Drug+resistance
-
 What Fuels a “Domino Effect” in Cancer Drug Resistance?
KAIST researchers have identified mechanisms that relay prior acquired resistance to the first-line chemotherapy to the second-line targeted therapy, fueling a “domino effect” in cancer drug resistance. Their study featured in the February 7 edition of Science Advances suggests a new strategy for improving the second-line setting of cancer treatment for patients who showed resistance to anti-cancer drugs.
Resistance to cancer drugs is often managed in the clinic by chemotherapy and targeted therapy. Unlike chemotherapy that works by repressing fast-proliferating cells, targeted therapy blocks a single oncogenic pathway to halt tumor growth. In many cases, targeted therapy is engaged as a maintenance therapy or employed in the second-line after front-line chemotherapy.
A team of researchers led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering and the KAIST Institute for Health Science and Technology (KIHST) has discovered an unexpected resistance signature that occurs between chemotherapy and targeted therapy. The team further identified a set of integrated mechanisms that promotes this kind of sequential therapy resistance.
“There have been multiple clinical accounts reflecting that targeted therapies tend to be least successful in patients who have exhausted all standard treatments,” said the first author of the paper Mark Borris D. Aldonza. He continued, “These accounts ignited our hypothesis that failed responses to some chemotherapies might speed up the evolution of resistance to other drugs, particularly those with specific targets.”
Aldonza and his colleagues extracted large amounts of drug-resistance information from the open-source database the Genomics of Drug Sensitivity in Cancer (GDSC), which contains thousands of drug response data entries from various human cancer cell lines. Their big data analysis revealed that cancer cell lines resistant to chemotherapies classified as anti-mitotic drugs (AMDs), toxins that inhibit overacting cell division, are also resistant to a class of targeted therapies called epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs).
In all of the cancer types analyzed, more than 84 percent of those resistant to AMDs, representatively ‘paclitaxel’, were also resistant to at least nine EGFR-TKIs. In lung, pancreatic, and breast cancers where paclitaxel is often used as a first-line, standard-of-care regimen, greater than 92 percent showed resistance to EGFR-TKIs. Professor Kim said, “It is surprising to see that such collateral resistance can occur specifically between two chemically different classes of drugs.”
To figure out how failed responses to paclitaxel leads to resistance to EGFR-TKIs, the team validated co-resistance signatures that they found in the database by generating and analyzing a subset of slow-doubling, paclitaxel-resistant cancer models called ‘persisters’.
The results demonstrated that paclitaxel-resistant cancers remodel their stress response by first becoming more stem cell-like, evolving the ability to self-renew to adapt to more stressful conditions like drug exposures. More surprisingly, when the researchers characterized the metabolic state of the cells, EGFR-TKI persisters derived from paclitaxel-resistant cancer cells showed high dependencies to energy-producing processes such as glycolysis and glutaminolysis.
“We found that, without an energy stimulus like glucose, these cells transform to becoming more senescent, a characteristic of cells that have arrested cell division. However, this senescence is controlled by stem cell factors, which the paclitaxel-resistant cancers use to escape from this arrested state given a favorable condition to re-grow,” said Aldonza.
Professor Kim explained, “Before this research, there was no reason to expect that acquiring the cancer stem cell phenotype that dramatically leads to a cascade of changes in cellular states affecting metabolism and cell death is linked with drug-specific sequential resistance between two classes of therapies.”
He added, “The expansion of our work to other working models of drug resistance in a much more clinically-relevant setting, perhaps in clinical trials, will take on increasing importance, as sequential treatment strategies will continue to be adapted to various forms of anti-cancer therapy regimens.”
This study was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF-2016R1C1B2009886), and the KAIST Future Systems Healthcare Project (KAISTHEALTHCARE42) funded by the Korean Ministry of Science and ICT (MSIT). Undergraduate student Aldonza participated in this research project and presented the findings as the lead author as part of the Undergraduate Research Participation (URP) Program at KAIST.
< Figure 1. Schematic overview of the study. >
< Figure 2. Big data analysis revealing co-resistance signatures between classes of anti-cancer drugs. >
Publication:
Aldonza et al. (2020) Prior acquired resistance to paclitaxel relays diverse EGFR-targeted therapy persistence mechanisms. Science Advances, Vol. 6, No. 6, eaav7416. Available online at http://dx.doi.org/10.1126/sciadv.aav7416
Profile: Prof. Yoosik Kim, MA, PhD
ysyoosik@kaist.ac.kr
https://qcbio.kaist.ac.kr/
Assistant Professor
Bio Network Analysis Laboratory
Department of Chemical and Biomolecular Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon, Republic of Korea
Profile: Mark Borris D. Aldonza
borris@kaist.ac.kr
Undergraduate Student
Department of Biological Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon, Republic of Korea
(END)
2020.02.10 View 16860
What Fuels a “Domino Effect” in Cancer Drug Resistance?
