Nature
-
 A research paper by Professor Myung-Chul Choi reviewed in Science (February 28, 2014)
A research paper entitled “Transformation of taxol-stabilized microtubules into inverted tubulin tubules triggered by a tubulin conformation switch” was published in
Nature Materials
this year, dated January 19, 2014.
Professor Myung-Chul Choi and Dr. Chae-Yeon Song from the Department of Bio and Brain Engineering at KAIST co-authored the paper together with researchers from the University of California in Santa Barbara and the Hebrew University of Jerusalem.
Science,
dated February 28, 2014, has recently reviewed the paper in its section called "Perspectives."
2014.03.13 View 9743
A research paper by Professor Myung-Chul Choi reviewed in Science (February 28, 2014)
A research paper entitled “Transformation of taxol-stabilized microtubules into inverted tubulin tubules triggered by a tubulin conformation switch” was published in
Nature Materials
this year, dated January 19, 2014.
Professor Myung-Chul Choi and Dr. Chae-Yeon Song from the Department of Bio and Brain Engineering at KAIST co-authored the paper together with researchers from the University of California in Santa Barbara and the Hebrew University of Jerusalem.
Science,
dated February 28, 2014, has recently reviewed the paper in its section called "Perspectives."
2014.03.13 View 9743 -
 Spillover Phenomenon Identified Using Model Catalyst System
Researchers at KAIST have identified spillover phenomenon, which has remained controversial since its discovery in the early 1960s.
KAIST Department of Chemical and Biomolecular Engineering’s Professor Min-Gi Choi and his team has explained the "spillover phenomenon," using their own model catalyst system where platinum is selectively located within the amorphous aluminosilicate.
The research results were published on the 25th February online edition of Nature Communications.
Spillover refers to a phenomenon that occurs when hydrogen atoms that have been activated on the surface of metals, such as platinum, move to the surface of the catalyst. It was predicted that this phenomenon can be used to design a catalyst with high activity and stability, and thus has been actively studied over the last 50 years.
However, many cases of the known catalysts involved competing reactions on the exposed metal surface, which made it impossible to directly identify the presence and formation mechanism of spillover.
The catalysts developed by the researchers at KAIST used platinum nanoparticles covered with aluminosilicate. This only allowed the hydrogen molecules to pass through and has effectively blocked the competing reactions, enabling the research team to study the spillover phenomenon.
Through various catalyst structure and reactivity analysis, as well as computer modeling, the team has discovered that Brönsted acid sites present on the aluminosilicate plays a crucial role in spillover phenomenon.
In addition, the spillover-based hydrogenation catalyst proposed by the research team showed very high hydrogenation and dehydrogenation activity. The ability of the catalyst to significantly inhibit unwanted hydrogenolysis reaction during the petrochemical processes also suggested a large industrial potential.
Professor Min-Gi Choi said, “This particular catalyst, which can trigger the reaction only by spillover phenomenon, can be properly designed to exceed the capacity of the conventional metal catalysts. The future goal is to make a catalyst with much higher activity and selectivity.”
The research was conducted through funds subsidized by SK Innovation and Ministry of Science, ICT and Future Planning.
The senior research fellow of SK Innovation Seung-Hun Oh said, “SK Innovation will continue to develop a new commercial catalyst based on the technology from this research.”
Juh-Wan Lim and Hye-Yeong Shin led the research as joint first authors under supervision of Professor Min-Gi Choi and computer modeling works were conducted by KAIST EEWS (environment, energy, water, and sustainability) graduate school’s Professor Hyeong-Jun Kim.
2014.03.03 View 11441
Spillover Phenomenon Identified Using Model Catalyst System
Researchers at KAIST have identified spillover phenomenon, which has remained controversial since its discovery in the early 1960s.
KAIST Department of Chemical and Biomolecular Engineering’s Professor Min-Gi Choi and his team has explained the "spillover phenomenon," using their own model catalyst system where platinum is selectively located within the amorphous aluminosilicate.
The research results were published on the 25th February online edition of Nature Communications.
Spillover refers to a phenomenon that occurs when hydrogen atoms that have been activated on the surface of metals, such as platinum, move to the surface of the catalyst. It was predicted that this phenomenon can be used to design a catalyst with high activity and stability, and thus has been actively studied over the last 50 years.
However, many cases of the known catalysts involved competing reactions on the exposed metal surface, which made it impossible to directly identify the presence and formation mechanism of spillover.
The catalysts developed by the researchers at KAIST used platinum nanoparticles covered with aluminosilicate. This only allowed the hydrogen molecules to pass through and has effectively blocked the competing reactions, enabling the research team to study the spillover phenomenon.
Through various catalyst structure and reactivity analysis, as well as computer modeling, the team has discovered that Brönsted acid sites present on the aluminosilicate plays a crucial role in spillover phenomenon.
In addition, the spillover-based hydrogenation catalyst proposed by the research team showed very high hydrogenation and dehydrogenation activity. The ability of the catalyst to significantly inhibit unwanted hydrogenolysis reaction during the petrochemical processes also suggested a large industrial potential.
Professor Min-Gi Choi said, “This particular catalyst, which can trigger the reaction only by spillover phenomenon, can be properly designed to exceed the capacity of the conventional metal catalysts. The future goal is to make a catalyst with much higher activity and selectivity.”
The research was conducted through funds subsidized by SK Innovation and Ministry of Science, ICT and Future Planning.
The senior research fellow of SK Innovation Seung-Hun Oh said, “SK Innovation will continue to develop a new commercial catalyst based on the technology from this research.”
Juh-Wan Lim and Hye-Yeong Shin led the research as joint first authors under supervision of Professor Min-Gi Choi and computer modeling works were conducted by KAIST EEWS (environment, energy, water, and sustainability) graduate school’s Professor Hyeong-Jun Kim.
2014.03.03 View 11441 -
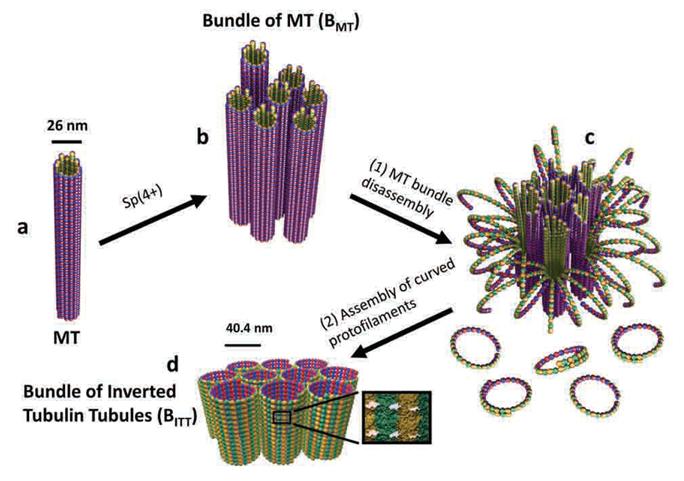 A Molecular Switch Controlling Self-Assembly of Protein Nanotubes Discovered
International collaborative research among South Korea, United States, and Israel research institutionsThe key to the treatment of cancer and brain disease mechanism The molecular switch that controls the self-assembly structure of the protein nanotubes, which plays crucial role in cell division and intracellular transport of materials, has been discovered. KAIST Bio and Brain Engineering Department’s Professor Myeong-Cheol Choi and Professor Chae-Yeon Song conducted the research, in collaboration with the University of California in Santa Barbara, U.S., and Hebrew University in Israel. The findings of the research were published in Nature Materials on the 19th. Microtubules are tube shaped and composed of protein that plays a key role in cell division, cytoskeleton, and intercellular material transport and is only 25nm in diameter (1/100,000 thickness of a human hair). Conventionally, cancer treatment focused on disrupting the formation of microtubules to suppress the division of cancer cells. In addition Alzheimer’s is known to be caused by the diminishing of structural integrity of microtubules responsible for intercellular material transport which leads to failure in signal transfer. The research team utilized synchrotron x-ray scattering and transmission electron microscope to analyze the self assemble structure of protein nanotubes to subnanometer accuracy. As a result, the microtubules were found to assemble into 25nm thickness tubules by stacking protein blocks 4 x 5 x 8nm in dimension. In the process, the research team discovered the molecular switch that controls the shape of these protein blocks. In addition the research team was successful in creating a new protein tube structure. Professor Choi commented that they were successful in introducing a new paradigm that suggests the possibility of controlling the complex biological functions of human’s biological system with the simple use of physical principles. He commented further that it is anticipated that the findings will allow for the application of bio nanotubes in engineering and that this is a small step in finding the mechanism behind cancer treatment and neural diseases.
