candidate Tae Soo Kim (first author), MS candidate DaEun Choi, and Ph.D. candidate Yoonseo Choi from the School of Computing received an honorable mention award for their paper, “Stylette: styling the Web with Natural Language”.
The research team developed a novel interface technology that allows nonexperts who are unfamiliar with technical jargon to edit website features through speech. People often find it difficult to use or find the information they need from various websites due to accessibility issues, device-related constraints, inconvenient design, style preferences, etc. However, it is not easy for laymen to edit website features without expertise in programming or design, and most end up just putting up with the inconveniences. But what if the system could read the intentions of its users from their everyday language like “emphasize this part a little more”, or “I want a more modern design”, and edit the features automatically?
Based on this question, Professor Kim’s research team developed ‘Stylette’, a system in which AI analyses its users’ speech expressed in their natural language and automatically recommends a new style that best fits their intentions. The research team created a new system by putting together language AI, visual AI, and user interface technologies. On the linguistic side, a large-scale language model AI converts the intentions of the users expressed through their everyday language into adequate style elements. On the visual side, computer vision AI compares 1.7 million existing web design features and recommends a style adequate for the current website. In an experiment where 40 nonexperts were asked to edit a website design, the subjects that used this system showed double the success rate in a time span that was 35% shorter compared to the control group.
It is meaningful that this research proposed a practical case in which AI technology constructs intuitive interactions with users. The developed technology can be applied to existing design applications and web browsers in a plug-in format, and can be utilized to improve websites or for advertisements by collecting the natural intention data of users on a large scale.
2022.06.13 View 7897 -
 Now You Can See Floral Scents!
Optical interferometry visualizes how often lilies emit volatile organic compounds
Have you ever thought about when flowers emit their scents?
KAIST mechanical engineers and biological scientists directly visualized how often a lily releases a floral scent using a laser interferometry method. These measurement results can provide new insights for understanding and further exploring the biosynthesis and emission mechanisms of floral volatiles.
Why is it important to know this? It is well known that the fragrance of flowers affects their interactions with pollinators, microorganisms, and florivores. For instance, many flowering plants can tune their scent emission rates when pollinators are active for pollination. Petunias and the wild tobacco Nicotiana attenuata emit floral scents at night to attract night-active pollinators. Thus, visualizing scent emissions can help us understand the ecological evolution of plant-pollinator interactions.
Many groups have been trying to develop methods for scent analysis. Mass spectrometry has been one widely used method for investigating the fragrance of flowers. Although mass spectrometry reveals the quality and quantity of floral scents, it is impossible to directly measure the releasing frequency. A laser-based gas detection system and a smartphone-based detection system using chemo-responsive dyes have also been used to measure volatile organic compounds (VOCs) in real-time, but it is still hard to measure the time-dependent emission rate of floral scents.
However, the KAIST research team co-led by Professor Hyoungsoo Kim from the Department of Mechanical Engineering and Professor Sang-Gyu Kim from the Department of Biological Sciences measured a refractive index difference between the vapor of the VOCs of lilies and the air to measure the emission frequency. The floral scent vapor was detected and the refractive index of air was 1.0 while that of the major floral scent of a linalool lily was 1.46.
Professor Hyoungsoo Kim said, “We expect this technology to be further applicable to various industrial sectors such as developing it to detect hazardous substances in a space.” The research team also plans to identify the DNA mechanism that controls floral scent secretion.
The current work entitled “Real-time visualization of scent accumulation reveals the frequency of floral scent emissions” was published in ‘Frontiers in Plant Science’ on April 18, 2022. (https://doi.org/10.3389/fpls.2022.835305).
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2021R1A2C2007835), the Rural Development Administration (PJ016403), and the KAIST-funded Global Singularity Research PREP-Program.
