Department+of+Bio
-
 Finding Human Thermal Comfort with a Watch-type Sweat Rate Sensor
(from left: Professor Young-Ho Cho and Researcher SungHyun Yoon)
KAIST developed a watch-type sweat rate sensor. This subminiature device can detect human thermal comfort accurately and steadily by measuring an individual’s sweat rate.
It is natural to sweat more in the summer and less in the winter; however, an individual’s sweat rate may vary in a given environment. Therefore, sweat can be an excellent proxy for sensing core body temperature.
Conventional sweat rate sensors using natural ventilation require bulky external devices, such as pumps and ice condensers. They are usually for physiological experiments, hence they need a manual ventilation process or high power, bulky thermos-pneumatic actuators to lift sweat rate detection chambers above skin for continuous measurement. There is also a small sweat rate sensor, but it needs a long recovery period.
To overcome these problems, Professor Young-Ho Cho and his team from the Department of Bio and Brain Engineering developed a lightweight, watch-type sweat sensor. The team integrated miniaturized thermos-pneumatic actuators for automatic natural ventilation, which allows sweat to be measured continuously.
This watch-type sensor measures sweat rate with the humidity rising rate when the chamber is closed during skin contact. Since the team integrated thermos-pneumatic actuators, the chamber no longer needs to be separated manually from skin after each measurement in order for the chamber to ventilate the collected humidity.
Moreover, this sensor is wind-resistant enough to be used for portable and wearable devices. The team identified that the sensor operates steadily with air velocity ranging up to 1.5m/s, equivalent to the average human walking speed.
Although this subminiature sensor (35mm x 25mm) only weighs 30 grams, it operates continuously for more than four hours using the conventional wrist watch batteries.
The team plans to utilize this technology for developing a new concept of cognitive air-conditioning systems recognizing Human thermal status directly; while the conventional air-conditioning systems measuring air temperature and humidity.
Professor Cho said, “Our sensor for human thermal comfort monitoring can be applied to customized or smart air conditioners. Furthermore, there will be more demands for both physical and mental healthcare, hence this technology will serve as a new platform for personalized emotional communion between humans and devices.”
This research, led by researchers Jai Kyoung Sim and SungHyun Yoon, was published in Scientific Reports on January 19, 2018.
Figure1. The fabricated watch-type sweat rate sensor for human thermal comfort monitoring
Figure 2. Views of the watch-type sweat rate sensor
Figure 3. Operation of the watch-type sweat rate sensor
2018.02.08 View 9324
Finding Human Thermal Comfort with a Watch-type Sweat Rate Sensor
(from left: Professor Young-Ho Cho and Researcher SungHyun Yoon)
KAIST developed a watch-type sweat rate sensor. This subminiature device can detect human thermal comfort accurately and steadily by measuring an individual’s sweat rate.
It is natural to sweat more in the summer and less in the winter; however, an individual’s sweat rate may vary in a given environment. Therefore, sweat can be an excellent proxy for sensing core body temperature.
Conventional sweat rate sensors using natural ventilation require bulky external devices, such as pumps and ice condensers. They are usually for physiological experiments, hence they need a manual ventilation process or high power, bulky thermos-pneumatic actuators to lift sweat rate detection chambers above skin for continuous measurement. There is also a small sweat rate sensor, but it needs a long recovery period.
To overcome these problems, Professor Young-Ho Cho and his team from the Department of Bio and Brain Engineering developed a lightweight, watch-type sweat sensor. The team integrated miniaturized thermos-pneumatic actuators for automatic natural ventilation, which allows sweat to be measured continuously.
This watch-type sensor measures sweat rate with the humidity rising rate when the chamber is closed during skin contact. Since the team integrated thermos-pneumatic actuators, the chamber no longer needs to be separated manually from skin after each measurement in order for the chamber to ventilate the collected humidity.
Moreover, this sensor is wind-resistant enough to be used for portable and wearable devices. The team identified that the sensor operates steadily with air velocity ranging up to 1.5m/s, equivalent to the average human walking speed.
Although this subminiature sensor (35mm x 25mm) only weighs 30 grams, it operates continuously for more than four hours using the conventional wrist watch batteries.
The team plans to utilize this technology for developing a new concept of cognitive air-conditioning systems recognizing Human thermal status directly; while the conventional air-conditioning systems measuring air temperature and humidity.
Professor Cho said, “Our sensor for human thermal comfort monitoring can be applied to customized or smart air conditioners. Furthermore, there will be more demands for both physical and mental healthcare, hence this technology will serve as a new platform for personalized emotional communion between humans and devices.”
This research, led by researchers Jai Kyoung Sim and SungHyun Yoon, was published in Scientific Reports on January 19, 2018.
Figure1. The fabricated watch-type sweat rate sensor for human thermal comfort monitoring
Figure 2. Views of the watch-type sweat rate sensor
Figure 3. Operation of the watch-type sweat rate sensor
2018.02.08 View 9324 -
 Technology to Find Optimum Drug Target for Cancer Developed
(Professor Kwang-Hyun Cho (right) and lead author Dr. Minsoo Choi)
A KAIST research team led by Professor Kwang-Hyun Cho of the Department of Bio and Brain Engineering developed technology to find the optimum drug target according to the type of cancer cell. The team used systems biology to analyze molecular network dynamics that reflect genetic mutations in cancer cells and to predict drug response. The technology could contribute greatly to future anti-cancer drug development.
There are many types of genetic variations found in cancer cells, including gene mutations and copy number variations. These variations differ in cancer cells even within the same type of cancer, and thus the drug response varies cell by cell. Cancer researchers worked towards identifying frequently occurring genetic variations in cancer patients and, in particular, the mutations that can be used as an index for specific drugs. Previous studies focused on identifying a single genetic mutation or creating an analysis of the structural characteristics of a gene network. However, this approach was limited in its inability to explain the biological properties of cancer which are induced by various gene and protein interactions in cancer cells, which result in differences in drug response.
Gene mutations in cancer cells not only affect the function of the affected gene, but also other genes that interact with the mutated gene and proteins. As a consequence, one mutation could lead to changes in the dynamical properties of the molecular network. Therefore, the responses to anti-cancer drugs by cancer cells differ. The current treatment approach that ignores molecular network dynamics and targets a few cancer-related genes is only effective on a fraction of patients, while many other patients exhibit resistance to the drug.
Professor Cho’s team integrated a large-scale computer simulation using super-computing and cellular experiments to analyze changes in molecular network dynamics in cancer cells.
This led to development of technology to find the optimum drug target according to the type of cancer cells by predicting drug response. This technology was applied to the molecular network of known tumor suppressor p53. The team used large-scale cancer cell genomic data available from The Cancer Cell Line Encyclopedia (CCLE) to construct different molecular networks specific to the characteristics of genetic variations.
Perturbation analysis on drug response in each molecular network was used to quantify changes in cancer cells from drug response and similar networks were clustered. Then, computer simulations were used to analyze the synergetic effects in terms of efficacy and combination to predict the level of drug response. Based on the simulation results from various cancer cell lines including lung, breast, bone, skin, kidney, and ovary cancers were used in drug response experiments for compare analysis.
This technique can be applied in any molecular network to identify the optimum drug target for personalized medicine.