2014.02.03 View 11564
A Molecular Switch Controlling Self-Assembly of Protein Nanotubes Discovered
International collaborative research among South Korea, United States, and Israel research institutionsThe key to the treatment of cancer and brain disease mechanism The molecular switch that controls the self-assembly structure of the protein nanotubes, which plays crucial role in cell division and intracellular transport of materials, has been discovered. KAIST Bio and Brain Engineering Department’s Professor Myeong-Cheol Choi and Professor Chae-Yeon Song conducted the research, in collaboration with the University of California in Santa Barbara, U.S., and Hebrew University in Israel. The findings of the research were published in Nature Materials on the 19th. Microtubules are tube shaped and composed of protein that plays a key role in cell division, cytoskeleton, and intercellular material transport and is only 25nm in diameter (1/100,000 thickness of a human hair). Conventionally, cancer treatment focused on disrupting the formation of microtubules to suppress the division of cancer cells. In addition Alzheimer’s is known to be caused by the diminishing of structural integrity of microtubules responsible for intercellular material transport which leads to failure in signal transfer. The research team utilized synchrotron x-ray scattering and transmission electron microscope to analyze the self assemble structure of protein nanotubes to subnanometer accuracy. As a result, the microtubules were found to assemble into 25nm thickness tubules by stacking protein blocks 4 x 5 x 8nm in dimension. In the process, the research team discovered the molecular switch that controls the shape of these protein blocks. In addition the research team was successful in creating a new protein tube structure. Professor Choi commented that they were successful in introducing a new paradigm that suggests the possibility of controlling the complex biological functions of human’s biological system with the simple use of physical principles. He commented further that it is anticipated that the findings will allow for the application of bio nanotubes in engineering and that this is a small step in finding the mechanism behind cancer treatment and neural diseases.
2014.02.03 View 11564 -
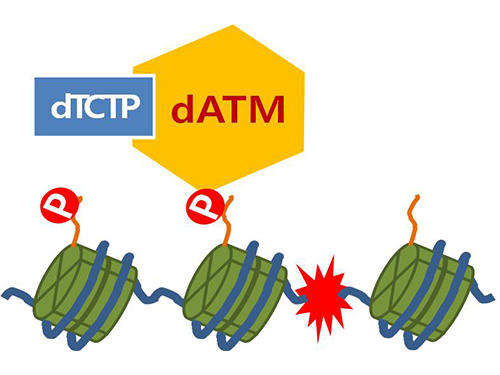 Mechanism in regulation of cancer-related key enzyme, ATM, for DNA damage and repair revealed
Professor Kwang-Wook Choi
A research team led by Professor Kwang-Wook Choi and Dr. Seong-Tae Hong from the Department of Biological Sciences at KAIST has successfully investigated the operational mechanism of the protein Ataxia Telangiectasia Mutated (ATM), an essential protein to the function of a crucial key enzyme that repairs the damaged DNA which stores biometric information. The results were published on December 19th Nature Communications online edition.
All organisms, including humans, constantly strive to protect the information within their DNA from damages posed by a number of factors, such as carbonized materials in our daily food intake, radioactive materials such as radon emitting from the cement of buildings or ultraviolet of the sunlight, which could be a trigger for cancer.
In order to keep the DNA information safe, the organisms are always carrying out complex and sophisticated DNA repair work, which involves the crucial DNA damage repair protein ATM. Consequently, a faulty ATM leads to higher risks of cancer.
Until now, academia predicted that the Translationally Controlled Tumor Protein (TCTP) will play an important role in regulating the function of ATM. However, since most of main research regarding TCTP has only been conducted in cultured cells, it was unable to identify exactly what mechanisms TCTP employs to control ATM.
The KAIST research team identified that TCTP can combine with ATM or increase the enzymatic activity of ATM. In addition, Drosophilia, one of the most widely used model organisms for molecular genetics, has been used to identify that TCTP and ATM play a very important role in repairing the DNA damaged by radiation. This information has allowed the researchers to establish TCTP’s essential function in maintaining the DNA information in cell cultures and even in higher organisms, and to provide specific and important clues to the regulation of ATM by TCTP.
Professor Kwang-Wook Choi said, “Our research is a good example that basic research using Drosophilia can make important contributions to understanding the process of diseases, such as cancer, and to developing adequate treatment.”
The research has been funded by the Ministry of Science, ICT and Future Planning, Republic of Korea, and the National Research Foundation of Korea.
Figure 1. When the amount of TCTP protein is reduced, cells of the Drosophila's eye are abnormally deformed by radiation. Scale bars = 200mm
Figure 2. When the amount of TCTP protein is reduced, the chromosomes of Drosophilia are easily broken by radiation. Scale bars = 10 mm.
Figure 3. When gene expressions of TCTP and ATM are reduced, large defects occur in the normal development of the eye. (Left: normal Drosophilia's eye, right: development-deficient eye)
Figure 4. ATM marks the position of the broken DNA, with TCTP helping to facilitate this reaction. DNA (blue line) within the cell nucleus is coiled around the histone protein (green cylinder). When DNA is broken, ATM protein attaches a phosphate group (P). Multiple DNA repair protein recognizes the phosphate as a signal that requires repair and gathers at the site.
2014.01.07 View 15257
Mechanism in regulation of cancer-related key enzyme, ATM, for DNA damage and repair revealed
Professor Kwang-Wook Choi
A research team led by Professor Kwang-Wook Choi and Dr. Seong-Tae Hong from the Department of Biological Sciences at KAIST has successfully investigated the operational mechanism of the protein Ataxia Telangiectasia Mutated (ATM), an essential protein to the function of a crucial key enzyme that repairs the damaged DNA which stores biometric information. The results were published on December 19th Nature Communications online edition.
All organisms, including humans, constantly strive to protect the information within their DNA from damages posed by a number of factors, such as carbonized materials in our daily food intake, radioactive materials such as radon emitting from the cement of buildings or ultraviolet of the sunlight, which could be a trigger for cancer.
In order to keep the DNA information safe, the organisms are always carrying out complex and sophisticated DNA repair work, which involves the crucial DNA damage repair protein ATM. Consequently, a faulty ATM leads to higher risks of cancer.
Until now, academia predicted that the Translationally Controlled Tumor Protein (TCTP) will play an important role in regulating the function of ATM. However, since most of main research regarding TCTP has only been conducted in cultured cells, it was unable to identify exactly what mechanisms TCTP employs to control ATM.
The KAIST research team identified that TCTP can combine with ATM or increase the enzymatic activity of ATM. In addition, Drosophilia, one of the most widely used model organisms for molecular genetics, has been used to identify that TCTP and ATM play a very important role in repairing the DNA damaged by radiation. This information has allowed the researchers to establish TCTP’s essential function in maintaining the DNA information in cell cultures and even in higher organisms, and to provide specific and important clues to the regulation of ATM by TCTP.
Professor Kwang-Wook Choi said, “Our research is a good example that basic research using Drosophilia can make important contributions to understanding the process of diseases, such as cancer, and to developing adequate treatment.”
The research has been funded by the Ministry of Science, ICT and Future Planning, Republic of Korea, and the National Research Foundation of Korea.
Figure 1. When the amount of TCTP protein is reduced, cells of the Drosophila's eye are abnormally deformed by radiation. Scale bars = 200mm
Figure 2. When the amount of TCTP protein is reduced, the chromosomes of Drosophilia are easily broken by radiation. Scale bars = 10 mm.
