Jae+Kyoung+Kim
-
 “One Experiment Is All It Takes”: KAIST Team Revolutionizes Drug Interaction Testing, Replacing 60,000 Studies
A groundbreaking new method developed by researchers at KAIST and Chungnam National University could drastically streamline drug interaction testing — replacing dozens of traditional experiments with just one.
The research, led by Professor Jae Kyoung Kim of KAIST Department of Mathematical Sciences & IBS Biomedical Mathematics Group and Professor Sang Kyum Kim of Chungnam National University's College of Pharmacy, introduces a novel analysis technique called 50-BOA, published in Nature Communications on June 5, 2025.
< Photo 1. (From left) Professor Sang Kyum Kim (Chungnam National University College of Pharmacy, co-corresponding author), Dr. Yun Min Song (IBS Biomedical Mathematics Group, formerly KAIST Department of Mathematical Sciences, co-first author), undergraduate student Hyeong Jun Jang (KAIST, co-first author), Professor Jae Kyoung Kim (KAIST and IBS Biomedical Mathematics Group, co-corresponding author) (Top left in the bubble) Professor Hwi-yeol Yun (Chungnam National University College of Pharmacy, co-author) >
For decades, scientists have had to repeat drug inhibition experiments across a wide range of concentrations to estimate inhibition constants — a process seen in over 60,000 scientific publications. But the KAIST-led team discovered that a single, well-chosen inhibitor concentration can yield even more accurate results.
< Figure 1. Graphical summary of 50-BOA. 50-BOA improves the accuracy and efficiency of inhibition constant estimation by using only a single inhibitor concentration instead of the traditionally used method of employing multiple inhibitor concentrations. >
“This approach challenges long-standing assumptions in experimental pharmacology,” says Prof. Kim. “It shows how mathematics can fundamentally redesign life science experiments.”
By mathematically analyzing the sources of error in conventional methods, the team found that over half the data typically collected adds no value or even skews results. Their new method not only cuts experimental effort by over 75%, but also enhances reproducibility and accuracy.
To help researchers adopt the method quickly, the team developed a user-friendly tool that takes simple Excel files as input, now freely available on GitHub:
☞ https://github.com/Mathbiomed/50-BOA
< Figure 2. The MATLAB and R package of 50-BOA at GitHub >
The work holds promise for faster and more reliable drug development, especially in assessing potential interactions in combination therapies. The U.S. FDA already emphasizes the importance of accurate enzyme inhibition assessment during early-stage drug evaluation — and this method could soon become a new gold standard.
2025.06.16 View 1658
“One Experiment Is All It Takes”: KAIST Team Revolutionizes Drug Interaction Testing, Replacing 60,000 Studies
A groundbreaking new method developed by researchers at KAIST and Chungnam National University could drastically streamline drug interaction testing — replacing dozens of traditional experiments with just one.
The research, led by Professor Jae Kyoung Kim of KAIST Department of Mathematical Sciences & IBS Biomedical Mathematics Group and Professor Sang Kyum Kim of Chungnam National University's College of Pharmacy, introduces a novel analysis technique called 50-BOA, published in Nature Communications on June 5, 2025.
< Photo 1. (From left) Professor Sang Kyum Kim (Chungnam National University College of Pharmacy, co-corresponding author), Dr. Yun Min Song (IBS Biomedical Mathematics Group, formerly KAIST Department of Mathematical Sciences, co-first author), undergraduate student Hyeong Jun Jang (KAIST, co-first author), Professor Jae Kyoung Kim (KAIST and IBS Biomedical Mathematics Group, co-corresponding author) (Top left in the bubble) Professor Hwi-yeol Yun (Chungnam National University College of Pharmacy, co-author) >
For decades, scientists have had to repeat drug inhibition experiments across a wide range of concentrations to estimate inhibition constants — a process seen in over 60,000 scientific publications. But the KAIST-led team discovered that a single, well-chosen inhibitor concentration can yield even more accurate results.
< Figure 1. Graphical summary of 50-BOA. 50-BOA improves the accuracy and efficiency of inhibition constant estimation by using only a single inhibitor concentration instead of the traditionally used method of employing multiple inhibitor concentrations. >
“This approach challenges long-standing assumptions in experimental pharmacology,” says Prof. Kim. “It shows how mathematics can fundamentally redesign life science experiments.”
By mathematically analyzing the sources of error in conventional methods, the team found that over half the data typically collected adds no value or even skews results. Their new method not only cuts experimental effort by over 75%, but also enhances reproducibility and accuracy.
To help researchers adopt the method quickly, the team developed a user-friendly tool that takes simple Excel files as input, now freely available on GitHub:
☞ https://github.com/Mathbiomed/50-BOA
< Figure 2. The MATLAB and R package of 50-BOA at GitHub >
The work holds promise for faster and more reliable drug development, especially in assessing potential interactions in combination therapies. The U.S. FDA already emphasizes the importance of accurate enzyme inhibition assessment during early-stage drug evaluation — and this method could soon become a new gold standard.