The research team suggests that the technology can analyze varying drug response due to the heterogeneity of cancer cells by considering the overall modulatory interactions rather than focusing only on a specific gene or protein. Further, the technology aids the prediction of causes of drug resistance and thus the identification of the optimum drug target to inhibit the resistance. This could be core source technology that can be used in drug repositioning, a process of applying existing drugs to new disease targets.
Professor Cho said, “Genetic variations in cancer cells are the cause of diverse drug response, but a complete analysis had not yet been made.” He continued, “Systems biology allowed the simulation of drug responses by cancer cell molecular networks to identify fundamental principles of drug response and optimum drug targets using a new conceptual approach.”
This research was published in Nature Communications on December 5 and was funded by Ministry of Science and ICT and National Research Foundation of Korea.
(Figure 1. Drug response prediction for each cancer cell type from computer simulation and cellular experiment verification for comparison)
(Figure 2. Drug response prediction based on cancer cell molecular network dynamics and clustering of cancer cells by their molecular networks)
(Figure 3. Identification of drug target for each cancer cell type by cellular molecular network analysis and establishment for personalized medicine strategy for each cancer patient)
2017.12.15 View 7592
Technology to Find Optimum Drug Target for Cancer Developed
(Professor Kwang-Hyun Cho (right) and lead author Dr. Minsoo Choi)
A KAIST research team led by Professor Kwang-Hyun Cho of the Department of Bio and Brain Engineering developed technology to find the optimum drug target according to the type of cancer cell. The team used systems biology to analyze molecular network dynamics that reflect genetic mutations in cancer cells and to predict drug response. The technology could contribute greatly to future anti-cancer drug development.
There are many types of genetic variations found in cancer cells, including gene mutations and copy number variations. These variations differ in cancer cells even within the same type of cancer, and thus the drug response varies cell by cell. Cancer researchers worked towards identifying frequently occurring genetic variations in cancer patients and, in particular, the mutations that can be used as an index for specific drugs. Previous studies focused on identifying a single genetic mutation or creating an analysis of the structural characteristics of a gene network. However, this approach was limited in its inability to explain the biological properties of cancer which are induced by various gene and protein interactions in cancer cells, which result in differences in drug response.
Gene mutations in cancer cells not only affect the function of the affected gene, but also other genes that interact with the mutated gene and proteins. As a consequence, one mutation could lead to changes in the dynamical properties of the molecular network. Therefore, the responses to anti-cancer drugs by cancer cells differ. The current treatment approach that ignores molecular network dynamics and targets a few cancer-related genes is only effective on a fraction of patients, while many other patients exhibit resistance to the drug.
Professor Cho’s team integrated a large-scale computer simulation using super-computing and cellular experiments to analyze changes in molecular network dynamics in cancer cells.
This led to development of technology to find the optimum drug target according to the type of cancer cells by predicting drug response. This technology was applied to the molecular network of known tumor suppressor p53. The team used large-scale cancer cell genomic data available from The Cancer Cell Line Encyclopedia (CCLE) to construct different molecular networks specific to the characteristics of genetic variations.
Perturbation analysis on drug response in each molecular network was used to quantify changes in cancer cells from drug response and similar networks were clustered. Then, computer simulations were used to analyze the synergetic effects in terms of efficacy and combination to predict the level of drug response. Based on the simulation results from various cancer cell lines including lung, breast, bone, skin, kidney, and ovary cancers were used in drug response experiments for compare analysis.
This technique can be applied in any molecular network to identify the optimum drug target for personalized medicine.
The research team suggests that the technology can analyze varying drug response due to the heterogeneity of cancer cells by considering the overall modulatory interactions rather than focusing only on a specific gene or protein. Further, the technology aids the prediction of causes of drug resistance and thus the identification of the optimum drug target to inhibit the resistance. This could be core source technology that can be used in drug repositioning, a process of applying existing drugs to new disease targets.
Professor Cho said, “Genetic variations in cancer cells are the cause of diverse drug response, but a complete analysis had not yet been made.” He continued, “Systems biology allowed the simulation of drug responses by cancer cell molecular networks to identify fundamental principles of drug response and optimum drug targets using a new conceptual approach.”
This research was published in Nature Communications on December 5 and was funded by Ministry of Science and ICT and National Research Foundation of Korea.
(Figure 1. Drug response prediction for each cancer cell type from computer simulation and cellular experiment verification for comparison)
(Figure 2. Drug response prediction based on cancer cell molecular network dynamics and clustering of cancer cells by their molecular networks)
(Figure 3. Identification of drug target for each cancer cell type by cellular molecular network analysis and establishment for personalized medicine strategy for each cancer patient)
2017.12.15 View 7592 -
 Mutant Gene Network in Colon Cancer Identified
The principles of the gene network for colon tumorigenesis have been identified by a KAIST research team. The principles will be used to find the molecular target for effective anti-cancer drugs in the future. Further, this research gained attention for using a systems biology approach, which is an integrated research area of IT and BT.
The KAIST research team led by Professor Kwang-Hyun Cho for the Department of Bio and Brain Engineering succeeded in the identification. Conducted by Dr. Dongkwan Shin and student researchers Jonghoon Lee and Jeong-Ryeol Gong, the research was published in Nature Communications online on November 2.
Human cancer is caused by genetic mutations. The frequency of the mutations differs by the type of cancer; for example, only around 10 mutations are found in leukemia and childhood cancer, but an average of 50 mutations are found in adult solid cancers and even hundreds of mutations are found in cancers due to external factors, such as with lung cancer.
Cancer researchers around the world are working to identify frequently found genetic mutations in patients, and in turn identify important cancer-inducing genes (called ‘driver genes’) to develop targets for anti-cancer drugs. However, gene mutations not only affect their own functions but also affect other genes through interactions. Therefore, there are limitations in current treatments targeting a few cancer-inducing genes without further knowledge on gene networks, hence current drugs are only effective in a few patients and often induce drug resistance.
Professor Cho’s team used large-scale genomic data from cancer patients to construct a mathematical model on the cooperative effects of multiple genetic mutations found in gene interaction networks. The basis of the model construction was The Cancer Genome Atlas (TCGA) presented at the International Cancer Genome Consortium. The team successfully quantified the effects of mutations in gene networks to group colon cancer patients by clinical characteristics.
Further, the critical transition phenomenon that occurs in tumorigenesis was identified using large-scale computer simulation analysis, which was the first hidden gene network principle to be identified. Critical transition is the phenomenon in which the state of matter is suddenly changed through phase transition. It was not possible to identify the presence of transition phenomenon in the past, as it was difficult to track the sequence of gene mutations during tumorigenesis.
The research team used a systems biology-based research method to find that colon cancer tumorigenesis shows a critical transition phenomenon if the known driver gene mutations follow sequentially. Using the developed mathematical model, it can be possible to develop a new anti-cancer targeting drug that most effectively inhibits the effects of many gene mutations found in cancer patients. In particular, not only driver genes, but also other passenger genes affected by the gene mutations, could be evaluated to find the most effective drug targets.
Professor Cho said, “Little was known about the contribution of many gene mutations during tumorigenesis.” He continued, “In this research, a systems biology approach identified the principle of gene networks for the first time to suggest the possibility of anti-cancer drug target identification from a new perspective.”
This research was funded by the Ministry of Science and ICT and the National Research Foundation of Korea.