-Publication:H. Kim, G. Lee, J. Song, and S.-G. Kim, "Real-time visualization of scent accumulation reveals the frequency of floral scent emissions," Frontiers in Plant Science 18, 835305 (2022) (https://doi.org/10.3389/fpls.2022.835305)
-Profile:Professor Hyoungsoo Kimhttp://fil.kaist.ac.kr
@MadeInH on TwitterDepartment of Mechanical EngineeringKAIST
Professor Sang-Gyu Kimhttps://sites.google.com/view/kimlab/home Department of Biological SciencesKAIST
2022.05.25 View 8836
Now You Can See Floral Scents!
Optical interferometry visualizes how often lilies emit volatile organic compounds
Have you ever thought about when flowers emit their scents?
KAIST mechanical engineers and biological scientists directly visualized how often a lily releases a floral scent using a laser interferometry method. These measurement results can provide new insights for understanding and further exploring the biosynthesis and emission mechanisms of floral volatiles.
Why is it important to know this? It is well known that the fragrance of flowers affects their interactions with pollinators, microorganisms, and florivores. For instance, many flowering plants can tune their scent emission rates when pollinators are active for pollination. Petunias and the wild tobacco Nicotiana attenuata emit floral scents at night to attract night-active pollinators. Thus, visualizing scent emissions can help us understand the ecological evolution of plant-pollinator interactions.
Many groups have been trying to develop methods for scent analysis. Mass spectrometry has been one widely used method for investigating the fragrance of flowers. Although mass spectrometry reveals the quality and quantity of floral scents, it is impossible to directly measure the releasing frequency. A laser-based gas detection system and a smartphone-based detection system using chemo-responsive dyes have also been used to measure volatile organic compounds (VOCs) in real-time, but it is still hard to measure the time-dependent emission rate of floral scents.
However, the KAIST research team co-led by Professor Hyoungsoo Kim from the Department of Mechanical Engineering and Professor Sang-Gyu Kim from the Department of Biological Sciences measured a refractive index difference between the vapor of the VOCs of lilies and the air to measure the emission frequency. The floral scent vapor was detected and the refractive index of air was 1.0 while that of the major floral scent of a linalool lily was 1.46.
Professor Hyoungsoo Kim said, “We expect this technology to be further applicable to various industrial sectors such as developing it to detect hazardous substances in a space.” The research team also plans to identify the DNA mechanism that controls floral scent secretion.
The current work entitled “Real-time visualization of scent accumulation reveals the frequency of floral scent emissions” was published in ‘Frontiers in Plant Science’ on April 18, 2022. (https://doi.org/10.3389/fpls.2022.835305).
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2021R1A2C2007835), the Rural Development Administration (PJ016403), and the KAIST-funded Global Singularity Research PREP-Program.
-Publication:H. Kim, G. Lee, J. Song, and S.-G. Kim, "Real-time visualization of scent accumulation reveals the frequency of floral scent emissions," Frontiers in Plant Science 18, 835305 (2022) (https://doi.org/10.3389/fpls.2022.835305)
-Profile:Professor Hyoungsoo Kimhttp://fil.kaist.ac.kr
@MadeInH on TwitterDepartment of Mechanical EngineeringKAIST
Professor Sang-Gyu Kimhttps://sites.google.com/view/kimlab/home Department of Biological SciencesKAIST
2022.05.25 View 8836 -
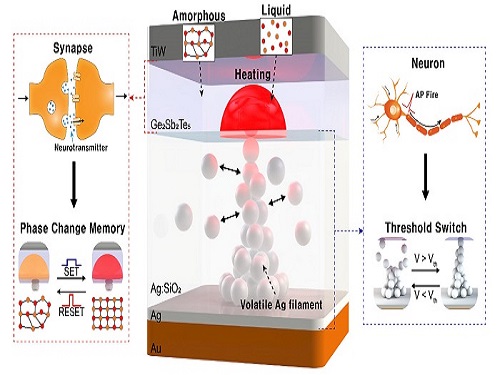 Neuromorphic Memory Device Simulates Neurons and Synapses
Simultaneous emulation of neuronal and synaptic properties promotes the development of brain-like artificial intelligence
Researchers have reported a nano-sized neuromorphic memory device that emulates neurons and synapses simultaneously in a unit cell, another step toward completing the goal of neuromorphic computing designed to rigorously mimic the human brain with semiconductor devices.