2025.06.16 View 1658 -
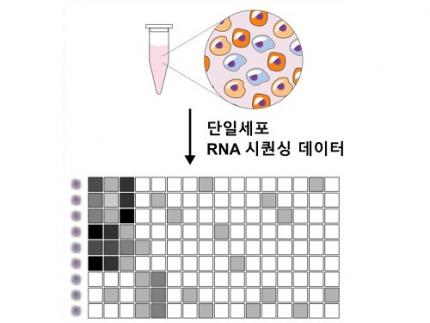 Revolutionary 'scLENS' Unveiled to Decode Complex Single-Cell Genomic Data
Unlocking biological information from complex single-cell genomic data has just become easier and more precise, thanks to the innovative 'scLENS' tool developed by the Biomedical Mathematics Group within the IBS Center for Mathematical and Computational Sciences led by Chief Investigator Jae Kyoung Kim, who is also a professor at KAIST. This new finding represents a significant leap forward in the field of single-cell transcriptomics.
Single-cell genomic analysis is an advanced technique that measures gene expression at the individual cell level, revealing cellular changes and interactions that are not observable with traditional genomic analysis methods. When applied to cancer tissues, this analysis can delineate the composition of diverse cell types within a tumor, providing insights into how cancer progresses and identifying key genes involved during each stage of progression.
Despite the immense potential of single-cell genomic analysis, handling the vast amount of data that it generates has always been challenging. The amount of data covers the expression of tens of thousands of genes across hundreds to thousands of individual cells. This not only results in large datasets but also introduces noise-related distortions, which arise in part due to current measurement limitations.
< Figure 1. Overview of scLENS (single-cell Low-dimensional embedding using the effective Noise Subtract) >
(Left) Current dimensionality reduction methods for scRNA-seq data involve conventional data preprocessing steps, such as log normalization, followed by manual selection of signals from the scaled data. However, this study reveals that the high levels of sparsity and variability in scRNA-seq data can lead to signal distortion during the data preprocessing, compromising the accuracy of downstream analyses.
(Right) To address this issue, the researchers integrated L2 normalization into the conventional preprocessing pipeline, effectively mitigating signal distortion. Moreover, they developed a novel signal detection algorithm that eliminates the need for user intervention by leveraging random matrix theory-based noise filtering and signal robustness testing. By incorporating these techniques, scLENS enables accurate and automated analysis of scRNA-seq data, overcoming the limitations of existing dimensionality reduction methods.
Corresponding author Jae Kyoung Kim highlighted, “There has been a remarkable advancement in experimental technologies for analyzing single-cell transcriptomes over the past decade. However, due to limitations in data analysis methods, there has been a struggle to fully utilize valuable data obtained through extensive cost and time."
Researchers have developed numerous analysis methods over the years to discern biological signals from this noise. However, the accuracy of these methods has been less than satisfactory. A critical issue is that determining signal and noise thresholds often depends on subjective decisions from the users.
The newly developed scLENS tool harnesses Random Matrix Theory and Signal robustness test to automatically differentiate signals from noise without relying on subjective user input.
First author Hyun Kim stated, "Previously, users had to arbitrarily decide the threshold for signal and noise, which compromised the reproducibility of analysis results and introduced subjectivity. scLENS eliminates this problem by automatically detecting signals using only the inherent structure of the data."
During the development of scLENS, researchers identified the fundamental reasons for inaccuracies in existing analysis methods. They found that commonly used data preprocessing methods distort both biological signals and noise. The new preprocessing approach that scLENS offers is free from such distortions.
By resolving issues related to noise threshold determined by subjective user choice and signal distortion in conventional data preprocessing, scLENS significantly outperforms existing methods in accuracy. Additionally, scLENS automates the laborious process of signal dimension selection, allowing researchers to extract biological signals conveniently and automatically.
CI Kim added, "scLENS solves major issues in single-cell transcriptome data analysis, substantially improving the accuracy and efficiency throughout the analysis process. This is a prime example of how fundamental mathematical theories can drive innovation in life sciences research, allowing researchers to more quickly and accurately answer biological questions and uncover secrets of life that were previously hidden."
This research was published in the international journal 'Nature Communications' on April 27.
Terminology
* Single-cell RNA sequencing (scRNA-seq): A technique used to measure gene expression levels in individual cells, providing insights into cell heterogeneity and rare cell types.
* Dimensionality reduction: A method to reduce the number of features or variables in a dataset while preserving the most important information, making data analysis more manageable and interpretable.
* Random matrix theory: A mathematical framework used to model and analyze the properties of large, random matrices, which can be applied to filter out noise in high-dimensional data.
* Signal robustness test: Among the signals, this test selects signals that are robust to the slight perturbation in data because real biological signals should be invariant for such slight modification in the data.