KAIST researchers have identified mechanisms that relay prior acquired resistance to the first-line chemotherapy to the second-line targeted therapy, fueling a “domino effect” in cancer drug resistance. Their study featured in the February 7 edition of Science Advances suggests a new strategy for improving the second-line setting of cancer treatment for patients who showed resistance to anti-cancer drugs.
Resistance to cancer drugs is often managed in the clinic by chemotherapy and targeted therapy. Unlike chemotherapy that works by repressing fast-proliferating cells, targeted therapy blocks a single oncogenic pathway to halt tumor growth. In many cases, targeted therapy is engaged as a maintenance therapy or employed in the second-line after front-line chemotherapy.
A team of researchers led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering and the KAIST Institute for Health Science and Technology (KIHST) has discovered an unexpected resistance signature that occurs between chemotherapy and targeted therapy. The team further identified a set of integrated mechanisms that promotes this kind of sequential therapy resistance.
“There have been multiple clinical accounts reflecting that targeted therapies tend to be least successful in patients who have exhausted all standard treatments,” said the first author of the paper Mark Borris D. Aldonza. He continued, “These accounts ignited our hypothesis that failed responses to some chemotherapies might speed up the evolution of resistance to other drugs, particularly those with specific targets.”
Aldonza and his colleagues extracted large amounts of drug-resistance information from the open-source database the Genomics of Drug Sensitivity in Cancer (GDSC), which contains thousands of drug response data entries from various human cancer cell lines. Their big data analysis revealed that cancer cell lines resistant to chemotherapies classified as anti-mitotic drugs (AMDs), toxins that inhibit overacting cell division, are also resistant to a class of targeted therapies called epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs).
In all of the cancer types analyzed, more than 84 percent of those resistant to AMDs, representatively ‘paclitaxel’, were also resistant to at least nine EGFR-TKIs. In lung, pancreatic, and breast cancers where paclitaxel is often used as a first-line, standard-of-care regimen, greater than 92 percent showed resistance to EGFR-TKIs. Professor Kim said, “It is surprising to see that such collateral resistance can occur specifically between two chemically different classes of drugs.”
To figure out how failed responses to paclitaxel leads to resistance to EGFR-TKIs, the team validated co-resistance signatures that they found in the database by generating and analyzing a subset of slow-doubling, paclitaxel-resistant cancer models called ‘persisters’.
The results demonstrated that paclitaxel-resistant cancers remodel their stress response by first becoming more stem cell-like, evolving the ability to self-renew to adapt to more stressful conditions like drug exposures. More surprisingly, when the researchers characterized the metabolic state of the cells, EGFR-TKI persisters derived from paclitaxel-resistant cancer cells showed high dependencies to energy-producing processes such as glycolysis and glutaminolysis.
“We found that, without an energy stimulus like glucose, these cells transform to becoming more senescent, a characteristic of cells that have arrested cell division. However, this senescence is controlled by stem cell factors, which the paclitaxel-resistant cancers use to escape from this arrested state given a favorable condition to re-grow,” said Aldonza.
Professor Kim explained, “Before this research, there was no reason to expect that acquiring the cancer stem cell phenotype that dramatically leads to a cascade of changes in cellular states affecting metabolism and cell death is linked with drug-specific sequential resistance between two classes of therapies.”
He added, “The expansion of our work to other working models of drug resistance in a much more clinically-relevant setting, perhaps in clinical trials, will take on increasing importance, as sequential treatment strategies will continue to be adapted to various forms of anti-cancer therapy regimens.”
This study was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF-2016R1C1B2009886), and the KAIST Future Systems Healthcare Project (KAISTHEALTHCARE42) funded by the Korean Ministry of Science and ICT (MSIT). Undergraduate student Aldonza participated in this research project and presented the findings as the lead author as part of the Undergraduate Research Participation (URP) Program at KAIST.