Figure1. Formation of giant clusters via mutation propagation
Figure2. Critical transition phenomenon by cooperative effect of mutations in tumorigenesis
2017.11.10 View 8668
Mutant Gene Network in Colon Cancer Identified
The principles of the gene network for colon tumorigenesis have been identified by a KAIST research team. The principles will be used to find the molecular target for effective anti-cancer drugs in the future. Further, this research gained attention for using a systems biology approach, which is an integrated research area of IT and BT.
The KAIST research team led by Professor Kwang-Hyun Cho for the Department of Bio and Brain Engineering succeeded in the identification. Conducted by Dr. Dongkwan Shin and student researchers Jonghoon Lee and Jeong-Ryeol Gong, the research was published in Nature Communications online on November 2.
Human cancer is caused by genetic mutations. The frequency of the mutations differs by the type of cancer; for example, only around 10 mutations are found in leukemia and childhood cancer, but an average of 50 mutations are found in adult solid cancers and even hundreds of mutations are found in cancers due to external factors, such as with lung cancer.
Cancer researchers around the world are working to identify frequently found genetic mutations in patients, and in turn identify important cancer-inducing genes (called ‘driver genes’) to develop targets for anti-cancer drugs. However, gene mutations not only affect their own functions but also affect other genes through interactions. Therefore, there are limitations in current treatments targeting a few cancer-inducing genes without further knowledge on gene networks, hence current drugs are only effective in a few patients and often induce drug resistance.
Professor Cho’s team used large-scale genomic data from cancer patients to construct a mathematical model on the cooperative effects of multiple genetic mutations found in gene interaction networks. The basis of the model construction was The Cancer Genome Atlas (TCGA) presented at the International Cancer Genome Consortium. The team successfully quantified the effects of mutations in gene networks to group colon cancer patients by clinical characteristics.
Further, the critical transition phenomenon that occurs in tumorigenesis was identified using large-scale computer simulation analysis, which was the first hidden gene network principle to be identified. Critical transition is the phenomenon in which the state of matter is suddenly changed through phase transition. It was not possible to identify the presence of transition phenomenon in the past, as it was difficult to track the sequence of gene mutations during tumorigenesis.
The research team used a systems biology-based research method to find that colon cancer tumorigenesis shows a critical transition phenomenon if the known driver gene mutations follow sequentially. Using the developed mathematical model, it can be possible to develop a new anti-cancer targeting drug that most effectively inhibits the effects of many gene mutations found in cancer patients. In particular, not only driver genes, but also other passenger genes affected by the gene mutations, could be evaluated to find the most effective drug targets.
Professor Cho said, “Little was known about the contribution of many gene mutations during tumorigenesis.” He continued, “In this research, a systems biology approach identified the principle of gene networks for the first time to suggest the possibility of anti-cancer drug target identification from a new perspective.”
This research was funded by the Ministry of Science and ICT and the National Research Foundation of Korea.
Figure1. Formation of giant clusters via mutation propagation
Figure2. Critical transition phenomenon by cooperative effect of mutations in tumorigenesis
2017.11.10 View 8668 -
 Photoacoustic Imaging and Photothermal Cancer Therapy Using BR Nanoparticles
(Professor Sangyong Jon and PhD Candidate Dong Yun Lee)
Sangyong Jon, a professor in the Department of Biological Sciences at KAIST, and his team developed combined photoacoustic imaging and photothermal therapy for cancer by using Bilirubin (BR) nanoparticles.
The research team applied the properties of a bile pigment called BR, which exerts potent antioxidant and anti-inflammatory effects, to this research.
The team expects this research, which shows high biocompatibility as well as outstanding photoacoustic imaging and photothermal therapy, to be an appropriate system in the field of treatment for cancer.
In the past, the research team developed a PEGylated bilirubin-based nanoparticle system by combining water-insoluble BR with water-soluble Polyethylene Glycol (PEG).
This technology facilitated BR exerting antioxidants yet prevented them from being accumulated in the body. Its efficiency and safety was identified in an animal disease model, for conditions such as inflammatory bowel disease, islet cell transportation, and asthma.
Differing from previous research methods, this research applied the different physicochemical properties of BR to cancer treatment.
When the causative agent of jaundice, yellow BR, is exposed to a certain wavelength of blue light, the agent becomes a photonic nanomaterial as it responses to the light. This light-responsive nanomaterial can be used to cure jaundice because it allows for active excretion in infants.
Secondly, the team identified that BR is a major component of black pigment gallstones which can be often found in gall bladders or bile ducts under certain pathological conditions. The findings show that BR forms black pigment gallstones without the role of an intermediate or cation, such as calcium and copper. The research team combined cisplatin, a platinum metal-based anticancer drug, with BR so that BR nanoparticles changed the solution color from yellow to purple.
The team also examined the possibility of cisplatin-chelated BR nanoparticles as a probe for photoacoustic images. They found that considerable photoacoustic activity was shown when it was exposed to near infrared light. In fact, the photoacoustic signal was increased significantly in tumors of animals with colorectal cancer when the nanoparticles were administered to it intravenously. The team expects a more accurate diagnosis of tumors through this technology.
Moreover, the team assessed the photothermal effects of cisplatin-chelated BR nanoparticles. The research showed that the temperature of tumors increased by 25 degrees Celsius within five minutes when they were exposed to near infrared light, due to the photothermal effect. After two weeks, their size was reduced compared to that of other groups, and sometimes the tumors were even necrotized.
Professor Jon said, “Existing substances have a low biocompatibility and limitation for clinical therapy because they are artificially oriented; therefore, they might have toxicity. I am hoping that these cisplatin-chelated BR-based nanoparticles will provide a new platform for preclinical, translational research and clinical adaptation of the photoacoustic imaging and photothermal therapy.”
The paper (Dong Yun Lee as a first author) was published online in the renowned journal in the field of applied chemistry, Angewandte Chemi International Edition, on September 4. This research was sponsored by the National Research Foundation of Korea.
(Schematic diagram of the research)
(From left: Bilirubin nanoparticles, cisplatin-chelated Bilirubin nanoparticles)
2017.09.26 View 9719
Photoacoustic Imaging and Photothermal Cancer Therapy Using BR Nanoparticles
(Professor Sangyong Jon and PhD Candidate Dong Yun Lee)
Sangyong Jon, a professor in the Department of Biological Sciences at KAIST, and his team developed combined photoacoustic imaging and photothermal therapy for cancer by using Bilirubin (BR) nanoparticles.
The research team applied the properties of a bile pigment called BR, which exerts potent antioxidant and anti-inflammatory effects, to this research.
The team expects this research, which shows high biocompatibility as well as outstanding photoacoustic imaging and photothermal therapy, to be an appropriate system in the field of treatment for cancer.
In the past, the research team developed a PEGylated bilirubin-based nanoparticle system by combining water-insoluble BR with water-soluble Polyethylene Glycol (PEG).
This technology facilitated BR exerting antioxidants yet prevented them from being accumulated in the body. Its efficiency and safety was identified in an animal disease model, for conditions such as inflammatory bowel disease, islet cell transportation, and asthma.
Differing from previous research methods, this research applied the different physicochemical properties of BR to cancer treatment.
When the causative agent of jaundice, yellow BR, is exposed to a certain wavelength of blue light, the agent becomes a photonic nanomaterial as it responses to the light. This light-responsive nanomaterial can be used to cure jaundice because it allows for active excretion in infants.