2024.05.09 View 6297
Revolutionary 'scLENS' Unveiled to Decode Complex Single-Cell Genomic Data
Unlocking biological information from complex single-cell genomic data has just become easier and more precise, thanks to the innovative 'scLENS' tool developed by the Biomedical Mathematics Group within the IBS Center for Mathematical and Computational Sciences led by Chief Investigator Jae Kyoung Kim, who is also a professor at KAIST. This new finding represents a significant leap forward in the field of single-cell transcriptomics.
Single-cell genomic analysis is an advanced technique that measures gene expression at the individual cell level, revealing cellular changes and interactions that are not observable with traditional genomic analysis methods. When applied to cancer tissues, this analysis can delineate the composition of diverse cell types within a tumor, providing insights into how cancer progresses and identifying key genes involved during each stage of progression.
Despite the immense potential of single-cell genomic analysis, handling the vast amount of data that it generates has always been challenging. The amount of data covers the expression of tens of thousands of genes across hundreds to thousands of individual cells. This not only results in large datasets but also introduces noise-related distortions, which arise in part due to current measurement limitations.
< Figure 1. Overview of scLENS (single-cell Low-dimensional embedding using the effective Noise Subtract) >
(Left) Current dimensionality reduction methods for scRNA-seq data involve conventional data preprocessing steps, such as log normalization, followed by manual selection of signals from the scaled data. However, this study reveals that the high levels of sparsity and variability in scRNA-seq data can lead to signal distortion during the data preprocessing, compromising the accuracy of downstream analyses.
(Right) To address this issue, the researchers integrated L2 normalization into the conventional preprocessing pipeline, effectively mitigating signal distortion. Moreover, they developed a novel signal detection algorithm that eliminates the need for user intervention by leveraging random matrix theory-based noise filtering and signal robustness testing. By incorporating these techniques, scLENS enables accurate and automated analysis of scRNA-seq data, overcoming the limitations of existing dimensionality reduction methods.
Corresponding author Jae Kyoung Kim highlighted, “There has been a remarkable advancement in experimental technologies for analyzing single-cell transcriptomes over the past decade. However, due to limitations in data analysis methods, there has been a struggle to fully utilize valuable data obtained through extensive cost and time."
Researchers have developed numerous analysis methods over the years to discern biological signals from this noise. However, the accuracy of these methods has been less than satisfactory. A critical issue is that determining signal and noise thresholds often depends on subjective decisions from the users.
The newly developed scLENS tool harnesses Random Matrix Theory and Signal robustness test to automatically differentiate signals from noise without relying on subjective user input.
First author Hyun Kim stated, "Previously, users had to arbitrarily decide the threshold for signal and noise, which compromised the reproducibility of analysis results and introduced subjectivity. scLENS eliminates this problem by automatically detecting signals using only the inherent structure of the data."
During the development of scLENS, researchers identified the fundamental reasons for inaccuracies in existing analysis methods. They found that commonly used data preprocessing methods distort both biological signals and noise. The new preprocessing approach that scLENS offers is free from such distortions.
By resolving issues related to noise threshold determined by subjective user choice and signal distortion in conventional data preprocessing, scLENS significantly outperforms existing methods in accuracy. Additionally, scLENS automates the laborious process of signal dimension selection, allowing researchers to extract biological signals conveniently and automatically.
CI Kim added, "scLENS solves major issues in single-cell transcriptome data analysis, substantially improving the accuracy and efficiency throughout the analysis process. This is a prime example of how fundamental mathematical theories can drive innovation in life sciences research, allowing researchers to more quickly and accurately answer biological questions and uncover secrets of life that were previously hidden."
This research was published in the international journal 'Nature Communications' on April 27.
Terminology
* Single-cell RNA sequencing (scRNA-seq): A technique used to measure gene expression levels in individual cells, providing insights into cell heterogeneity and rare cell types.
* Dimensionality reduction: A method to reduce the number of features or variables in a dataset while preserving the most important information, making data analysis more manageable and interpretable.
* Random matrix theory: A mathematical framework used to model and analyze the properties of large, random matrices, which can be applied to filter out noise in high-dimensional data.
* Signal robustness test: Among the signals, this test selects signals that are robust to the slight perturbation in data because real biological signals should be invariant for such slight modification in the data.
2024.05.09 View 6297 -
 Afternoon chemotherapy proved to deliver more desirable results for female lymphoma patients
Chemotherapy is a commonly used regimen for cancer treatment, but it is also a double-edged sword. While the drugs are highly effective at killing cancer cells, they are also notorious for killing healthy cells in the body. As such, minimizing the drug’s damage to the patient’s body is necessary for improving the prognosis of chemotherapy.
Recently, “chrono-chemotherapy” have been gaining interest in the research community. As the name suggests, the aim is timing the delivery of the drugs when the body is least vulnerable to their harmful effects and while the cancer cells are at their most vulnerable.