Neuromorphic computing aims to realize artificial intelligence (AI) by mimicking the mechanisms of neurons and synapses that make up the human brain. Inspired by the cognitive functions of the human brain that current computers cannot provide, neuromorphic devices have been widely investigated. However, current Complementary Metal-Oxide Semiconductor (CMOS)-based neuromorphic circuits simply connect artificial neurons and synapses without synergistic interactions, and the concomitant implementation of neurons and synapses still remains a challenge. To address these issues, a research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering implemented the biological working mechanisms of humans by introducing the neuron-synapse interactions in a single memory cell, rather than the conventional approach of electrically connecting artificial neuronal and synaptic devices.
Similar to commercial graphics cards, the artificial synaptic devices previously studied often used to accelerate parallel computations, which shows clear differences from the operational mechanisms of the human brain. The research team implemented the synergistic interactions between neurons and synapses in the neuromorphic memory device, emulating the mechanisms of the biological neural network. In addition, the developed neuromorphic device can replace complex CMOS neuron circuits with a single device, providing high scalability and cost efficiency.
The human brain consists of a complex network of 100 billion neurons and 100 trillion synapses. The functions and structures of neurons and synapses can flexibly change according to the external stimuli, adapting to the surrounding environment. The research team developed a neuromorphic device in which short-term and long-term memories coexist using volatile and non-volatile memory devices that mimic the characteristics of neurons and synapses, respectively. A threshold switch device is used as volatile memory and phase-change memory is used as a non-volatile device. Two thin-film devices are integrated without intermediate electrodes, implementing the functional adaptability of neurons and synapses in the neuromorphic memory.
Professor Keon Jae Lee explained, "Neurons and synapses interact with each other to establish cognitive functions such as memory and learning, so simulating both is an essential element for brain-inspired artificial intelligence. The developed neuromorphic memory device also mimics the retraining effect that allows quick learning of the forgotten information by implementing a positive feedback effect between neurons and synapses.”
This result entitled “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse” was published in the May 19, 2022 issue of Nature Communications.
-Publication:Sang Hyun Sung, Tae Jin Kim, Hyera Shin, Tae Hong Im, and Keon Jae Lee (2022) “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse,” Nature Communications May 19, 2022 (DOI: 10.1038/s41467-022-30432-2)
-Profile:Professor Keon Jae Leehttp://fand.kaist.ac.kr
Department of Materials Science and EngineeringKAIST
2022.05.20 View 12559
Neuromorphic Memory Device Simulates Neurons and Synapses
Simultaneous emulation of neuronal and synaptic properties promotes the development of brain-like artificial intelligence
Researchers have reported a nano-sized neuromorphic memory device that emulates neurons and synapses simultaneously in a unit cell, another step toward completing the goal of neuromorphic computing designed to rigorously mimic the human brain with semiconductor devices.
Neuromorphic computing aims to realize artificial intelligence (AI) by mimicking the mechanisms of neurons and synapses that make up the human brain. Inspired by the cognitive functions of the human brain that current computers cannot provide, neuromorphic devices have been widely investigated. However, current Complementary Metal-Oxide Semiconductor (CMOS)-based neuromorphic circuits simply connect artificial neurons and synapses without synergistic interactions, and the concomitant implementation of neurons and synapses still remains a challenge. To address these issues, a research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering implemented the biological working mechanisms of humans by introducing the neuron-synapse interactions in a single memory cell, rather than the conventional approach of electrically connecting artificial neuronal and synaptic devices.
Similar to commercial graphics cards, the artificial synaptic devices previously studied often used to accelerate parallel computations, which shows clear differences from the operational mechanisms of the human brain. The research team implemented the synergistic interactions between neurons and synapses in the neuromorphic memory device, emulating the mechanisms of the biological neural network. In addition, the developed neuromorphic device can replace complex CMOS neuron circuits with a single device, providing high scalability and cost efficiency.