Secondly, the team identified that BR is a major component of black pigment gallstones which can be often found in gall bladders or bile ducts under certain pathological conditions. The findings show that BR forms black pigment gallstones without the role of an intermediate or cation, such as calcium and copper. The research team combined cisplatin, a platinum metal-based anticancer drug, with BR so that BR nanoparticles changed the solution color from yellow to purple.
The team also examined the possibility of cisplatin-chelated BR nanoparticles as a probe for photoacoustic images. They found that considerable photoacoustic activity was shown when it was exposed to near infrared light. In fact, the photoacoustic signal was increased significantly in tumors of animals with colorectal cancer when the nanoparticles were administered to it intravenously. The team expects a more accurate diagnosis of tumors through this technology.
Moreover, the team assessed the photothermal effects of cisplatin-chelated BR nanoparticles. The research showed that the temperature of tumors increased by 25 degrees Celsius within five minutes when they were exposed to near infrared light, due to the photothermal effect. After two weeks, their size was reduced compared to that of other groups, and sometimes the tumors were even necrotized.
Professor Jon said, “Existing substances have a low biocompatibility and limitation for clinical therapy because they are artificially oriented; therefore, they might have toxicity. I am hoping that these cisplatin-chelated BR-based nanoparticles will provide a new platform for preclinical, translational research and clinical adaptation of the photoacoustic imaging and photothermal therapy.”
The paper (Dong Yun Lee as a first author) was published online in the renowned journal in the field of applied chemistry, Angewandte Chemi International Edition, on September 4. This research was sponsored by the National Research Foundation of Korea.
(Schematic diagram of the research)
(From left: Bilirubin nanoparticles, cisplatin-chelated Bilirubin nanoparticles)
2017.09.26 View 9719 -
 Unlocking the Keys to Parkinson's Disease
A KAIST research team has identified a new mechanism that causes the hallmark symptoms of Parkinson’s disease, namely tremors, rigidity, and loss of voluntary movement.
The discovery, made in collaboration with Nanyang Technological University in Singapore, presents a new perspective to three decades of conventional wisdom in Parkinson’s disease research. It also opens up new avenues that can help alleviate the motor problems suffered by patients of the disease, which reportedly number more than 10 million worldwide. The research was published in Neuron on August 30.
The research team was led by Professor Daesoo Kim from the Department of Biological Sciences at KAIST and Professor George Augustine from the Lee Kong Chian School of Medicine at NTU. Dr. Jeongjin Kim, a former postdoctoral fellow at KAIST who now works at the Korea Institute of Science and Technology (KIST), is the lead author.
It is known that Parkinson’s disease is caused by a lack of dopamine, a chemical in the brain that transmits neural signals. However, it remains unknown how the disease causes the motor
Smooth, voluntary movements, such as reaching for a cup of coffee, are controlled by the basal ganglia, which issue instructions via neurons (nerve cells that process and transmit information in the brain) in the thalamus to the cortex. These instructions come in two types: one that triggers a response (excitatory signals) and the other that suppresses a response (inhibitory signals). Proper balance between the two controls movement.
A low level of dopamine causes the basal ganglia to severely inhibit target neurons in the thalamus, called an inhibition. Scientists have long assumed that this stronger inhibition causes the motor problems of Parkinson’s disease patients.
To test this assumption, the research team used optogenetic technology in an animal model to study the effects of this increased inhibition of the thalamus and ultimately movement. Optogenetics is the use of light to control the activity of specific types of neurons within the brain.
They found that when signals from the basal ganglia are more strongly activated by light, the target neurons in the thalamus paradoxically became hyperactive. Called rebound excitation, this hyperactivity produced abnormal muscular stiffness and tremor. Such motor problems are very similar to the symptoms of Parkinson’s disease patients. When this hyperactivity of thalamic neurons is suppressed by light, mice show normal movments without Parkinson’s disease symptoms. Reducing the levels of activity back to normal caused the motor symptoms to stop, proving that the hyperactivity caused the motor problems experienced by Parkinson’s disease patients.
Professor Kim at KAIST said, “This study overturns three decades of consensus on the provenance of Parkinsonian symptoms.” The lead author, Dr Jeongjin Kim said, “The therapeutic implications of this study for the treatment of Parkinsonian symptoms are profound. It may soon become possible to remedy movement disorders without using L-DOPA, a pre-cursor to dopamine.”
Professor Augustine at NTU added, “Our findings are a breakthrough, both for understanding how the brain normally controls the movement of our body and how this control goes awry during Parkinson’s disease and related dopamine-deficiency disorders.”
The study took five years to complete, and includes researchers from the Department of Bio & Brain Engineering at KAIST.
The research team will move forward by investigating how hyperactivity in neurons in the thalamus leads to abnormal movement, as well as developing therapeutic strategies for the disease by targeting this neural mechanism.
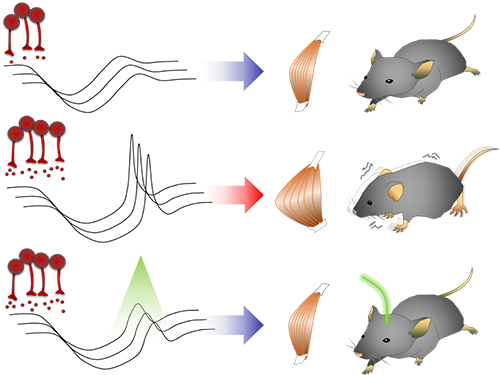
Figure abstract: Inhibitory inputs from the basal ganglia inhibit thalamic neurons (upper). In low-dopamine states, like PD, rebound firing follows inhibition and causes movement disorders (middle). The inhibition of rebound firing alleviates PD-like symptoms in a mouse model of PD.
2017.09.22 View 11371
Unlocking the Keys to Parkinson's Disease
A KAIST research team has identified a new mechanism that causes the hallmark symptoms of Parkinson’s disease, namely tremors, rigidity, and loss of voluntary movement.
The discovery, made in collaboration with Nanyang Technological University in Singapore, presents a new perspective to three decades of conventional wisdom in Parkinson’s disease research. It also opens up new avenues that can help alleviate the motor problems suffered by patients of the disease, which reportedly number more than 10 million worldwide. The research was published in Neuron on August 30.
The research team was led by Professor Daesoo Kim from the Department of Biological Sciences at KAIST and Professor George Augustine from the Lee Kong Chian School of Medicine at NTU. Dr. Jeongjin Kim, a former postdoctoral fellow at KAIST who now works at the Korea Institute of Science and Technology (KIST), is the lead author.
It is known that Parkinson’s disease is caused by a lack of dopamine, a chemical in the brain that transmits neural signals. However, it remains unknown how the disease causes the motor
Smooth, voluntary movements, such as reaching for a cup of coffee, are controlled by the basal ganglia, which issue instructions via neurons (nerve cells that process and transmit information in the brain) in the thalamus to the cortex. These instructions come in two types: one that triggers a response (excitatory signals) and the other that suppresses a response (inhibitory signals). Proper balance between the two controls movement.
A low level of dopamine causes the basal ganglia to severely inhibit target neurons in the thalamus, called an inhibition. Scientists have long assumed that this stronger inhibition causes the motor problems of Parkinson’s disease patients.