< Figure 1. Chrono-chemotherapy considering circadian rhythm >
Chrono-chemotherapy exploits the fact that human physiological processes, including cell proliferation and differentiation, are regulated by an endogenous timer called the circadian clock. However, this has not been widely exploited in real-world clinical settings because, as of now, there is no systematic method for finding the optimal chemotherapy delivery time.
This problem was tackled by an interdisciplinary team of researchers from South Korea. They were led by principal investigators Jae Kyoung Kim (a mathematician from the Biomedical Mathematics Group, Institute for Basic Science) and Youngil Koh (an oncologist at Seoul National University Hospital). The researchers studied a group of patients suffering from diffuse large B-cell lymphoma (DLBCL).
Terminology
* Diffuse large B-cell lymphoma (DLBCL): Lymphoma is a type of blood cancer caused by the malignant transformation of lymphoid tissue cells. Lymphoma is divided into Hodgkin's lymphoma and non-Hodgkin's lymphoma (malignant lymphoma), and diffuse large B-cell lymphoma accounts for about 30 to 40% of non-Hodgkin's lymphoma.
The research team noticed that DLBCL patients at Seoul National University Hospital received chemotherapy on two different schedules, with some patients receiving morning treatment (8:30 a.m.) and others taking the drugs in the afternoon (2:30 p.m.). All patients received the same cancer treatment (R-CHOP), which is a combination of targeted therapy and chemotherapy, four to six times in the morning or afternoon at intervals of about three weeks.
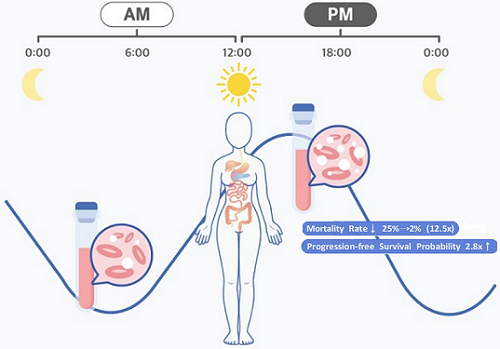
They analyzed 210 patients to investigate whether there was any difference between morning and afternoon treatments. It was found that female patients who received the afternoon treatment had a 12.5 times reduced mortality rate (25% to 2%), while the cancer recurrence after 60 months decreased by 2.8 times (37% to 13%). In addition, chemotherapy side effects such as neutropenia were more common in female patients who received the morning treatment.
Surprisingly, there was no differences found in treatment efficiency depending on the treatment schedule in the cases of male patients.
To understand the cause of the gender differences, the research team analyzed upto 14,000 blood samples from the Seoul National University Hospital Health Examination Center. It was found that in females, white blood cell counts tended to decrease in the morning and increase in the afternoon. This indicates that the bone marrow proliferation rate was higher in the morning than in the afternoon because there is a upto 12 hour delay between bone marrow proliferation and blood cell production.
This means that if a female patient receives chemotherapy in the morning when bone marrow is actively producing blood cells, the possibility of adverse side effects becomes greater. These results are consistent with the findings from recent randomized clinical trials that showed female colorectal cancer patients treated with irinotecan in the morning suffered from higher drug toxicities.
One confounding variable was the drug dose. Since the morning female patients suffered from greater adverse side effects, oftentimes the dose had to be reduced for these patients. On average, the drug dose was reduced by upto 10% compared to the dose intensity given to female patients receiving the afternoon treatment.
Unlike the female patients, it was found that male patients did not show a significant difference in white blood cell count and bone marrow cell proliferation activity throughout the day, which explains why the timing of the treatment had no impact.
Professor Youngil Koh said, “We plan to verify the conclusions of this study again with a large-scale follow-up study that completely controls for the confounding variables, and to confirm whether chrono-chemotherapy has similar effects on other cancers.”
CI Jae Kyoung Kim said, “Because the time of the internal circadian clock can vary greatly depending on the individual's sleep-wake patterns, we are currently developing a technology to estimate a patient’s circadian clock from their sleep pattern. We hope that this can be used to develop an individualized anti-cancer chronotherapy schedule.”
< Figure 2. Chemotherapy in the afternoon can improve treatment outcomes. >
The daily fluctuation of proliferative activity of bone marrow is larger in females than in males, and it becomes higher in the morning (left). Thus, chemotherapy in the morning strongly inhibits proliferative activity in female lymphoma patients, resulting in a higher incidence of adverse events such as neutropenia and infections. This forced the clinicians to reduce the dose intensity (center). Consequently, female patients undergoing the morning treatment showed a lower survival probability than those undergoing the afternoon treatment (right). Specifically, only ~13% of female patients treated in the afternoon had a worse outcome and ~2% of them died while ~37% of female patients treated in the morning had a worse outcome and ~25% of them died. Male patients did not show any difference in treatment outcomes depending on the chemotherapy delivery time.