The human brain consists of a complex network of 100 billion neurons and 100 trillion synapses. The functions and structures of neurons and synapses can flexibly change according to the external stimuli, adapting to the surrounding environment. The research team developed a neuromorphic device in which short-term and long-term memories coexist using volatile and non-volatile memory devices that mimic the characteristics of neurons and synapses, respectively. A threshold switch device is used as volatile memory and phase-change memory is used as a non-volatile device. Two thin-film devices are integrated without intermediate electrodes, implementing the functional adaptability of neurons and synapses in the neuromorphic memory.
Professor Keon Jae Lee explained, "Neurons and synapses interact with each other to establish cognitive functions such as memory and learning, so simulating both is an essential element for brain-inspired artificial intelligence. The developed neuromorphic memory device also mimics the retraining effect that allows quick learning of the forgotten information by implementing a positive feedback effect between neurons and synapses.”
This result entitled “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse” was published in the May 19, 2022 issue of Nature Communications.
-Publication:Sang Hyun Sung, Tae Jin Kim, Hyera Shin, Tae Hong Im, and Keon Jae Lee (2022) “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse,” Nature Communications May 19, 2022 (DOI: 10.1038/s41467-022-30432-2)
-Profile:Professor Keon Jae Leehttp://fand.kaist.ac.kr
Department of Materials Science and EngineeringKAIST
2022.05.20 View 12559 -
 Anonymous Donor Makes a Gift of Property Valued at 30 Billion KRW
The KAIST Development Foundation announced on May 9 that an anonymous donor in his 50s made a gift of real estate valued at 30 billion KRW. This is the first donation from an anonymous benefactor on such a grand scale. The benefactor expressed his wishes to fund scholarships for students in need and R&D for medical and bio sciences.
According to the Development Foundation official, the benefactor is reported to have said that he felt burdened that he earned much more than he needed and was looking for the right way to share his assets. The benefactor refused to hold an official donation ceremony and meeting with high-level university administrators.
The donor believes that KAIST is filled with young and dynamic energy, saying, “I would like to help KAIST move forward and create breakthroughs that will benefit the nation as well as all humanity.”
Before making up his mind to give his asset to KAIST, he had planned to establish his own social foundation but he changed his mind. “I decided that an investment in education would be the best investment,” he said. He explained that he was inspired by his KAIST graduate friend who is running a company. He was deeply motivated to help KAIST after witnessing the KAIST graduate’s passion for conducting his business.
After receiving the gift, KAIST President Kwang Hyung Lee was thankful for the full support and trust of the benefactor. “We will spare no effort to foster next-generation talents and advance science and technology in the field of biomedicine.”
2022.05.11 View 5257
Anonymous Donor Makes a Gift of Property Valued at 30 Billion KRW
The KAIST Development Foundation announced on May 9 that an anonymous donor in his 50s made a gift of real estate valued at 30 billion KRW. This is the first donation from an anonymous benefactor on such a grand scale. The benefactor expressed his wishes to fund scholarships for students in need and R&D for medical and bio sciences.
According to the Development Foundation official, the benefactor is reported to have said that he felt burdened that he earned much more than he needed and was looking for the right way to share his assets. The benefactor refused to hold an official donation ceremony and meeting with high-level university administrators.
The donor believes that KAIST is filled with young and dynamic energy, saying, “I would like to help KAIST move forward and create breakthroughs that will benefit the nation as well as all humanity.”
Before making up his mind to give his asset to KAIST, he had planned to establish his own social foundation but he changed his mind. “I decided that an investment in education would be the best investment,” he said. He explained that he was inspired by his KAIST graduate friend who is running a company. He was deeply motivated to help KAIST after witnessing the KAIST graduate’s passion for conducting his business.