Figure 3. When gene expressions of TCTP and ATM are reduced, large defects occur in the normal development of the eye. (Left: normal Drosophilia's eye, right: development-deficient eye)
Figure 4. ATM marks the position of the broken DNA, with TCTP helping to facilitate this reaction. DNA (blue line) within the cell nucleus is coiled around the histone protein (green cylinder). When DNA is broken, ATM protein attaches a phosphate group (P). Multiple DNA repair protein recognizes the phosphate as a signal that requires repair and gathers at the site.
2014.01.07 View 15257 -
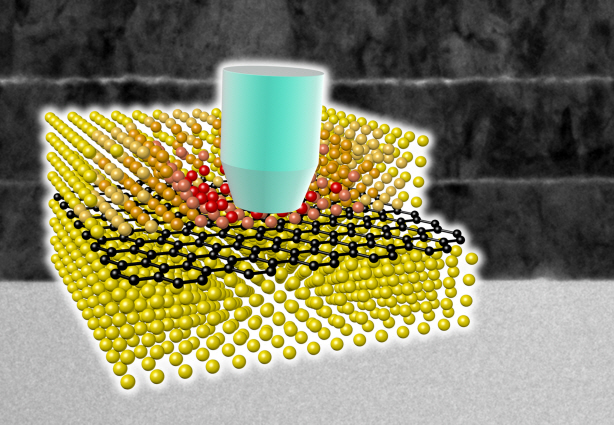 Ultra-High Strength Metamaterial Developed Using Graphene
New metamaterial has been developed, exhibiting hundreds of times greater strength than pure metals.
Professor Seung Min, Han and Yoo Sung, Jeong (Graduate School of Energy, Environment, Water, and Sustainability (EEWS)) and Professor Seok Woo, Jeon (Department of Material Science and Engineering) have developed a composite nanomaterial. The nanomaterial consists of graphene inserted in copper and nickel and exhibits strengths 500 times and 180 times, respectively, greater than that of pure metals. The result of the research was published on the July 2nd online edition in Nature Communications journal.
Graphene displays strengths 200 times greater than that of steel, is stretchable, and is flexible. The U.S. Army Armaments Research, Development and Engineering Center developed a graphene-metal nanomaterial but failed to drastically improve the strength of the material.
To maximize the strength increased by the addition of graphene, the KAIST research team created a layered structure of metal and graphene. Using CVD (Chemical Vapor Deposition), the team grew a single layer of graphene on a metal deposited substrate and then deposited another metal layer. They repeated this process to produce a metal-graphene multilayer composite material, utilizing a single layer of graphene. Micro-compression tests within Transmission Electronic Microscope and Molecular Dynamics simulations effectively showed the strength enhancing effect and the dislocation movement in grain boundaries of graphene on an atomic level.
The mechanical characteristics of the graphene layer within the metal-graphene composite material successfully blocked the dislocations and cracks from external damage from traveling inwards. Therefore the composite material displayed strength beyond conventional metal-metal multilayer materials. The copper-graphene multilayer material with an interplanar distance of 70nm exhibited 500 times greater (1.5GPa) strength than pure copper. Nickel-graphene multilayer material with an interplanar distance of 100nm showed 180 times greater (4.0GPa) strength than pure nickel.
It was found that there is a clear relationship between the interplanar distance and the strength of the multilayer material. A smaller interplanar distance made the dislocation movement more difficult and therefore increased the strength of the material. Professor Han, who led the research, commented, “the result is astounding as 0.00004% in weight of graphene increased the strength of the materials by hundreds of times” and “improvements based on this success, especially mass production with roll-to-roll process or metal sintering process in the production of ultra-high strength, lightweight parts for automobile and spacecraft, may become possible.” In addition, Professor Han mentioned that “the new material can be applied to coating materials for nuclear reactor construction or other structural materials requiring high reliability.”
The research project received support from National Research Foundation, Global Frontier Program, KAIST EEWS-KINC Program and KISTI Supercomputer and was a collaborative effort with KISTI (Korea Institute of Science and Technology Information), KBSI (Korea Basic Science Institute), Stanford University, and Columbia University.
A schematic diagram shows the structure of metal-graphene multi-layers. The metal-graphene multi-layered composite materials, containing a single-layered graphene, block the dislocation movement of graphene layers, resulting in a greater strength in the materials.
2013.08.23 View 18136
Ultra-High Strength Metamaterial Developed Using Graphene
New metamaterial has been developed, exhibiting hundreds of times greater strength than pure metals.
Professor Seung Min, Han and Yoo Sung, Jeong (Graduate School of Energy, Environment, Water, and Sustainability (EEWS)) and Professor Seok Woo, Jeon (Department of Material Science and Engineering) have developed a composite nanomaterial. The nanomaterial consists of graphene inserted in copper and nickel and exhibits strengths 500 times and 180 times, respectively, greater than that of pure metals. The result of the research was published on the July 2nd online edition in Nature Communications journal.
Graphene displays strengths 200 times greater than that of steel, is stretchable, and is flexible. The U.S. Army Armaments Research, Development and Engineering Center developed a graphene-metal nanomaterial but failed to drastically improve the strength of the material.
To maximize the strength increased by the addition of graphene, the KAIST research team created a layered structure of metal and graphene. Using CVD (Chemical Vapor Deposition), the team grew a single layer of graphene on a metal deposited substrate and then deposited another metal layer. They repeated this process to produce a metal-graphene multilayer composite material, utilizing a single layer of graphene. Micro-compression tests within Transmission Electronic Microscope and Molecular Dynamics simulations effectively showed the strength enhancing effect and the dislocation movement in grain boundaries of graphene on an atomic level.
The mechanical characteristics of the graphene layer within the metal-graphene composite material successfully blocked the dislocations and cracks from external damage from traveling inwards. Therefore the composite material displayed strength beyond conventional metal-metal multilayer materials. The copper-graphene multilayer material with an interplanar distance of 70nm exhibited 500 times greater (1.5GPa) strength than pure copper. Nickel-graphene multilayer material with an interplanar distance of 100nm showed 180 times greater (4.0GPa) strength than pure nickel.
It was found that there is a clear relationship between the interplanar distance and the strength of the multilayer material. A smaller interplanar distance made the dislocation movement more difficult and therefore increased the strength of the material. Professor Han, who led the research, commented, “the result is astounding as 0.00004% in weight of graphene increased the strength of the materials by hundreds of times” and “improvements based on this success, especially mass production with roll-to-roll process or metal sintering process in the production of ultra-high strength, lightweight parts for automobile and spacecraft, may become possible.” In addition, Professor Han mentioned that “the new material can be applied to coating materials for nuclear reactor construction or other structural materials requiring high reliability.”
The research project received support from National Research Foundation, Global Frontier Program, KAIST EEWS-KINC Program and KISTI Supercomputer and was a collaborative effort with KISTI (Korea Institute of Science and Technology Information), KBSI (Korea Basic Science Institute), Stanford University, and Columbia University.
A schematic diagram shows the structure of metal-graphene multi-layers. The metal-graphene multi-layered composite materials, containing a single-layered graphene, block the dislocation movement of graphene layers, resulting in a greater strength in the materials.