2023.01.27 View 8131
Afternoon chemotherapy proved to deliver more desirable results for female lymphoma patients
Chemotherapy is a commonly used regimen for cancer treatment, but it is also a double-edged sword. While the drugs are highly effective at killing cancer cells, they are also notorious for killing healthy cells in the body. As such, minimizing the drug’s damage to the patient’s body is necessary for improving the prognosis of chemotherapy.
Recently, “chrono-chemotherapy” have been gaining interest in the research community. As the name suggests, the aim is timing the delivery of the drugs when the body is least vulnerable to their harmful effects and while the cancer cells are at their most vulnerable.
< Figure 1. Chrono-chemotherapy considering circadian rhythm >
Chrono-chemotherapy exploits the fact that human physiological processes, including cell proliferation and differentiation, are regulated by an endogenous timer called the circadian clock. However, this has not been widely exploited in real-world clinical settings because, as of now, there is no systematic method for finding the optimal chemotherapy delivery time.
This problem was tackled by an interdisciplinary team of researchers from South Korea. They were led by principal investigators Jae Kyoung Kim (a mathematician from the Biomedical Mathematics Group, Institute for Basic Science) and Youngil Koh (an oncologist at Seoul National University Hospital). The researchers studied a group of patients suffering from diffuse large B-cell lymphoma (DLBCL).
Terminology
* Diffuse large B-cell lymphoma (DLBCL): Lymphoma is a type of blood cancer caused by the malignant transformation of lymphoid tissue cells. Lymphoma is divided into Hodgkin's lymphoma and non-Hodgkin's lymphoma (malignant lymphoma), and diffuse large B-cell lymphoma accounts for about 30 to 40% of non-Hodgkin's lymphoma.
The research team noticed that DLBCL patients at Seoul National University Hospital received chemotherapy on two different schedules, with some patients receiving morning treatment (8:30 a.m.) and others taking the drugs in the afternoon (2:30 p.m.). All patients received the same cancer treatment (R-CHOP), which is a combination of targeted therapy and chemotherapy, four to six times in the morning or afternoon at intervals of about three weeks.
They analyzed 210 patients to investigate whether there was any difference between morning and afternoon treatments. It was found that female patients who received the afternoon treatment had a 12.5 times reduced mortality rate (25% to 2%), while the cancer recurrence after 60 months decreased by 2.8 times (37% to 13%). In addition, chemotherapy side effects such as neutropenia were more common in female patients who received the morning treatment.
Surprisingly, there was no differences found in treatment efficiency depending on the treatment schedule in the cases of male patients.
To understand the cause of the gender differences, the research team analyzed upto 14,000 blood samples from the Seoul National University Hospital Health Examination Center. It was found that in females, white blood cell counts tended to decrease in the morning and increase in the afternoon. This indicates that the bone marrow proliferation rate was higher in the morning than in the afternoon because there is a upto 12 hour delay between bone marrow proliferation and blood cell production.
This means that if a female patient receives chemotherapy in the morning when bone marrow is actively producing blood cells, the possibility of adverse side effects becomes greater. These results are consistent with the findings from recent randomized clinical trials that showed female colorectal cancer patients treated with irinotecan in the morning suffered from higher drug toxicities.
One confounding variable was the drug dose. Since the morning female patients suffered from greater adverse side effects, oftentimes the dose had to be reduced for these patients. On average, the drug dose was reduced by upto 10% compared to the dose intensity given to female patients receiving the afternoon treatment.
Unlike the female patients, it was found that male patients did not show a significant difference in white blood cell count and bone marrow cell proliferation activity throughout the day, which explains why the timing of the treatment had no impact.
Professor Youngil Koh said, “We plan to verify the conclusions of this study again with a large-scale follow-up study that completely controls for the confounding variables, and to confirm whether chrono-chemotherapy has similar effects on other cancers.”
CI Jae Kyoung Kim said, “Because the time of the internal circadian clock can vary greatly depending on the individual's sleep-wake patterns, we are currently developing a technology to estimate a patient’s circadian clock from their sleep pattern. We hope that this can be used to develop an individualized anti-cancer chronotherapy schedule.”
< Figure 2. Chemotherapy in the afternoon can improve treatment outcomes. >
The daily fluctuation of proliferative activity of bone marrow is larger in females than in males, and it becomes higher in the morning (left). Thus, chemotherapy in the morning strongly inhibits proliferative activity in female lymphoma patients, resulting in a higher incidence of adverse events such as neutropenia and infections. This forced the clinicians to reduce the dose intensity (center). Consequently, female patients undergoing the morning treatment showed a lower survival probability than those undergoing the afternoon treatment (right). Specifically, only ~13% of female patients treated in the afternoon had a worse outcome and ~2% of them died while ~37% of female patients treated in the morning had a worse outcome and ~25% of them died. Male patients did not show any difference in treatment outcomes depending on the chemotherapy delivery time.