To test this assumption, the research team used optogenetic technology in an animal model to study the effects of this increased inhibition of the thalamus and ultimately movement. Optogenetics is the use of light to control the activity of specific types of neurons within the brain.
They found that when signals from the basal ganglia are more strongly activated by light, the target neurons in the thalamus paradoxically became hyperactive. Called rebound excitation, this hyperactivity produced abnormal muscular stiffness and tremor. Such motor problems are very similar to the symptoms of Parkinson’s disease patients. When this hyperactivity of thalamic neurons is suppressed by light, mice show normal movments without Parkinson’s disease symptoms. Reducing the levels of activity back to normal caused the motor symptoms to stop, proving that the hyperactivity caused the motor problems experienced by Parkinson’s disease patients.
Professor Kim at KAIST said, “This study overturns three decades of consensus on the provenance of Parkinsonian symptoms.” The lead author, Dr Jeongjin Kim said, “The therapeutic implications of this study for the treatment of Parkinsonian symptoms are profound. It may soon become possible to remedy movement disorders without using L-DOPA, a pre-cursor to dopamine.”
Professor Augustine at NTU added, “Our findings are a breakthrough, both for understanding how the brain normally controls the movement of our body and how this control goes awry during Parkinson’s disease and related dopamine-deficiency disorders.”
The study took five years to complete, and includes researchers from the Department of Bio & Brain Engineering at KAIST.
The research team will move forward by investigating how hyperactivity in neurons in the thalamus leads to abnormal movement, as well as developing therapeutic strategies for the disease by targeting this neural mechanism.
Figure abstract: Inhibitory inputs from the basal ganglia inhibit thalamic neurons (upper). In low-dopamine states, like PD, rebound firing follows inhibition and causes movement disorders (middle). The inhibition of rebound firing alleviates PD-like symptoms in a mouse model of PD.
2017.09.22 View 11371 -
 Discovery of an Optimal Drug Combination: Overcoming Resistance to Targeted Drugs for Liver Cancer
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences. Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
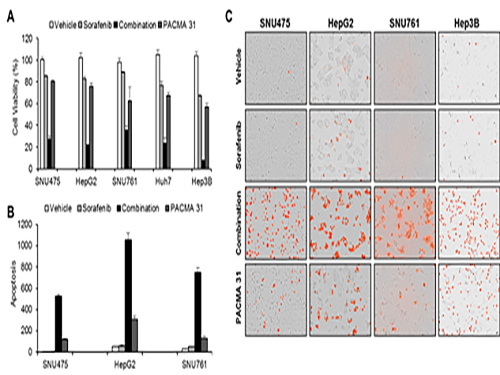
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)
(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)
(Figure 3. ER stress network model)
2017.08.30 View 13118
Discovery of an Optimal Drug Combination: Overcoming Resistance to Targeted Drugs for Liver Cancer
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences. Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)
(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)
(Figure 3. ER stress network model)
2017.08.30 View 13118 -
 Study Identifies the Novel Molecular Signal for Triggering Septic Shock
Professor Seyun Kim’s team at the Department of Biological Sciences reported the mechanism by which cellular signaling transduction networks are precisely controlled in mediating innate immune responses, such as sepsis, by the enzyme IPMK (Inositol polyphosphate multikinase) which is essential for inositol biosynthesis metabolism.
In collaboration with Professor Hyun Seong Roh at Seoul National University, the study’s first author, Eunha Kim, a Ph.D. candidate in Department of Biological Sciences, performed a series of cellular, biochemical, and physiological experiments searching for the new function of IPMK enzymes in macrophages. The research findings were published in Science Advances on April 21.
Professor Kim’s team has been investigating various inositol metabolites and their biosynthesis metabolism for several years and has multilaterally identified the signaling actions of IPMK for controlling cellular growth and energy homeostasis.
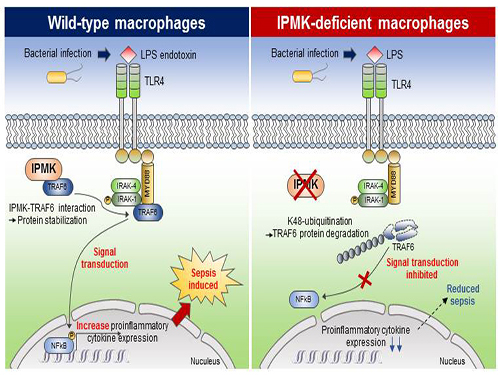
This research showed that the specific deletion of IPMK enzymes in macrophages could significantly reduce levels of inflammation and increase survival rates in mice when they were challenged by microbial septic shock and endotoxins. This suggests a role for IPMK enzymes in mediating innate inflammatory responses that are directly related to a host’s defense against pathogenic bacterial infection.
The team further discovered that IPMK enzymes directly bind to TRAF6 proteins, a key player in immune signaling, thus protecting TRAF6 proteins from ubiquitination reactions that are involved in protein degradation. In addition, Kim and his colleagues successfully verified this IPMK-dependent immune control by employing short peptides which can specifically interfere with the binding between IPMK enzymes and TRAF6 proteins in macrophage cells.
This research revealed a novel function of IPMK enzymes in the fine tuning of innate immune signaling networks, suggesting a new direction for developing therapeutics targeting serious medical conditions such as neuroinflammation, type 2 diabetes, as well as polymicrobial sepsis that are developed from uncontrolled host immune responses. This research was funded by the Ministry of Science, ICT and Future Planning.
(Figure: Deletion of IPMK (inositol polyphosphate multikinase) in macrophages reduces the stability of TRAF6 protein which is the key to innate immune signaling, thereby blocking excessive inflammation in response to pathological bacterial infection.)
2017.05.11 View 10040
Study Identifies the Novel Molecular Signal for Triggering Septic Shock
Professor Seyun Kim’s team at the Department of Biological Sciences reported the mechanism by which cellular signaling transduction networks are precisely controlled in mediating innate immune responses, such as sepsis, by the enzyme IPMK (Inositol polyphosphate multikinase) which is essential for inositol biosynthesis metabolism.
In collaboration with Professor Hyun Seong Roh at Seoul National University, the study’s first author, Eunha Kim, a Ph.D. candidate in Department of Biological Sciences, performed a series of cellular, biochemical, and physiological experiments searching for the new function of IPMK enzymes in macrophages. The research findings were published in Science Advances on April 21.
Professor Kim’s team has been investigating various inositol metabolites and their biosynthesis metabolism for several years and has multilaterally identified the signaling actions of IPMK for controlling cellular growth and energy homeostasis.
This research showed that the specific deletion of IPMK enzymes in macrophages could significantly reduce levels of inflammation and increase survival rates in mice when they were challenged by microbial septic shock and endotoxins. This suggests a role for IPMK enzymes in mediating innate inflammatory responses that are directly related to a host’s defense against pathogenic bacterial infection.
The team further discovered that IPMK enzymes directly bind to TRAF6 proteins, a key player in immune signaling, thus protecting TRAF6 proteins from ubiquitination reactions that are involved in protein degradation. In addition, Kim and his colleagues successfully verified this IPMK-dependent immune control by employing short peptides which can specifically interfere with the binding between IPMK enzymes and TRAF6 proteins in macrophage cells.