2013.08.23 View 18136 -
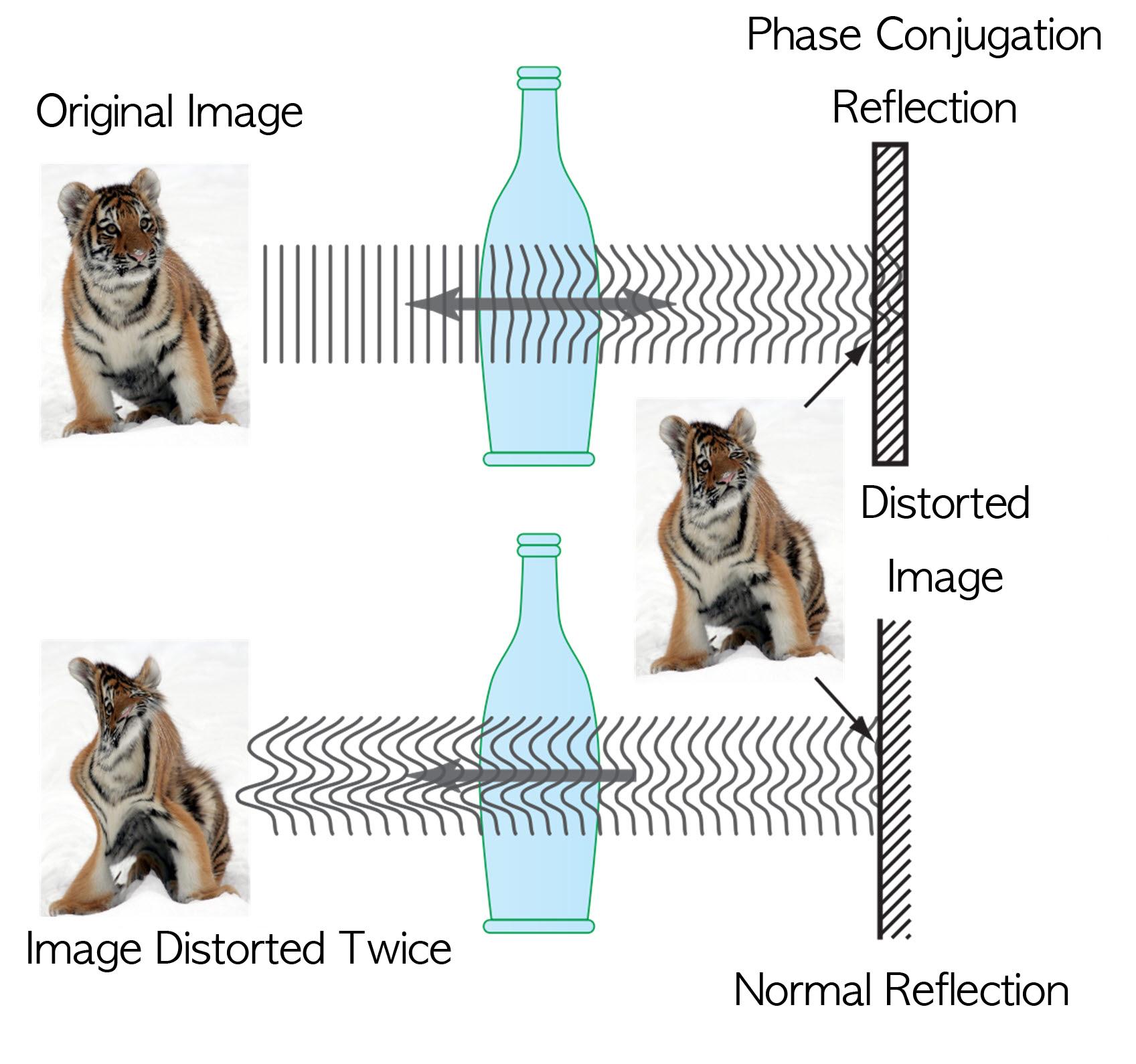 Technology Developed to Control Light Scattering Using Holography
Published on May 29th Nature Scientific Reports online
Recently, a popular article demonstrated that an opaque glass becomes transparent as transparent tape is applied to the glass. The scientific principle is that light is less scattered as the rough surface of the opaque glass is filled by transparent tape, thereby making things behind the opaque glass look clearer.
Professor Yong-Keun Park from KAIST’s Department of Physics, in a joint research with MIT Spectroscopy Lab, has developed a technology to easily control light scattering using holography. Their results are published on Nature’s Scientific Reports May 29th online edition.
This technology allows us to see things behind visual obstructions such as cloud and smoke, or even human skin that is highly scattering, optically thick materials. The research team applied the holography technology that records both the direction and intensity of light, and controlled light scattering of obstacles lied between an observer and a target image. The team was able to retrieve the original image by recording the information of scattered light and reflecting the light precisely to the other side.This phenomenon is known as “phase conjugation” in physics. Professor Park’s team applied phase conjugation and digital holography to observe two-dimensional image behind a highly scattering wall. “This technology will be utilized in many fields of physics, optics, nanotechnology, medical science, and even military science,” said Professor Park. “This is different from what is commonly known as penetrating camera or invisible clothes.” He nevertheless drew the line at over-interpreting the technology, “Currently, the significance is on the development of the technology itself that allows us to accurately control the scattering of light."
Figure I. Observed Images
Figure II. Light Scattering Control
2013.07.19 View 9894
Technology Developed to Control Light Scattering Using Holography
Published on May 29th Nature Scientific Reports online
Recently, a popular article demonstrated that an opaque glass becomes transparent as transparent tape is applied to the glass. The scientific principle is that light is less scattered as the rough surface of the opaque glass is filled by transparent tape, thereby making things behind the opaque glass look clearer.
Professor Yong-Keun Park from KAIST’s Department of Physics, in a joint research with MIT Spectroscopy Lab, has developed a technology to easily control light scattering using holography. Their results are published on Nature’s Scientific Reports May 29th online edition.
This technology allows us to see things behind visual obstructions such as cloud and smoke, or even human skin that is highly scattering, optically thick materials. The research team applied the holography technology that records both the direction and intensity of light, and controlled light scattering of obstacles lied between an observer and a target image. The team was able to retrieve the original image by recording the information of scattered light and reflecting the light precisely to the other side.This phenomenon is known as “phase conjugation” in physics. Professor Park’s team applied phase conjugation and digital holography to observe two-dimensional image behind a highly scattering wall. “This technology will be utilized in many fields of physics, optics, nanotechnology, medical science, and even military science,” said Professor Park. “This is different from what is commonly known as penetrating camera or invisible clothes.” He nevertheless drew the line at over-interpreting the technology, “Currently, the significance is on the development of the technology itself that allows us to accurately control the scattering of light."
Figure I. Observed Images
Figure II. Light Scattering Control
2013.07.19 View 9894 -
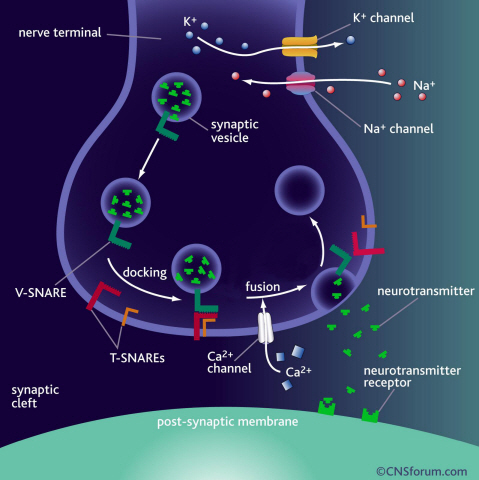 Neurotransmitter protein structure and operation principle identified
Professor Tae-Young Yoon
- Real-time measurement of structural change of bio-membrane fusion protein
- A new clue to degenerative brain diseases research
KAIST Physics Department’s Professor Tae-Young Yoon has successfully identified the hidden structure and operation mechanism of the SNARE protein, which has a central role in transporting neurotransmitters between neurons, using magnetic nanotweezers. SNARE protein’s cell membrane fusion function is closely related to degenerative brain diseases or neurological disorders such as Alzheimer’s. Hence, this research may provide a clue to the disease’s prevention and treatment.
Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. The SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters. The academia speculated the SNARE protein would regulate the exchange of neurotransmitters, but its precise function and structure has been unknown. Professor Yoon’s research team developed an experimental technique using nanotweezers to measure physical changes to nanometer level by pulling and releasing each protein with force of 1 pN (piconewton). The research identified the existence of hidden SNARE protein"s intermediate structure. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been identified.
Professor Yoon’s research team developed an experimental technique using magnetic nanotweezers to measure physical changes of proteins to nanometer level by pulling and releasing each protein with force of 1 pN. The research identified the existence of hidden SNARE protein"s intermediate structure and its formation. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been discovered.