2023.01.27 View 8131 -
 Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 11634
Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 11634 -
 Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
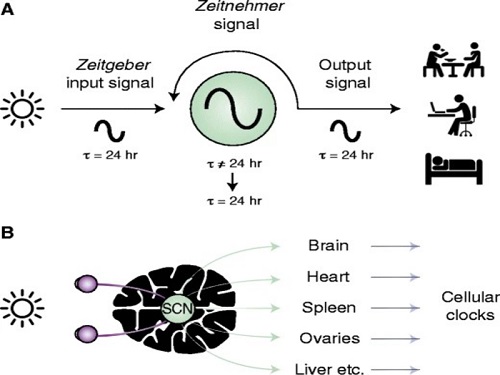
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 11659
Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 11659 -
 Professor Jae Kyoung Kim to Lead a New Mathematical Biology Research Group at IBS
Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences was appointed as the third Chief Investigator (CI) of the Pioneer Research Center (PRC) for Mathematical and Computational Sciences at the Institute for Basic Science (IBS). Professor Kim will launch and lead a new research group that will be devoted to resolving various biological conundrums from a mathematical perspective. His appointment began on March 1, 2021.
Professor Kim, a rising researcher in the field of mathematical biology, has received attention from both the mathematical and biological communities at the international level. Professor Kim puts novel and unremitting efforts into understanding biological systems such as cell-to-cell interactions mathematically and designing mathematical models for identifying causes of diseases and developing therapeutic medicines.
Through active joint research with biologists, mathematician Kim has addressed many challenges that have remained unsolved in biology and published papers in a number of leading international journals in related fields. His notable works based on mathematical modelling include having designed a biological circuit that can maintain a stable circadian rhythm (Science, 2015) and unveiling the principles of how the biological clock in the body maintains a steady speed for the first time in over 60 years (Molecular Cell, 2015). Recently, through a joint research project with Pfizer, Professor Kim identified what causes the differences between animal and clinical test results during drug development explaining why drugs have different efficacies in different people (Molecular Systems Biology, 2019).
The new IBS biomedical mathematics research group led by Professor Kim will further investigate the causes of unstable circadian rhythms and sleeping patterns. The team will aim to present a new paradigm in treatments for sleep disorders.
Professor Kim said, “We are all so familiar with sleep behaviors, but the exact mechanisms behind how such behaviors occur are still unknown. Through cooperation with biomedical scientists, our group will do its best to discover the complicated, fundamental mechanisms of sleep, and investigate the causes and cures of sleep disorders.”
Every year, the IBS selects young and promising researchers and appoints them as CIs. A maximum of five selected CIs can form each independent research group within the IBS PRC, and receive research funds of 1 billion to 1.5 billion KRW over five years.
(END)
2021.03.18 View 11243
Professor Jae Kyoung Kim to Lead a New Mathematical Biology Research Group at IBS
Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences was appointed as the third Chief Investigator (CI) of the Pioneer Research Center (PRC) for Mathematical and Computational Sciences at the Institute for Basic Science (IBS). Professor Kim will launch and lead a new research group that will be devoted to resolving various biological conundrums from a mathematical perspective. His appointment began on March 1, 2021.
Professor Kim, a rising researcher in the field of mathematical biology, has received attention from both the mathematical and biological communities at the international level. Professor Kim puts novel and unremitting efforts into understanding biological systems such as cell-to-cell interactions mathematically and designing mathematical models for identifying causes of diseases and developing therapeutic medicines.
Through active joint research with biologists, mathematician Kim has addressed many challenges that have remained unsolved in biology and published papers in a number of leading international journals in related fields. His notable works based on mathematical modelling include having designed a biological circuit that can maintain a stable circadian rhythm (Science, 2015) and unveiling the principles of how the biological clock in the body maintains a steady speed for the first time in over 60 years (Molecular Cell, 2015). Recently, through a joint research project with Pfizer, Professor Kim identified what causes the differences between animal and clinical test results during drug development explaining why drugs have different efficacies in different people (Molecular Systems Biology, 2019).
The new IBS biomedical mathematics research group led by Professor Kim will further investigate the causes of unstable circadian rhythms and sleeping patterns. The team will aim to present a new paradigm in treatments for sleep disorders.
Professor Kim said, “We are all so familiar with sleep behaviors, but the exact mechanisms behind how such behaviors occur are still unknown. Through cooperation with biomedical scientists, our group will do its best to discover the complicated, fundamental mechanisms of sleep, and investigate the causes and cures of sleep disorders.”
Every year, the IBS selects young and promising researchers and appoints them as CIs. A maximum of five selected CIs can form each independent research group within the IBS PRC, and receive research funds of 1 billion to 1.5 billion KRW over five years.