This research revealed a novel function of IPMK enzymes in the fine tuning of innate immune signaling networks, suggesting a new direction for developing therapeutics targeting serious medical conditions such as neuroinflammation, type 2 diabetes, as well as polymicrobial sepsis that are developed from uncontrolled host immune responses. This research was funded by the Ministry of Science, ICT and Future Planning.
(Figure: Deletion of IPMK (inositol polyphosphate multikinase) in macrophages reduces the stability of TRAF6 protein which is the key to innate immune signaling, thereby blocking excessive inflammation in response to pathological bacterial infection.)
2017.05.11 View 10040 -
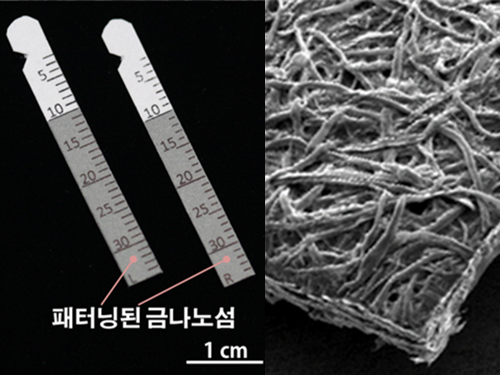 Gout Diagnostic Strip Using a Single Teardrop
A novel diagnostic strip for gout patients using a single teardrop has been announced by KAIST research team. This technology analyzes biological molecules in tears for a non-invasive diagnosis, significantly reducing the time and expense previously required for a diagnosis.
The research team under Professor Ki-Hun Jeong of the Department of Bio and Brain Engineering succeeded in developing an affordable and elaborate gout diagnostic strip by depositing metal nanoparticles on paper. This technology can not only be used in diagnostic medicine and drug testing, but also in various other areas such as field diagnoses that require prompt and accurate detection of a certain substance.
Gout induces pain in joints due to needle-shaped uric acid crystal build up. In general, therapeutic treatments exist to administer pain relief, stimulate uric acid discharge, and uric acid depressant. Such treatments work for temporary relief, but there have significant limitations. Thus, patients are required to regularly check uric acid concentrations, as well as control their diets. Therefore, simpler ways to measure uric acid would greatly benefit gout control and its prevention in a more affordable and convenient manner.
Existing gout diagnostic techniques include measuring uric acid concentrations from blood samples or observing uric acid crystals from joint synovial fluid under a microscope. These existing methods are invasive and time consuming. To overcome their limitations, the research team uniformly deposited gold nanoislands with nanoplasnomics properties on the surface of paper that can easily collect tears.
Nanoplasnomics techniques collect light on the surface of a metal nanostructure, and can be applied to disease and health diagnostic indicators as well as for genetic material detection. Further, metals such as gold absorb stronger light when it is irradiated, and thus can maximize light concentration on board surfaces while maintaining the properties of paper. The developed metal nanostructure production technology allows the flexible manufacturing of nanostructures on a large surface, which in turn allows flexible control of light concentrations.
The research team grafted surface-enhanced Raman spectroscopy on paper diagnostic strips to allow uric acid concentration measurements in teardrops without additional indicators. The measured concentration in teardrops can be compared to blood uric acid concentrations for diagnosing gout.
Professor Jeong explained, “Based on these research results, our strip will make it possible to conduct low-cost, no indicator, supersensitive biological molecule analysis and fast field diagnosis using tears.” He continued, “Tears, as well as various other bodily fluids, can be used to contribute to disease diagnosis and physiological functional research.”
Ph.D. candidate Moonseong Park participated in the research as the first author of the paper that was published in the online edition of ACS Nano on December 14, 2016. Park said, “The strip will allow fast and simple field diagnosis, and can be produced on a large scale using the existing semiconductor process.”
(Figure 1. Optical image of paper gout diagnostic strip covered with gold)
(Figure 2. Scanning delectron microscopic image of paper gout diagnostic strip)
(Figure 3. Scanning electron microscope image of cellulos fiber coated with gold nanoislands)
(Figure 4. Gout diagnosis using tears)
2017.04.27 View 9665
Gout Diagnostic Strip Using a Single Teardrop
A novel diagnostic strip for gout patients using a single teardrop has been announced by KAIST research team. This technology analyzes biological molecules in tears for a non-invasive diagnosis, significantly reducing the time and expense previously required for a diagnosis.
The research team under Professor Ki-Hun Jeong of the Department of Bio and Brain Engineering succeeded in developing an affordable and elaborate gout diagnostic strip by depositing metal nanoparticles on paper. This technology can not only be used in diagnostic medicine and drug testing, but also in various other areas such as field diagnoses that require prompt and accurate detection of a certain substance.
Gout induces pain in joints due to needle-shaped uric acid crystal build up. In general, therapeutic treatments exist to administer pain relief, stimulate uric acid discharge, and uric acid depressant. Such treatments work for temporary relief, but there have significant limitations. Thus, patients are required to regularly check uric acid concentrations, as well as control their diets. Therefore, simpler ways to measure uric acid would greatly benefit gout control and its prevention in a more affordable and convenient manner.
Existing gout diagnostic techniques include measuring uric acid concentrations from blood samples or observing uric acid crystals from joint synovial fluid under a microscope. These existing methods are invasive and time consuming. To overcome their limitations, the research team uniformly deposited gold nanoislands with nanoplasnomics properties on the surface of paper that can easily collect tears.
Nanoplasnomics techniques collect light on the surface of a metal nanostructure, and can be applied to disease and health diagnostic indicators as well as for genetic material detection. Further, metals such as gold absorb stronger light when it is irradiated, and thus can maximize light concentration on board surfaces while maintaining the properties of paper. The developed metal nanostructure production technology allows the flexible manufacturing of nanostructures on a large surface, which in turn allows flexible control of light concentrations.
The research team grafted surface-enhanced Raman spectroscopy on paper diagnostic strips to allow uric acid concentration measurements in teardrops without additional indicators. The measured concentration in teardrops can be compared to blood uric acid concentrations for diagnosing gout.
Professor Jeong explained, “Based on these research results, our strip will make it possible to conduct low-cost, no indicator, supersensitive biological molecule analysis and fast field diagnosis using tears.” He continued, “Tears, as well as various other bodily fluids, can be used to contribute to disease diagnosis and physiological functional research.”
Ph.D. candidate Moonseong Park participated in the research as the first author of the paper that was published in the online edition of ACS Nano on December 14, 2016. Park said, “The strip will allow fast and simple field diagnosis, and can be produced on a large scale using the existing semiconductor process.”
(Figure 1. Optical image of paper gout diagnostic strip covered with gold)
(Figure 2. Scanning delectron microscopic image of paper gout diagnostic strip)
(Figure 3. Scanning electron microscope image of cellulos fiber coated with gold nanoislands)
(Figure 4. Gout diagnosis using tears)
2017.04.27 View 9665 -
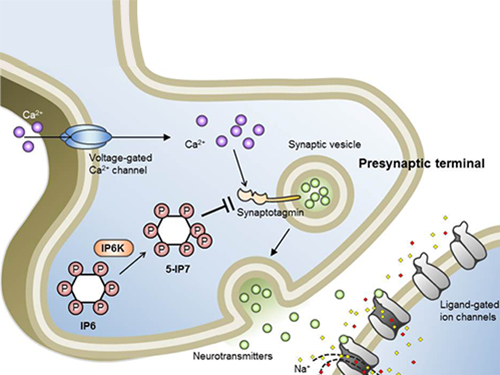 Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin.
Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team.
The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30, 2016.
This interdisciplinary research project was conducted by six research teams from four different countries and covered a wide scope of academic fields, from neurobiology to super resolution optic imaging.
Inositol pyrophosphates such as 5-diphosphoinositol pentakisphos-phate (5-IP7), which naturally occur in corns and beans, are essential metabolites in the body. In particular, inositol hexakisphosphate (IP6) has anti-cancer properties and is thought to have an important role in cell signaling.
Inositol pentakisphosphate (IP7) differs from IP6 by having an additional phosphate group, which was first discovered 20 years ago. IP7 has recently been identified as playing a key role in diabetes and obesity.
Psychopathy and neurodegenerative diseases are known to result from the disrupted balance of inositol pyrophosphates. However, the role and the mechanism of action of IP7 in brain neurons and nerve transmission remained unknown.
Professor Kim’s team has worked on inositol pyrophosphates for several years and discovered that very small quantities of IP7 control cell-signaling transduction. Professor Yoon of Yonsei University identified IP7 as a much stronger inhibitor of neuron signaling compared to IP6. In particular, IP7 directly suppresses synaptotagmin, one of the key proteins in neuron signaling. Moreover, Professor Kim of Kyung Hee University observed IP7 inhibition in sea horse neurons.
Together, the joint research team identified inositol pyrophosphates as the key switch metabolite of brain-signaling transduction.
The researchers hope that future research on synaptotagmin and IP7 will reveal the mechanism of neuron-signal transduction and thus enable the treatment of neurological disorders.
These research findings were the result of cooperation of various science and technology institutes: KAIST, Yonsei-IBS (Institute for Basic Science), Kyung Hee University, Sungkyunkwan University, KIST, University of Zurich in Switzerland, and Albert-Ludwigs-University Freiburg in Germany.
Schematic Image of Controlling the Synaptic Exocytotic Pathway by 5-IP7 , Helping the Understanding of the Signaling Mechanisms of Inositol Pyrophosphates
2016.07.21 View 12960
Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin.
Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team.
The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30, 2016.
This interdisciplinary research project was conducted by six research teams from four different countries and covered a wide scope of academic fields, from neurobiology to super resolution optic imaging.
Inositol pyrophosphates such as 5-diphosphoinositol pentakisphos-phate (5-IP7), which naturally occur in corns and beans, are essential metabolites in the body. In particular, inositol hexakisphosphate (IP6) has anti-cancer properties and is thought to have an important role in cell signaling.
Inositol pentakisphosphate (IP7) differs from IP6 by having an additional phosphate group, which was first discovered 20 years ago. IP7 has recently been identified as playing a key role in diabetes and obesity.
Psychopathy and neurodegenerative diseases are known to result from the disrupted balance of inositol pyrophosphates. However, the role and the mechanism of action of IP7 in brain neurons and nerve transmission remained unknown.
Professor Kim’s team has worked on inositol pyrophosphates for several years and discovered that very small quantities of IP7 control cell-signaling transduction. Professor Yoon of Yonsei University identified IP7 as a much stronger inhibitor of neuron signaling compared to IP6. In particular, IP7 directly suppresses synaptotagmin, one of the key proteins in neuron signaling. Moreover, Professor Kim of Kyung Hee University observed IP7 inhibition in sea horse neurons.
Together, the joint research team identified inositol pyrophosphates as the key switch metabolite of brain-signaling transduction.
The researchers hope that future research on synaptotagmin and IP7 will reveal the mechanism of neuron-signal transduction and thus enable the treatment of neurological disorders.
These research findings were the result of cooperation of various science and technology institutes: KAIST, Yonsei-IBS (Institute for Basic Science), Kyung Hee University, Sungkyunkwan University, KIST, University of Zurich in Switzerland, and Albert-Ludwigs-University Freiburg in Germany.
Schematic Image of Controlling the Synaptic Exocytotic Pathway by 5-IP7 , Helping the Understanding of the Signaling Mechanisms of Inositol Pyrophosphates
2016.07.21 View 12960 -
 Nikon Instruments Korea Donates a Fund to KAIST's Department of Biological Sciences
Representatives from Nikon Instruments Korea Co., Ltd., a producer of microscopes and measuring instruments, visited the KAIST campus on September 25, 2015, and donated USD 9,000 to the Department of Biological Sciences at KAIST.
A small ceremony to mark the donation took place at the department’s conference room.
In the picture from left to right were Professor Won-Do Heo, Department Head Byung-Ha Oh, Professor Sangyong Jon, President Sam-Sup Jang of Korea Instech, and Director Ik-Soo Yoo of Nikon Instruments Korea.
The department announced that the fund would be used to build its new research center to house the state-of-the-art research equipment and tools for the development of new medicine.
2015.10.03 View 5948
Nikon Instruments Korea Donates a Fund to KAIST's Department of Biological Sciences
Representatives from Nikon Instruments Korea Co., Ltd., a producer of microscopes and measuring instruments, visited the KAIST campus on September 25, 2015, and donated USD 9,000 to the Department of Biological Sciences at KAIST.
A small ceremony to mark the donation took place at the department’s conference room.
In the picture from left to right were Professor Won-Do Heo, Department Head Byung-Ha Oh, Professor Sangyong Jon, President Sam-Sup Jang of Korea Instech, and Director Ik-Soo Yoo of Nikon Instruments Korea.
The department announced that the fund would be used to build its new research center to house the state-of-the-art research equipment and tools for the development of new medicine.
2015.10.03 View 5948 -
 Brain Cognitive Engineering Experts from Korea and Abroad Gather at KAIST
The symposium presents recent and future research trends in brain and cognitive engineering.
KAIST hosted the Brain Cognitive Engineering Symposium on September 24, 2015, at the Dream Hall of the Chung Moon Soul building on campus. Around 100 experts in the field of neuroscience participated.
Organized by the Department of Bio and Brain Engineering at KAIST, the symposium celebrated the establishment of the Brain Cognitive Engineering Program at the university and examined the recent research trends in neuroscience.
Six neuroscience experts presented their research and held discussions.
Professor Paul M. Thompson of the University of Southern California (USC), a renowned scientist in neurology imaging genetics, gave a speech entitled “The ENIGMA Project: Mapping Disease and Genetic Effects on the Human Brain in 30,000 People Worldwide.”
Professor Jae-seung Jeong of KAIST’s Department of Bio and Brain Engineering, Director Sung-Gi Kim of IBS Center for Neuroscience Imaging Research, Professor Sung-Hwan Lee of Korea University’s Department of Brain Engineering, Professor Cheil-Moon of DGIST’s Department of Brain and Cognitive Science, and Professor Jun-Tani of KAIST’s Department of Electrical Engineering also participated in the symposium.
Participants discussed the most recent findings in the field of brain science such as the education and research trends of brain cognitive engineering, trends of the world’s brain integrated science, the prospects of brain cognitive engineering program, brain activities that induce blood flow and fMRI, activity production in the brain cortex model as well as the development of functional hierarchy for the motor visual perception, and the neurorobotics research.
Professor Jeong said that “this symposium is a place for examination of the most recent research findings in the field of neuroscience as well as for discussion of its education,”and that “it would be an important opportunity for learning research on brain’s basic mechanisms as well as its applications.”