Professor Yoon said, “Ground breaking research results have been produced. A simple experimental technique of applying the smallest possible forces to proteins (with tweezers) to see their hidden structure and formation process can produce the same result as real observation has been developed.” He continued, “This technique will be very important in researching biological object with physical experimental technique. It will be a vital foundation to consilient research of different academia in the future.”
This research was a joint project of Physics Department’s Professor Tae-Young Yoon, KAIST, and Biomedical Engineering Institute’s Professor Yeon-Kyun Shin at KIST. KAIST Physics Department’s Professor Yong-Hoon Cho, Ph.D. candidate Do-Yong Lee and KIAS Computational Sciences Department’s Professor Chang-Bong Hyun participated. The research was published on Nature Communications on April 16th.
a) Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. A SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters.
b) A schematic diagram using magnetic nanotweezers to measure protein structure changes on molecular level. The nanotweezers exert an exquisite pull and release of each protein with a force of 1 pN to measure physical changes to nanometer level in real-time to observe the hidden intermediate structure and operation principles of bio-membrane fusion protein.
2013.05.25 View 10303
Neurotransmitter protein structure and operation principle identified
Professor Tae-Young Yoon
- Real-time measurement of structural change of bio-membrane fusion protein
- A new clue to degenerative brain diseases research
KAIST Physics Department’s Professor Tae-Young Yoon has successfully identified the hidden structure and operation mechanism of the SNARE protein, which has a central role in transporting neurotransmitters between neurons, using magnetic nanotweezers. SNARE protein’s cell membrane fusion function is closely related to degenerative brain diseases or neurological disorders such as Alzheimer’s. Hence, this research may provide a clue to the disease’s prevention and treatment.
Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. The SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters. The academia speculated the SNARE protein would regulate the exchange of neurotransmitters, but its precise function and structure has been unknown. Professor Yoon’s research team developed an experimental technique using nanotweezers to measure physical changes to nanometer level by pulling and releasing each protein with force of 1 pN (piconewton). The research identified the existence of hidden SNARE protein"s intermediate structure. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been identified.
Professor Yoon’s research team developed an experimental technique using magnetic nanotweezers to measure physical changes of proteins to nanometer level by pulling and releasing each protein with force of 1 pN. The research identified the existence of hidden SNARE protein"s intermediate structure and its formation. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been discovered.
Professor Yoon said, “Ground breaking research results have been produced. A simple experimental technique of applying the smallest possible forces to proteins (with tweezers) to see their hidden structure and formation process can produce the same result as real observation has been developed.” He continued, “This technique will be very important in researching biological object with physical experimental technique. It will be a vital foundation to consilient research of different academia in the future.”
This research was a joint project of Physics Department’s Professor Tae-Young Yoon, KAIST, and Biomedical Engineering Institute’s Professor Yeon-Kyun Shin at KIST. KAIST Physics Department’s Professor Yong-Hoon Cho, Ph.D. candidate Do-Yong Lee and KIAS Computational Sciences Department’s Professor Chang-Bong Hyun participated. The research was published on Nature Communications on April 16th.
a) Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. A SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters.
b) A schematic diagram using magnetic nanotweezers to measure protein structure changes on molecular level. The nanotweezers exert an exquisite pull and release of each protein with a force of 1 pN to measure physical changes to nanometer level in real-time to observe the hidden intermediate structure and operation principles of bio-membrane fusion protein.
2013.05.25 View 10303 -
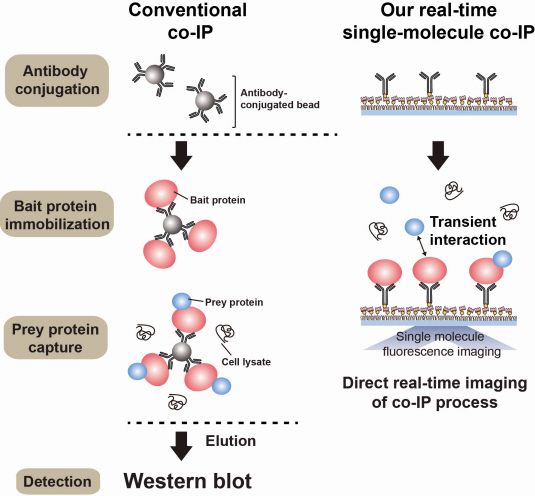 The new era of personalized cancer diagnosis and treatment
Professor Tae-Young Yoon
- Succeeded in observing carcinogenic protein at the molecular level
- “Paved the way to customized cancer treatment through accurate analysis of carcinogenic protein”
The joint KAIST research team of Professor Tae Young Yoon of the Department of Physics and Professor Won Do Huh of the Department of Biological Sciences have developed the technology to monitor characteristics of carcinogenic protein in cancer tissue – for the first time in the world.
The technology makes it possible to analyse the mechanism of cancer development through a small amount of carcinogenic protein from a cancer patient. Therefore, a personalised approach to diagnosis and treatment using the knowledge of the specific mechanism of cancer development in the patient may be possible in the future.
Until recently, modern medicine could only speculate on the cause of cancer through statistics. Although developed countries, such as the United States, are known to use a large sequencing technology that analyses the patient’s DNA, identification of the interactions between proteins responsible for causing cancer remained an unanswered question for a long time in medicine.
Firstly, Professor Yoon’s research team has developed a fluorescent microscope that can observe even a single molecule. Then, the “Immunoprecipitation method”, a technology to extract a specific protein exploiting the high affinity between antigens and antibodies was developed. Using this technology and the microscope, “Real-Time Single Molecule co-Immunoprecipitation Method” was created. In this way, the team succeeded in observing the interactions between carcinogenic and other proteins at a molecular level, in real time.
To validate the developed technology, the team investigated Ras, a carcinogenic protein; its mutation statistically is known to cause around 30% of cancers.
The experimental results confirmed that 30-50% of Ras protein was expressed in mouse tumour and human cancer cells. In normal cells, less than 5% of Ras protein was expressed. Thus, the experiment showed that unusual increase in activation of Ras protein induces cancer.
The increase in the ratio of active Ras protein can be inferred from existing research data but the measurement of specific numerical data has never been done before.
The team suggested a new molecular level diagnosis technique of identifying the progress of cancer in patients through measuring the percentage of activated carcinogenic protein in cancer tissue.
Professor Yoon Tae-young said, “This newly developed technology does not require a separate procedure of protein expression or refining, hence the existing proteins in real biological tissues or cancer cells can be observed directly.” He also said, “Since carcinogenic protein can be analyzed accurately, it has opened up the path to customized cancer treatment in the future.”
“Since the observation is possible on a molecular level, the technology confers the advantage that researchers can carry out various examinations on a small sample of the cancer patient.” He added, “The clinical trial will start in December 2012 and in a few years customized cancer diagnosis and treatment will be possible.”
Meanwhile, the research has been published in Nature Communications (February 19). Many researchers from various fields have participated, regardless of the differences in their speciality, and successfully produced interdisciplinary research. Professor Tae Young Yoon of the Department of Physics and Professors Dae Sik Lim and Won Do Huh of Biological Sciences at KAIST, and Professor Chang Bong Hyun of Computational Science of KIAS contributed to developing the technique.
Figure 1: Schematic diagram of observed interactions at the molecular level in real time using fluorescent microscope. The carcinogenic protein from a mouse tumour is fixed on the microchip, and its molecular characteristics are observed live.
Figure 2: Molecular interaction data using a molecular level fluorescent microscope. A signal in the form of spike is shown when two proteins combine. This is monitored live using an Electron Multiplying Charge Coupled Device (EMCCD). It shows signal results in bright dots.
An organism has an immune system as a defence mechanism to foreign intruders. The immune system is activated when unwanted pathogens or foreign protein are in the body. Antibodies form in recognition of the specific antigen to protect itself. Organisms evolved to form antibodies with high specificity to a certain antigen. Antibodies only react to its complementary antigens. The field of molecular biology uses the affinity between antigens and antibodies to extract specific proteins; a technology called immunoprecipitation. Even in a mixture of many proteins, the protein sought can be extracted using antibodies. Thus immunoprecipitation is widely used to detect pathogens or to extract specific proteins.