After receiving the gift, KAIST President Kwang Hyung Lee was thankful for the full support and trust of the benefactor. “We will spare no effort to foster next-generation talents and advance science and technology in the field of biomedicine.”
2022.05.11 View 5257 -
 Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 9338
Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 9338 -
 'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media
Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
Their results were made available online on Jan. 18 in Biosensors and Bioelectronics, ahead of publication in the journal’s April issue.
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. “By using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model, we demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures,” said Professor Sungho Jo from the School of Computing.
Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample — the spectral fingerprint — allowing researchers to identify its molecules. The surface-enhanced version places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. “Moreover, strong signals of surrounding media are also enhanced to overwhelm target signals, requiring time-consuming and tedious bacterial separation steps,” said Professor Yeon Sik Jung from the Department of Materials Science and Engineering.
To parse through the noisy signals, the researchers implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such an ability is critical for analyzing one-dimensional spectral data, according to Professor Jo.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that were almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” Professor Jo said. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
The National R&D Program, through a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, supported this research.
-PublicationEojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deepneural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
-ProfileProfessor Yeon Sik JungDepartment of Materials Science and EngineeringKAIST
Professor Sungho JoSchool of ComputingKAIST
2022.03.04 View 21244
'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media
Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
Their results were made available online on Jan. 18 in Biosensors and Bioelectronics, ahead of publication in the journal’s April issue.
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. “By using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model, we demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures,” said Professor Sungho Jo from the School of Computing.
Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample — the spectral fingerprint — allowing researchers to identify its molecules. The surface-enhanced version places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. “Moreover, strong signals of surrounding media are also enhanced to overwhelm target signals, requiring time-consuming and tedious bacterial separation steps,” said Professor Yeon Sik Jung from the Department of Materials Science and Engineering.
To parse through the noisy signals, the researchers implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such an ability is critical for analyzing one-dimensional spectral data, according to Professor Jo.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that were almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” Professor Jo said. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
The National R&D Program, through a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, supported this research.
-PublicationEojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deepneural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
-ProfileProfessor Yeon Sik JungDepartment of Materials Science and EngineeringKAIST
Professor Sungho JoSchool of ComputingKAIST
2022.03.04 View 21244 -
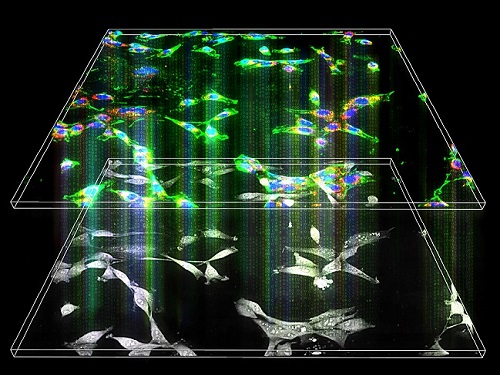 Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 10170
Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 10170 -
 Professor Sung-Ju Lee’s Team Wins the Best Paper and the Methods Recognition Awards at the ACM CSCW
A research team led by Professor Sung-Ju Lee at the School of Electrical Engineering won the Best Paper Award and the Methods Recognition Award from ACM CSCW (International Conference on Computer-Supported Cooperative Work and Social Computing) 2021 for their paper “Reflect, not Regret: Understanding Regretful Smartphone Use with App Feature-Level Analysis”.
Founded in 1986, CSCW has been a premier conference on HCI (Human Computer Interaction) and Social Computing. This year, 340 full papers were presented and the best paper awards are given to the top 1% papers of the submitted. Methods Recognition, which is a new award, is given “for strong examples of work that includes well developed, explained, or implemented methods, and methodological innovation.”
Hyunsung Cho (KAIST alumus and currently a PhD candidate at Carnegie Mellon University), Daeun Choi (KAIST undergraduate researcher), Donghwi Kim (KAIST PhD Candidate), Wan Ju Kang (KAIST PhD Candidate), and Professor Eun Kyoung Choe (University of Maryland and KAIST alumna) collaborated on this research.