2015.09.25 View 9903
Brain Cognitive Engineering Experts from Korea and Abroad Gather at KAIST
The symposium presents recent and future research trends in brain and cognitive engineering.
KAIST hosted the Brain Cognitive Engineering Symposium on September 24, 2015, at the Dream Hall of the Chung Moon Soul building on campus. Around 100 experts in the field of neuroscience participated.
Organized by the Department of Bio and Brain Engineering at KAIST, the symposium celebrated the establishment of the Brain Cognitive Engineering Program at the university and examined the recent research trends in neuroscience.
Six neuroscience experts presented their research and held discussions.
Professor Paul M. Thompson of the University of Southern California (USC), a renowned scientist in neurology imaging genetics, gave a speech entitled “The ENIGMA Project: Mapping Disease and Genetic Effects on the Human Brain in 30,000 People Worldwide.”
Professor Jae-seung Jeong of KAIST’s Department of Bio and Brain Engineering, Director Sung-Gi Kim of IBS Center for Neuroscience Imaging Research, Professor Sung-Hwan Lee of Korea University’s Department of Brain Engineering, Professor Cheil-Moon of DGIST’s Department of Brain and Cognitive Science, and Professor Jun-Tani of KAIST’s Department of Electrical Engineering also participated in the symposium.
Participants discussed the most recent findings in the field of brain science such as the education and research trends of brain cognitive engineering, trends of the world’s brain integrated science, the prospects of brain cognitive engineering program, brain activities that induce blood flow and fMRI, activity production in the brain cortex model as well as the development of functional hierarchy for the motor visual perception, and the neurorobotics research.
Professor Jeong said that “this symposium is a place for examination of the most recent research findings in the field of neuroscience as well as for discussion of its education,”and that “it would be an important opportunity for learning research on brain’s basic mechanisms as well as its applications.”
2015.09.25 View 9903 -
 KAIST to support the Genetic Donguibogam Research Project for global market entry of a new natural drug produced by Green Cross Corporation HS
In the wake of the spread of the Middle East Respiratory Syndrome (MERS), sales of immune-enhancing products in Korea such as red and white ginseng have risen dramatically. Ginseng is one of Korea’s major health supplement it exports, but due to the lack of precise scientific knowledge of its mechanism, sales of ginseng account for less than 2% of the global market share.
The Genetic Donguibogam Research Project represents a group of research initiatives to study genes and environmental factors that contribute to diseases and to discover alternative treatments through Eastern medicine. The project is being led by KAIST’s Department of Bio & Brain Engineering Professor Do-Heon Lee.
Professor Lee and Chief Executive Officer Young-Hyo Yoo of Green Cross Corporation HS, a Korean pharmaceutical company, signed a memorandum of understanding (MOU), as well as a non-disclosure agreement (NDA) to develop a naturally derived drug with an enhanced ginsenoside, pharmacological compounds of ginseng, for the global market entry of BST204 on June 10, 2015.
Donguibogam is the traditional Korean source for the principles and practice of Eastern medicine, which was compiled by the royal physician Heo Jun and first published in 1613 during the Joseon Dynasty of Korea.
Cooperating with Green Cross Co., HS, KAIST researchers will use a multi-component, multi-target (MCMT)-based development platform to produce the new natural drug, BST204. This cooperation is expected to assist the entry of the drug into the European market.
Green Cross Co., HS has applied a bio-conversion technique to ginseng to develop BST204, which is a drug with enhanced active constituent of aginsenosides. The drug is the first produced by any Korean pharmaceutical company to complete the first phase of clinical trials in Germany and is about to start the second phase of trials.
Professor Do-Heon Lee, the Director of the project said, “Genetic Donguibogam Research Project seeks to create new innovative healthcare material for the future using integrated fundamental technologies such as virtual human body computer modelling and multi-omics to explain the mechanism in which natural ingredients affect the human body.” He continued, “Especially, by employing the virtual human body computer modelling, we can develop an innovative new technology that will greatly assist Korean pharmaceutical industry and make it the platform technology in entering global markets.”
Young-Hyo Yoo, the CEO of Green Cross Co., HS, said, “For a new naturally derived drug to be acknowledged in the global market, such as Europe and the US, its mechanism, as well as its effectiveness and safety, should be proven. However, it is difficult and costly to explain the mechanism in which the complex composition of a natural substance influences the body. Innovative technology is needed to solve this problem.”
Professor Do-Heon Lee (left in the picture), the Director of Genetic Donguibogam Research Project, stands abreast Young-Hyo Yoo (right in the picture), the CEO of Green Cross Co., HS.
2015.06.10 View 10509
KAIST to support the Genetic Donguibogam Research Project for global market entry of a new natural drug produced by Green Cross Corporation HS
In the wake of the spread of the Middle East Respiratory Syndrome (MERS), sales of immune-enhancing products in Korea such as red and white ginseng have risen dramatically. Ginseng is one of Korea’s major health supplement it exports, but due to the lack of precise scientific knowledge of its mechanism, sales of ginseng account for less than 2% of the global market share.
The Genetic Donguibogam Research Project represents a group of research initiatives to study genes and environmental factors that contribute to diseases and to discover alternative treatments through Eastern medicine. The project is being led by KAIST’s Department of Bio & Brain Engineering Professor Do-Heon Lee.
Professor Lee and Chief Executive Officer Young-Hyo Yoo of Green Cross Corporation HS, a Korean pharmaceutical company, signed a memorandum of understanding (MOU), as well as a non-disclosure agreement (NDA) to develop a naturally derived drug with an enhanced ginsenoside, pharmacological compounds of ginseng, for the global market entry of BST204 on June 10, 2015.
Donguibogam is the traditional Korean source for the principles and practice of Eastern medicine, which was compiled by the royal physician Heo Jun and first published in 1613 during the Joseon Dynasty of Korea.
Cooperating with Green Cross Co., HS, KAIST researchers will use a multi-component, multi-target (MCMT)-based development platform to produce the new natural drug, BST204. This cooperation is expected to assist the entry of the drug into the European market.
Green Cross Co., HS has applied a bio-conversion technique to ginseng to develop BST204, which is a drug with enhanced active constituent of aginsenosides. The drug is the first produced by any Korean pharmaceutical company to complete the first phase of clinical trials in Germany and is about to start the second phase of trials.
Professor Do-Heon Lee, the Director of the project said, “Genetic Donguibogam Research Project seeks to create new innovative healthcare material for the future using integrated fundamental technologies such as virtual human body computer modelling and multi-omics to explain the mechanism in which natural ingredients affect the human body.” He continued, “Especially, by employing the virtual human body computer modelling, we can develop an innovative new technology that will greatly assist Korean pharmaceutical industry and make it the platform technology in entering global markets.”
Young-Hyo Yoo, the CEO of Green Cross Co., HS, said, “For a new naturally derived drug to be acknowledged in the global market, such as Europe and the US, its mechanism, as well as its effectiveness and safety, should be proven. However, it is difficult and costly to explain the mechanism in which the complex composition of a natural substance influences the body. Innovative technology is needed to solve this problem.”
Professor Do-Heon Lee (left in the picture), the Director of Genetic Donguibogam Research Project, stands abreast Young-Hyo Yoo (right in the picture), the CEO of Green Cross Co., HS.
2015.06.10 View 10509