Technology co-IP is a well-known example that uses immunoprecipitation. The research on interactions between proteins uses co-IP in general. The basis of fixing the antigen on the antibody to extract antigen protein is the same as immunoprecipitation. Then, researchers inject and observe its reaction with the partner protein to observe the interactions and precipitate the antibodies. If the reaction occurs, the partner protein will be found with the antibodies in the precipitations. If not, then the partner protein will not be found. This shows that the two proteins interact.
However, the traditional co-IP can be used to infer the interactions between the two proteins although the information of the dynamics on how the reaction occurs is lost. To overcome these shortcomings, the Real-Time Single Molecule co-IP Method enables observation on individual protein level in real time. Therefore, the significance of the new technique is in making observation of interactions more direct and quantitative.
Additional Figure 1: Comparison between Conventional co-IP and Real-Time Single Molecule co-IP
2013.04.01 View 20926
The new era of personalized cancer diagnosis and treatment
Professor Tae-Young Yoon
- Succeeded in observing carcinogenic protein at the molecular level
- “Paved the way to customized cancer treatment through accurate analysis of carcinogenic protein”
The joint KAIST research team of Professor Tae Young Yoon of the Department of Physics and Professor Won Do Huh of the Department of Biological Sciences have developed the technology to monitor characteristics of carcinogenic protein in cancer tissue – for the first time in the world.
The technology makes it possible to analyse the mechanism of cancer development through a small amount of carcinogenic protein from a cancer patient. Therefore, a personalised approach to diagnosis and treatment using the knowledge of the specific mechanism of cancer development in the patient may be possible in the future.
Until recently, modern medicine could only speculate on the cause of cancer through statistics. Although developed countries, such as the United States, are known to use a large sequencing technology that analyses the patient’s DNA, identification of the interactions between proteins responsible for causing cancer remained an unanswered question for a long time in medicine.
Firstly, Professor Yoon’s research team has developed a fluorescent microscope that can observe even a single molecule. Then, the “Immunoprecipitation method”, a technology to extract a specific protein exploiting the high affinity between antigens and antibodies was developed. Using this technology and the microscope, “Real-Time Single Molecule co-Immunoprecipitation Method” was created. In this way, the team succeeded in observing the interactions between carcinogenic and other proteins at a molecular level, in real time.
To validate the developed technology, the team investigated Ras, a carcinogenic protein; its mutation statistically is known to cause around 30% of cancers.
The experimental results confirmed that 30-50% of Ras protein was expressed in mouse tumour and human cancer cells. In normal cells, less than 5% of Ras protein was expressed. Thus, the experiment showed that unusual increase in activation of Ras protein induces cancer.
The increase in the ratio of active Ras protein can be inferred from existing research data but the measurement of specific numerical data has never been done before.
The team suggested a new molecular level diagnosis technique of identifying the progress of cancer in patients through measuring the percentage of activated carcinogenic protein in cancer tissue.
Professor Yoon Tae-young said, “This newly developed technology does not require a separate procedure of protein expression or refining, hence the existing proteins in real biological tissues or cancer cells can be observed directly.” He also said, “Since carcinogenic protein can be analyzed accurately, it has opened up the path to customized cancer treatment in the future.”
“Since the observation is possible on a molecular level, the technology confers the advantage that researchers can carry out various examinations on a small sample of the cancer patient.” He added, “The clinical trial will start in December 2012 and in a few years customized cancer diagnosis and treatment will be possible.”
Meanwhile, the research has been published in Nature Communications (February 19). Many researchers from various fields have participated, regardless of the differences in their speciality, and successfully produced interdisciplinary research. Professor Tae Young Yoon of the Department of Physics and Professors Dae Sik Lim and Won Do Huh of Biological Sciences at KAIST, and Professor Chang Bong Hyun of Computational Science of KIAS contributed to developing the technique.
Figure 1: Schematic diagram of observed interactions at the molecular level in real time using fluorescent microscope. The carcinogenic protein from a mouse tumour is fixed on the microchip, and its molecular characteristics are observed live.
Figure 2: Molecular interaction data using a molecular level fluorescent microscope. A signal in the form of spike is shown when two proteins combine. This is monitored live using an Electron Multiplying Charge Coupled Device (EMCCD). It shows signal results in bright dots.
An organism has an immune system as a defence mechanism to foreign intruders. The immune system is activated when unwanted pathogens or foreign protein are in the body. Antibodies form in recognition of the specific antigen to protect itself. Organisms evolved to form antibodies with high specificity to a certain antigen. Antibodies only react to its complementary antigens. The field of molecular biology uses the affinity between antigens and antibodies to extract specific proteins; a technology called immunoprecipitation. Even in a mixture of many proteins, the protein sought can be extracted using antibodies. Thus immunoprecipitation is widely used to detect pathogens or to extract specific proteins.
Technology co-IP is a well-known example that uses immunoprecipitation. The research on interactions between proteins uses co-IP in general. The basis of fixing the antigen on the antibody to extract antigen protein is the same as immunoprecipitation. Then, researchers inject and observe its reaction with the partner protein to observe the interactions and precipitate the antibodies. If the reaction occurs, the partner protein will be found with the antibodies in the precipitations. If not, then the partner protein will not be found. This shows that the two proteins interact.
However, the traditional co-IP can be used to infer the interactions between the two proteins although the information of the dynamics on how the reaction occurs is lost. To overcome these shortcomings, the Real-Time Single Molecule co-IP Method enables observation on individual protein level in real time. Therefore, the significance of the new technique is in making observation of interactions more direct and quantitative.
Additional Figure 1: Comparison between Conventional co-IP and Real-Time Single Molecule co-IP
2013.04.01 View 20926 -
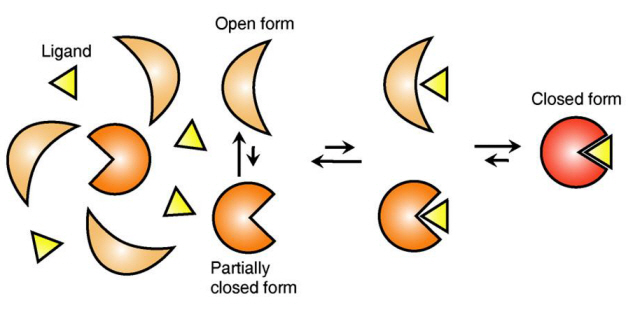 Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 13277
Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 13277 -
 An efficient strategy for developing microbial cell factories by employing synthetic small regulatory RNAs
A new metabolic engineering tool that allows fine control of gene expression level by employing synthetic small regulatory RNAs was developed to efficiently construct microbial cell factories producing desired chemicals and materials
Biotechnologists have been working hard to address the climate change and limited fossil resource issues through the development of sustainable processes for the production of chemicals, fuels and materials from renewable non-food biomass. One promising sustainable technology is the use of microbial cell factories for the efficient production of desired chemicals and materials. When microorganisms are isolated from nature, the performance in producing our desired product is rather poor. That is why metabolic engineering is performed to improve the metabolic and cellular characteristics to achieve enhanced production of desired product at high yield and productivity. Since the performance of microbial cell factory is very important in lowering the overall production cost of the bioprocess, many different strategies and tools have been developed for the metabolic engineering of microorganisms.
One of the big challenges in metabolic engineering is to find the best platform organism and to find those genes to be engineered so as to maximize the production efficiency of the desired chemical. Even Escherichia coli, the most widely utilized simple microorganism, has thousands of genes, the expression of which is highly regulated and interconnected to finely control cellular and metabolic activities. Thus, the complexity of cellular genetic interactions is beyond our intuition and thus it is very difficult to find effective target genes to engineer. Together with gene amplification strategy, gene knockout strategy has been an essential tool in metabolic engineering to redirect the pathway fluxes toward our desired product formation. However, experiment to engineer many genes can be rather difficult due to the time and effort required; for example, gene deletion experiment can take a few weeks depending on the microorganisms. Furthermore, as certain genes are essential or play important roles for the survival of a microorganism, gene knockout experiments cannot be performed. Even worse, there are many different microbial strains one can employ. There are more than 50 different E. coli strains that metabolic engineer can consider to use. Since gene knockout experiment is hard-coded (that is, one should repeat the gene knockout experiments for each strain), the result cannot be easily transferred from one strain to another.