(END)
2021.03.18 View 11243 -
 Mystery Solved with Math: Cytoplasmic Traffic Jam Disrupts Sleep-Wake Cycles
KAIST mathematicians and their collaborators at Florida State University have identified the principle of how aging and diseases like dementia and obesity cause sleep disorders. A combination of mathematical modelling and experiments demonstrated that the cytoplasmic congestion caused by aging, dementia, and/or obesity disrupts the circadian rhythms in the human body and leads to irregular sleep-wake cycles. This finding suggests new treatment strategies for addressing unstable sleep-wake cycles.
Human bodies adjust sleep schedules in accordance with the ‘circadian rhythms’, which are regulated by our time keeping system, the ‘circadian clock’. This clock tells our body when to rest by generating the 24-hour rhythms of a protein called PERIOD (PER) (See Figure 1).
The amount of the PER protein increases for half of the day and then decreases for the remaining half. The principle is that the PER protein accumulating in the cytoplasm for several hours enters the cell nucleus all at once, hindering the transcription of PER genes and thereby reducing the amount of PER.
However, it has remained a mystery how thousands of PER molecules can simultaneously enter into the nucleus in a complex cell environment where a variety of materials co-exist and can interfere with the motion of PER. This would be like finding a way for thousands of employees from all over New York City to enter an office building at the same time every day.
A group of researchers led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences solved the mystery by developing a spatiotemporal and probabilistic model that describes the motion of PER molecules in a cell environment.
This study was conducted in collaboration with Professor Choogon Lee’s group from Florida State University, where the experiments were carried out, and the results were published in the Proceedings of the National Academy of Sciences (PNAS) last month.
The joint research team’s spatial stochastic model (See Figure 2) described the motion of PER molecules in cells and demonstrated that the PER molecule should be sufficiently condensed around the cell nucleus to be phosphorylated simultaneously and enter the nucleus together (See Figure 3 Left). Thanks to this phosphorylation synchronization switch, thousands of PER molecules can enter the nucleus at the same time every day and maintain stable circadian rhythms.
However, when aging and/or diseases including dementia and obesity cause the cytoplasm to become congested with increased cytoplasmic obstacles such as protein aggregates and fat vacuoles, it hinders the timely condensation of PER molecules around the cell nucleus (See Figure 3 Right). As a result, the phosphorylation synchronization switch does not work and PER proteins enter into the nucleus at irregular times, making the circadian rhythms and sleep-wake cycles unstable, the study revealed.
Professor Kim said, “As a mathematician, I am excited to help enable the advancement of new treatment strategies that can improve the lives of so many patients who suffer from irregular sleep-wake cycles. Taking these findings as an opportunity, I hope to see more active interchanges of ideas and collaboration between mathematical and biological sciences.”
This work was supported by the National Institutes of Health and the National Science Foundation in the US, and the International Human Frontiers Science Program Organization and the National Research Foundation of Korea.
Publication:
Beesley, S. and Kim, D. W, et al. (2020) Wake-sleep cycles are severely disrupted by diseases affecting cytoplasmic homeostasis. Proceedings of the National Academy of Sciences (PNAS), Vol. 117, No. 45, 28402-28411. Available online at https://doi.org/10.1073/pnas.2003524117
Profile:
Jae Kyoung Kim, Ph.D.
Associate Professor
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
@umichkim on Twitter
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
Profile:
Choogon Lee, Ph.D.
Associate Professor
clee@neuro.fsu.edu
https://med.fsu.edu/biosci/lee-lab
Department of Biomedical Sciences
Florida State University
Florida, USA
(END)
2020.12.11 View 12086
Mystery Solved with Math: Cytoplasmic Traffic Jam Disrupts Sleep-Wake Cycles
KAIST mathematicians and their collaborators at Florida State University have identified the principle of how aging and diseases like dementia and obesity cause sleep disorders. A combination of mathematical modelling and experiments demonstrated that the cytoplasmic congestion caused by aging, dementia, and/or obesity disrupts the circadian rhythms in the human body and leads to irregular sleep-wake cycles. This finding suggests new treatment strategies for addressing unstable sleep-wake cycles.
Human bodies adjust sleep schedules in accordance with the ‘circadian rhythms’, which are regulated by our time keeping system, the ‘circadian clock’. This clock tells our body when to rest by generating the 24-hour rhythms of a protein called PERIOD (PER) (See Figure 1).
The amount of the PER protein increases for half of the day and then decreases for the remaining half. The principle is that the PER protein accumulating in the cytoplasm for several hours enters the cell nucleus all at once, hindering the transcription of PER genes and thereby reducing the amount of PER.
However, it has remained a mystery how thousands of PER molecules can simultaneously enter into the nucleus in a complex cell environment where a variety of materials co-exist and can interfere with the motion of PER. This would be like finding a way for thousands of employees from all over New York City to enter an office building at the same time every day.
A group of researchers led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences solved the mystery by developing a spatiotemporal and probabilistic model that describes the motion of PER molecules in a cell environment.
This study was conducted in collaboration with Professor Choogon Lee’s group from Florida State University, where the experiments were carried out, and the results were published in the Proceedings of the National Academy of Sciences (PNAS) last month.