The authors developed a tool that tracks and analyzes which features of a mobile app (e.g., Instagram’s following post, following story, recommended post, post upload, direct messaging, etc.) are in use based on a smartphone’s User Interface (UI) layout. Utilizing this novel method, the authors revealed which feature usage patterns result in regretful smartphone use.
Professor Lee said, “Although many people enjoy the benefits of smartphones, issues have emerged from the overuse of smartphones. With this feature level analysis, users can reflect on their smartphone usage based on finer grained analysis and this could contribute to digital wellbeing.”
2021.11.22 View 7138
Professor Sung-Ju Lee’s Team Wins the Best Paper and the Methods Recognition Awards at the ACM CSCW
A research team led by Professor Sung-Ju Lee at the School of Electrical Engineering won the Best Paper Award and the Methods Recognition Award from ACM CSCW (International Conference on Computer-Supported Cooperative Work and Social Computing) 2021 for their paper “Reflect, not Regret: Understanding Regretful Smartphone Use with App Feature-Level Analysis”.
Founded in 1986, CSCW has been a premier conference on HCI (Human Computer Interaction) and Social Computing. This year, 340 full papers were presented and the best paper awards are given to the top 1% papers of the submitted. Methods Recognition, which is a new award, is given “for strong examples of work that includes well developed, explained, or implemented methods, and methodological innovation.”
Hyunsung Cho (KAIST alumus and currently a PhD candidate at Carnegie Mellon University), Daeun Choi (KAIST undergraduate researcher), Donghwi Kim (KAIST PhD Candidate), Wan Ju Kang (KAIST PhD Candidate), and Professor Eun Kyoung Choe (University of Maryland and KAIST alumna) collaborated on this research.
The authors developed a tool that tracks and analyzes which features of a mobile app (e.g., Instagram’s following post, following story, recommended post, post upload, direct messaging, etc.) are in use based on a smartphone’s User Interface (UI) layout. Utilizing this novel method, the authors revealed which feature usage patterns result in regretful smartphone use.
Professor Lee said, “Although many people enjoy the benefits of smartphones, issues have emerged from the overuse of smartphones. With this feature level analysis, users can reflect on their smartphone usage based on finer grained analysis and this could contribute to digital wellbeing.”
2021.11.22 View 7138 -
 Study Finds Player-Character Relationships Affected Game Satisfaction in the Last of Us Part II
Research analyzed player experiences with a polarizing game and found differences in how the players related to their characters
The action adventure game ‘The Last of Us’ was a big hit worldwide in 2014. However, its sequel, the Last of Us Part II divided opinions in the game community when it was released in 2020.
A research team from the Games and Life Lab in the Graduate School of Culture Technology at KAIST analyzed why the game players’ reviews were so polarized and found that player-character relationships influenced the game players’ satisfaction. This study, published in Frontiers in Psychology, will help developers of character-driven games foresee how different players will react to their games.
The team under Professor Young Yim Doh conducted in-depth interviews with 12 players from diverse nations, both those satisfied and dissatisfied with the game.
The team found that three elements affected the game players’ satisfaction. First, players’ satisfaction varied according to their tolerance of forced character switches. When a player is forced to switch their controlled figure in the game to another character that is introduced as the antagonist, most players initially had a negative reaction. The feeling of being forced to play in a way they didn’t want reduced their rights as a player. However, later on, some players viewed this character switch as an interesting transition and were more tolerant toward forced game play.
Second, the researchers found that the flexibility of character attachment is related to game satisfaction. Players who were unhappy about the game resisted building a relationship with the new antagonist character. Meanwhile, players who were happy about the game slowly formed an additional relationship with the new character. This led to the player feeling conflicting emotions, which satisfied players considered a meaningful experience of understanding a perspective of someone initially considered the enemy.