A paper published in Nature Biotechnology online today addresses this issue and suggests a new strategy for identifying gene targets to be knocked out or knocked down through the use of synthetic small RNA. A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), a prestigeous science and engineering university in Korea reported that synthetic small RNA can be employed for finely controlling the expression levels of multiple genes at the translation level. Already well-known for their systems metabolic engineering strategies, Professor Lee’s team added one more strategy to efficiently develop microbial cell factories for the production of chemicals and materials.
Gene expression works like this: the hard-coded blueprint (DNA) is transcribed into messenger RNA (mRNA), and the coding information in mRNA is read to produce protein by ribosomes. Conventional genetic engineering approaches have often targeted modification of the blueprint itself (DNA) to alter organism’s physiological characteristics. Again, engineering the blueprint itself takes much time and effort, and in addition, the results obtained cannot be transferred to another organism without repeating the whole set of experiments. This is why Professor Lee and his colleagues aimed at controlling the gene expression level at the translation stage through the use of synthetic small RNA. They created novel RNAs that can regulate the translation of multiple messenger RNAs (mRNA), and consequently varying the expression levels of multiple genes at the same time. Briefly, synthetic regulatory RNAs interrupt gene expression process from DNA to protein by destroying the messenger RNAs to different yet controllable extents. The advantages of taking this strategy of employing synthetic small regulatory RNAs include simple, easy and high-throughput identification of gene knockout or knockdown targets, fine control of gene expression levels, transferability to many different host strains, and possibility of identifying those gene targets that are essential.
As proof-of-concept demonstration of the usefulness of this strategy, Professor Lee and his colleagues applied it to develop engineered E. coli strains capable of producing an aromatic amino acid tyrosine, which is used for stress symptom relief, food supplements, and precursor for many drugs. They examined a large number of genes in multiple E. coli strains, and developed a highly efficient tyrosine producer. Also, they were able to show that this strategy can be employed to an already metabolically engineered E. coli strain for further improvement by demonstrating the development of highly efficient producer of cadaverine, an important platform chemical for nylon in the chemical industry.
This new strategy, being simple yet very powerful for systems metabolic engineering, is thus expected to facilitate the efficient development of microbial cell factories capable of producing chemicals, fuels and materials from renewable biomass.
Source: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park, and Sang Yup Lee, “Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs”, Nature Biotechnology, doi:10.1038/nbt.2461 (2013)
2013.03.19 View 11336
An efficient strategy for developing microbial cell factories by employing synthetic small regulatory RNAs
A new metabolic engineering tool that allows fine control of gene expression level by employing synthetic small regulatory RNAs was developed to efficiently construct microbial cell factories producing desired chemicals and materials
Biotechnologists have been working hard to address the climate change and limited fossil resource issues through the development of sustainable processes for the production of chemicals, fuels and materials from renewable non-food biomass. One promising sustainable technology is the use of microbial cell factories for the efficient production of desired chemicals and materials. When microorganisms are isolated from nature, the performance in producing our desired product is rather poor. That is why metabolic engineering is performed to improve the metabolic and cellular characteristics to achieve enhanced production of desired product at high yield and productivity. Since the performance of microbial cell factory is very important in lowering the overall production cost of the bioprocess, many different strategies and tools have been developed for the metabolic engineering of microorganisms.
One of the big challenges in metabolic engineering is to find the best platform organism and to find those genes to be engineered so as to maximize the production efficiency of the desired chemical. Even Escherichia coli, the most widely utilized simple microorganism, has thousands of genes, the expression of which is highly regulated and interconnected to finely control cellular and metabolic activities. Thus, the complexity of cellular genetic interactions is beyond our intuition and thus it is very difficult to find effective target genes to engineer. Together with gene amplification strategy, gene knockout strategy has been an essential tool in metabolic engineering to redirect the pathway fluxes toward our desired product formation. However, experiment to engineer many genes can be rather difficult due to the time and effort required; for example, gene deletion experiment can take a few weeks depending on the microorganisms. Furthermore, as certain genes are essential or play important roles for the survival of a microorganism, gene knockout experiments cannot be performed. Even worse, there are many different microbial strains one can employ. There are more than 50 different E. coli strains that metabolic engineer can consider to use. Since gene knockout experiment is hard-coded (that is, one should repeat the gene knockout experiments for each strain), the result cannot be easily transferred from one strain to another.
A paper published in Nature Biotechnology online today addresses this issue and suggests a new strategy for identifying gene targets to be knocked out or knocked down through the use of synthetic small RNA. A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), a prestigeous science and engineering university in Korea reported that synthetic small RNA can be employed for finely controlling the expression levels of multiple genes at the translation level. Already well-known for their systems metabolic engineering strategies, Professor Lee’s team added one more strategy to efficiently develop microbial cell factories for the production of chemicals and materials.
Gene expression works like this: the hard-coded blueprint (DNA) is transcribed into messenger RNA (mRNA), and the coding information in mRNA is read to produce protein by ribosomes. Conventional genetic engineering approaches have often targeted modification of the blueprint itself (DNA) to alter organism’s physiological characteristics. Again, engineering the blueprint itself takes much time and effort, and in addition, the results obtained cannot be transferred to another organism without repeating the whole set of experiments. This is why Professor Lee and his colleagues aimed at controlling the gene expression level at the translation stage through the use of synthetic small RNA. They created novel RNAs that can regulate the translation of multiple messenger RNAs (mRNA), and consequently varying the expression levels of multiple genes at the same time. Briefly, synthetic regulatory RNAs interrupt gene expression process from DNA to protein by destroying the messenger RNAs to different yet controllable extents. The advantages of taking this strategy of employing synthetic small regulatory RNAs include simple, easy and high-throughput identification of gene knockout or knockdown targets, fine control of gene expression levels, transferability to many different host strains, and possibility of identifying those gene targets that are essential.
As proof-of-concept demonstration of the usefulness of this strategy, Professor Lee and his colleagues applied it to develop engineered E. coli strains capable of producing an aromatic amino acid tyrosine, which is used for stress symptom relief, food supplements, and precursor for many drugs. They examined a large number of genes in multiple E. coli strains, and developed a highly efficient tyrosine producer. Also, they were able to show that this strategy can be employed to an already metabolically engineered E. coli strain for further improvement by demonstrating the development of highly efficient producer of cadaverine, an important platform chemical for nylon in the chemical industry.
This new strategy, being simple yet very powerful for systems metabolic engineering, is thus expected to facilitate the efficient development of microbial cell factories capable of producing chemicals, fuels and materials from renewable biomass.
Source: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park, and Sang Yup Lee, “Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs”, Nature Biotechnology, doi:10.1038/nbt.2461 (2013)
2013.03.19 View 11336 -
 New BioFactory Technique Developed using sRNAs
Professor Sang Yup Lee
- published on the online edition of Nature Biotechnology. “Expected as a new strategy for the bio industry that may replace the chemical industry.”-
KAIST Chemical & Biomolecular engineering department’s Professor Sang Yup Lee and his team has developed a new technology that utilizes the synthetic small regulatory RNAs (sRNAs) to implement the BioFactory in a larger scale with more effectiveness.
* BioFactory: Microbial-based production system which creates the desired compound in mass by manipulating the genes of the cell.
In order to solve the problems of modern society, such as environmental pollution caused by the exhaustion of fossil fuels and usage of petrochemical products, an eco-friendly and sustainable bio industry is on the rise. BioFactory development technology has especially attracted the attention world-wide, with its ability to produce bio-energy, pharmaceuticals, eco-friendly materials and more.
For the development of an excellent BioFactory, selection for the gene that produces the desired compounds must be accompanied by finding the microorganism with high production efficiency; however, the previous research method had a complicated and time-consuming problem of having to manipulate the genes of the microorganism one by one.