< Figure 1. Schematic overview of the study. >
< Figure 2. Big data analysis revealing co-resistance signatures between classes of anti-cancer drugs. >
Publication:
Aldonza et al. (2020) Prior acquired resistance to paclitaxel relays diverse EGFR-targeted therapy persistence mechanisms. Science Advances, Vol. 6, No. 6, eaav7416. Available online at http://dx.doi.org/10.1126/sciadv.aav7416
Profile: Prof. Yoosik Kim, MA, PhD
ysyoosik@kaist.ac.kr
https://qcbio.kaist.ac.kr/
Assistant Professor
Bio Network Analysis Laboratory
Department of Chemical and Biomolecular Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon, Republic of Korea
Profile: Mark Borris D. Aldonza
borris@kaist.ac.kr
Undergraduate Student
Department of Biological Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon, Republic of Korea
(END)
2020.02.10 View 16860 -
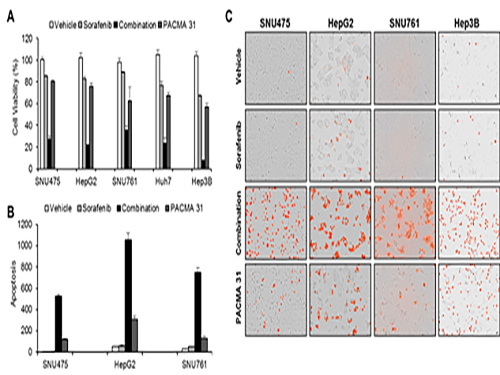 Discovery of an Optimal Drug Combination: Overcoming Resistance to Targeted Drugs for Liver Cancer
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences. Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)
(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)
(Figure 3. ER stress network model)
2017.08.30 View 13097
Discovery of an Optimal Drug Combination: Overcoming Resistance to Targeted Drugs for Liver Cancer
A KAIST research team presented a novel method for improving medication treatment for liver cancer using Systems Biology, combining research from information technology and the life sciences. Professor Kwang-Hyun Cho in the Department of Bio and Brain Engineering at KAIST conducted the research in collaboration with Professor Jung-Hwan Yoon in the Department of Internal Medicine at Seoul National University Hospital. This research was published in Hepatology in September 2017 (available online from August 24, 2017).
Liver cancer is the fifth and seventh most common cancer found in men and women throughout the world, which places it second in the cause of cancer deaths. In particular, Korea has 28.4 deaths from liver cancer per 100,000 persons, the highest death rate among OECD countries and twice that of Japan.
Each year in Korea, 16,000 people get liver cancer on average, yet the five-year survival rate stands below 12%. According to the National Cancer Information Center, lung cancer (17,399) took the highest portion of cancer-related deaths, followed by liver cancer (11,311) based on last year data.
Liver cancer is known to carry the highest social cost in comparison to other cancers and it causes the highest fatality in earlier age groups (40s-50s). In that sense, it is necessary to develop a new treatment that mitigates side effects yet elevates the survival rate.
There are ways in which liver cancer can be cured, such as surgery, embolization, and medication treatments; however, the options become limited for curing progressive cancer, a stage in which surgical methods cannot be executed.
Among anticancer medications, Sorafenib, a drug known for enhancing the survival rate of cancer patients, is a unique drug allowed for use as a targeted anticancer medication for progressive liver cancer patients. Its sales reached more than ten billion KRW annually in Korea, but its efficacy works on only about 20% of the treated patients. Also, acquired resistance to Sorafenib is emerging. Additionally, the action mechanism and resistance mechanism of Sorafenib is only vaguely identified.Although Sorafenib only extends the survival rate of terminal cancer patients less than three months on average, it is widely being used because drugs developed by global pharmaceutical companies failed to outperform its effectiveness.
Professor Cho’s research team analyzed the expression changes of genes in cell lines in response to Sorafenib in order to identify the effect and the resistance mechanism of Sorafenib.
As a result, the team discovered the resistance mechanism of Sorafenib using Systems Biology analysis. By combining computer simulations and biological experiments, it was revealed that protein disulfide isomerase (PDI) plays a crucial role in the resistance mechanism of Sorafenib and that its efficacy can be improved significantly by blocking PDI.
The research team used mice in the experiment and discovered the synergic effect of PDI inhibition with Sorafenib for reducing liver cancer cells, known as hepatocellular carcinoma. Also, more PDIs are shown in tissue from patients who possess a resistance to Sorafenib. From these findings, the team could identify the possibility of its clinical applications. The team also confirmed these findings from clinical data through a retrospective cohort study.
“Molecules that play an important role in cell lines are mostly put under complex regulation. For this reason, the existing biological research has a fundamental limitations for discovering its underlying principles,” Professor Cho said. “This research is a representative case of overcoming this limitation of traditional life science research by using a Systems Biology approach, combining IT and life science. It suggests the possibility of developing a new method that overcomes drug resistance with a network analysis of the targeted drug action mechanism of cancer.”
The research was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT.
(Figure 1. Simulation results from cellular experiments using hepatocellular carcinoma)
(Figure 2. Network analysis and computer simulation by using the endoplasmic reticulum (ER) stress network)
(Figure 3. ER stress network model)
2017.08.30 View 13097