Lastly, the satisfaction of the play depended on how much the players could accept a changing character image in the game. Dissatisfied players found inconsistencies in the characters’ behavior and did not accept the new information about the characters. Meanwhile, satisfied players tried to understand and accept the new information and actions.
“Previous research on narrative games focused more on the game design than on the players’ experiences. To understand why reactions to the game were very different across players, we focused our research on differences in the players’ psychological experiences with the game.” said lead author and Master’s candidate Valérie Erb.
Co-author Dr. Seyeon Lee added, “This suggests that there is no one way to satisfy all players in a character-based narrative game. To satisfy a game’s players, it is important to understand the different players in the player base, target the right player group, and manage expectations accordingly.”
This research was supported by the Year 2020 Culture Technology R&D Program by the Ministry of Culture, Sports and Tourism and the Korea Creative Content Agency.
-PublicationErb V, Lee S, and Doh YY (2021) “Player-Character Relationship and Game Satisfaction in Narrative Game: Focus on Player Experience of Character Switch in The Last of Us Part II”
Frontiers in Psychology. 12:709926. (https://doi.org/10.3389/fpsyg.2021.709926)
-ProfileProfessor Young Yim DohGames and Life LabGraduate School of Culture TechnologyKAIST
2021.11.15 View 5789
Study Finds Player-Character Relationships Affected Game Satisfaction in the Last of Us Part II
Research analyzed player experiences with a polarizing game and found differences in how the players related to their characters
The action adventure game ‘The Last of Us’ was a big hit worldwide in 2014. However, its sequel, the Last of Us Part II divided opinions in the game community when it was released in 2020.
A research team from the Games and Life Lab in the Graduate School of Culture Technology at KAIST analyzed why the game players’ reviews were so polarized and found that player-character relationships influenced the game players’ satisfaction. This study, published in Frontiers in Psychology, will help developers of character-driven games foresee how different players will react to their games.
The team under Professor Young Yim Doh conducted in-depth interviews with 12 players from diverse nations, both those satisfied and dissatisfied with the game.
The team found that three elements affected the game players’ satisfaction. First, players’ satisfaction varied according to their tolerance of forced character switches. When a player is forced to switch their controlled figure in the game to another character that is introduced as the antagonist, most players initially had a negative reaction. The feeling of being forced to play in a way they didn’t want reduced their rights as a player. However, later on, some players viewed this character switch as an interesting transition and were more tolerant toward forced game play.
Second, the researchers found that the flexibility of character attachment is related to game satisfaction. Players who were unhappy about the game resisted building a relationship with the new antagonist character. Meanwhile, players who were happy about the game slowly formed an additional relationship with the new character. This led to the player feeling conflicting emotions, which satisfied players considered a meaningful experience of understanding a perspective of someone initially considered the enemy.
Lastly, the satisfaction of the play depended on how much the players could accept a changing character image in the game. Dissatisfied players found inconsistencies in the characters’ behavior and did not accept the new information about the characters. Meanwhile, satisfied players tried to understand and accept the new information and actions.
“Previous research on narrative games focused more on the game design than on the players’ experiences. To understand why reactions to the game were very different across players, we focused our research on differences in the players’ psychological experiences with the game.” said lead author and Master’s candidate Valérie Erb.
Co-author Dr. Seyeon Lee added, “This suggests that there is no one way to satisfy all players in a character-based narrative game. To satisfy a game’s players, it is important to understand the different players in the player base, target the right player group, and manage expectations accordingly.”
This research was supported by the Year 2020 Culture Technology R&D Program by the Ministry of Culture, Sports and Tourism and the Korea Creative Content Agency.
-PublicationErb V, Lee S, and Doh YY (2021) “Player-Character Relationship and Game Satisfaction in Narrative Game: Focus on Player Experience of Character Switch in The Last of Us Part II”
Frontiers in Psychology. 12:709926. (https://doi.org/10.3389/fpsyg.2021.709926)
-ProfileProfessor Young Yim DohGames and Life LabGraduate School of Culture TechnologyKAIST
2021.11.15 View 5789