Professor Sang Yup Lee’s research team, including Dr. Dokyun Na and Dr. Seung Min Yoo, has produced the synthetic sRNAs and utilized it to overcome the technical limitations mentioned above.
In particular, unlike the existing method, this technology using synthetic sRNAs exhibits no strain specificity which can dramatically shorten the experiment that used to take months to just a few days.
The research team applied the synthetic small regulatory RNA technology to the production of the tyrosine*, which is used as the precursor of the medicinal compound, and cadaverine**, widely utilized in a variety of petrochemical products, and has succeeded developing BioFactory with the world’s highest yield rate (21.9g /L, 12.6g / L each).
*tyrosine: amino acid known to control stress and improve concentration
**cadaverine: base material used in many petrochemical products, such as polyurethane
Professor Sang Yup Lee highlighted the significance of this research: “it is expected the synthetic small regulatory RNA technology will stimulate the BioFactory development and also serve as a catalyst which can make the chemical industry, currently represented by its petroleum energy, transform into bio industry.”
The study was carried out with the support of Global Frontier Project (Intelligent Bio-Systems Design and Synthesis Research Unit (Chief Seon Chang Kim)) and the findings have been published on January 20th in the online edition of the worldwide journal Nature Biotechnology.
2013.02.21 View 13043
New BioFactory Technique Developed using sRNAs
Professor Sang Yup Lee
- published on the online edition of Nature Biotechnology. “Expected as a new strategy for the bio industry that may replace the chemical industry.”-
KAIST Chemical & Biomolecular engineering department’s Professor Sang Yup Lee and his team has developed a new technology that utilizes the synthetic small regulatory RNAs (sRNAs) to implement the BioFactory in a larger scale with more effectiveness.
* BioFactory: Microbial-based production system which creates the desired compound in mass by manipulating the genes of the cell.
In order to solve the problems of modern society, such as environmental pollution caused by the exhaustion of fossil fuels and usage of petrochemical products, an eco-friendly and sustainable bio industry is on the rise. BioFactory development technology has especially attracted the attention world-wide, with its ability to produce bio-energy, pharmaceuticals, eco-friendly materials and more.
For the development of an excellent BioFactory, selection for the gene that produces the desired compounds must be accompanied by finding the microorganism with high production efficiency; however, the previous research method had a complicated and time-consuming problem of having to manipulate the genes of the microorganism one by one.
Professor Sang Yup Lee’s research team, including Dr. Dokyun Na and Dr. Seung Min Yoo, has produced the synthetic sRNAs and utilized it to overcome the technical limitations mentioned above.
In particular, unlike the existing method, this technology using synthetic sRNAs exhibits no strain specificity which can dramatically shorten the experiment that used to take months to just a few days.
The research team applied the synthetic small regulatory RNA technology to the production of the tyrosine*, which is used as the precursor of the medicinal compound, and cadaverine**, widely utilized in a variety of petrochemical products, and has succeeded developing BioFactory with the world’s highest yield rate (21.9g /L, 12.6g / L each).
*tyrosine: amino acid known to control stress and improve concentration
**cadaverine: base material used in many petrochemical products, such as polyurethane
Professor Sang Yup Lee highlighted the significance of this research: “it is expected the synthetic small regulatory RNA technology will stimulate the BioFactory development and also serve as a catalyst which can make the chemical industry, currently represented by its petroleum energy, transform into bio industry.”
The study was carried out with the support of Global Frontier Project (Intelligent Bio-Systems Design and Synthesis Research Unit (Chief Seon Chang Kim)) and the findings have been published on January 20th in the online edition of the worldwide journal Nature Biotechnology.
2013.02.21 View 13043 -
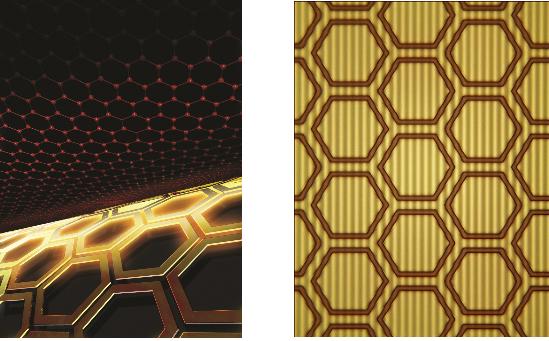 The control of light at the nano-level
Professor Min Bumki
Professor Min Bumki’s research team from the Department of Mechanical Engineering at KAIST have successfully gained control of the transmittance of light in optical devices using graphene* and artificial 2-dimensional metamaterials**.
* Graphene : a thin membrane composed of pure carbon, with atoms arranged in a regular hexagonal pattern
** Metamaterials : artificial materials engineered to have properties that may not be found in nature
The research results were published in the recent online edition (September 30th) of Nature Materials, a sister journal of the world renowned Nature journal, under the title ‘Terahertz waves with gate-controlled active graphene metamaterials’
Since the discovery of graphene in 2004 by Professors Novoselov and Geim from the University of Manchester (2010 Nobel Prize winners in Physics), it has been dubbed “the dream material” because of its outstanding physical properties.
Graphene has been especially praised for its ability to absorb approximately 2.3% of near infrared and visible rays due to its characteristic electron structure. This property allows graphene to be used as a transparent electrode, which is a vital electrical component used in touch screens and solar batteries. However, graphene’s optical transmittance was largely ignored by researchers due to its limited control using electrical methods and its small optical modulation in data transfer.
Professor Min’s team combined 0.34 nanometer-thick graphene with metamaterials to allow a more effective control of light transmittance and greater optical modulation. This graphene metamaterial can be integrated in to a thin and flexible macromolecule substrate which allows the control of transmittance using electric signals.
This research experimentally showed that graphene metamaterials can not only effective control optical transmittance, but can also be used in graphene optical memory devices using electrical hysteresis.
Professor Min said that “this research allows the effective control of light at the nanometer level” and that “this research will help in the development of microscopic optical modulators or memory disks”.
figure 1. The working drawing of graphene metamaterials
figure 2. Conceptual diagram (Left) and microscopic photo (right) of graphene metamaterials
2012.11.23 View 11698
The control of light at the nano-level
Professor Min Bumki
Professor Min Bumki’s research team from the Department of Mechanical Engineering at KAIST have successfully gained control of the transmittance of light in optical devices using graphene* and artificial 2-dimensional metamaterials**.
* Graphene : a thin membrane composed of pure carbon, with atoms arranged in a regular hexagonal pattern
** Metamaterials : artificial materials engineered to have properties that may not be found in nature
The research results were published in the recent online edition (September 30th) of Nature Materials, a sister journal of the world renowned Nature journal, under the title ‘Terahertz waves with gate-controlled active graphene metamaterials’
Since the discovery of graphene in 2004 by Professors Novoselov and Geim from the University of Manchester (2010 Nobel Prize winners in Physics), it has been dubbed “the dream material” because of its outstanding physical properties.
Graphene has been especially praised for its ability to absorb approximately 2.3% of near infrared and visible rays due to its characteristic electron structure. This property allows graphene to be used as a transparent electrode, which is a vital electrical component used in touch screens and solar batteries. However, graphene’s optical transmittance was largely ignored by researchers due to its limited control using electrical methods and its small optical modulation in data transfer.
Professor Min’s team combined 0.34 nanometer-thick graphene with metamaterials to allow a more effective control of light transmittance and greater optical modulation. This graphene metamaterial can be integrated in to a thin and flexible macromolecule substrate which allows the control of transmittance using electric signals.
This research experimentally showed that graphene metamaterials can not only effective control optical transmittance, but can also be used in graphene optical memory devices using electrical hysteresis.
Professor Min said that “this research allows the effective control of light at the nanometer level” and that “this research will help in the development of microscopic optical modulators or memory disks”.
figure 1. The working drawing of graphene metamaterials
figure 2. Conceptual diagram (Left) and microscopic photo (right) of graphene metamaterials
2012.11.23 View 11698