The joint research team’s spatial stochastic model (See Figure 2) described the motion of PER molecules in cells and demonstrated that the PER molecule should be sufficiently condensed around the cell nucleus to be phosphorylated simultaneously and enter the nucleus together (See Figure 3 Left). Thanks to this phosphorylation synchronization switch, thousands of PER molecules can enter the nucleus at the same time every day and maintain stable circadian rhythms.
However, when aging and/or diseases including dementia and obesity cause the cytoplasm to become congested with increased cytoplasmic obstacles such as protein aggregates and fat vacuoles, it hinders the timely condensation of PER molecules around the cell nucleus (See Figure 3 Right). As a result, the phosphorylation synchronization switch does not work and PER proteins enter into the nucleus at irregular times, making the circadian rhythms and sleep-wake cycles unstable, the study revealed.
Professor Kim said, “As a mathematician, I am excited to help enable the advancement of new treatment strategies that can improve the lives of so many patients who suffer from irregular sleep-wake cycles. Taking these findings as an opportunity, I hope to see more active interchanges of ideas and collaboration between mathematical and biological sciences.”
This work was supported by the National Institutes of Health and the National Science Foundation in the US, and the International Human Frontiers Science Program Organization and the National Research Foundation of Korea.
Publication:
Beesley, S. and Kim, D. W, et al. (2020) Wake-sleep cycles are severely disrupted by diseases affecting cytoplasmic homeostasis. Proceedings of the National Academy of Sciences (PNAS), Vol. 117, No. 45, 28402-28411. Available online at https://doi.org/10.1073/pnas.2003524117
Profile:
Jae Kyoung Kim, Ph.D.
Associate Professor
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
@umichkim on Twitter
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
Profile:
Choogon Lee, Ph.D.
Associate Professor
clee@neuro.fsu.edu
https://med.fsu.edu/biosci/lee-lab
Department of Biomedical Sciences
Florida State University
Florida, USA
(END)
2020.12.11 View 12086 -
 A Mathematical Model Reveals Long-Distance Cell Communication Mechanism
How can tens of thousands of people in a large football stadium all clap together with the same beat even though they can only hear the people near them clapping?
A combination of a partial differential equation and a synthetic circuit in microbes answers this question. An interdisciplinary collaborative team of Professor Jae Kyoung Kim at KAIST, Professor Krešimir Josić at the University of Houston, and Professor Matt Bennett at Rice University has identified how a large community can communicate with each other almost simultaneously even with very short distance signaling. The research was reported at Nature Chemical Biology.
Cells often communicate using signaling molecules, which can travel only a short distance. Nevertheless, the cells can also communicate over large distances to spur collective action. The team revealed a cell communication mechanism that quickly forms a network of local interactions to spur collective action, even in large communities.
The research team used an engineered transcriptional circuit of combined positive and negative feedback loops in E. coli, which can periodically release two types of signaling molecules: activator and repressor. As the signaling molecules travel over a short distance, cells can only talk to their nearest neighbors. However, cell communities synchronize oscillatory gene expression in spatially extended systems as long as the transcriptional circuit contains a positive feedback loop for the activator.
Professor Kim said that analyzing and understanding such high-dimensional dynamics was extremely difficult. He explained, “That’s why we used high-dimensional partial differential equation to describe the system based on the interactions among various types of molecules.” Surprisingly, the mathematical model accurately simulates the synthesis of the signaling molecules in the cell and their spatial diffusion throughout the chamber and their effect on neighboring cells.
The team simplified the high-dimensional system into a one-dimensional orbit, noting that the system repeats periodically. This allowed them to discover that cells can make one voice when they lowered their own voice and listened to the others. “It turns out the positive feedback loop reduces the distance between moving points and finally makes them move all together. That’s why you clap louder when you hear applause from nearby neighbors and everyone eventually claps together at almost the same time,” said Professor Kim.
Professor Kim added, “Math is a powerful as it simplifies complex thing so that we can find an essential underlying property. This finding would not have been possible without the simplification of complex systems using mathematics."
The National Institutes of Health, the National Science Foundation, the Robert A. Welch Foundation, the Hamill Foundation, the National Research Foundation of Korea, and the T.J. Park Science Fellowship of POSCO supported the research.
(Figure: Complex molecular interactions among microbial consortia is simplified as interactions among points on a limit cycle (right).)
2019.10.15 View 29923
A Mathematical Model Reveals Long-Distance Cell Communication Mechanism
How can tens of thousands of people in a large football stadium all clap together with the same beat even though they can only hear the people near them clapping?
A combination of a partial differential equation and a synthetic circuit in microbes answers this question. An interdisciplinary collaborative team of Professor Jae Kyoung Kim at KAIST, Professor Krešimir Josić at the University of Houston, and Professor Matt Bennett at Rice University has identified how a large community can communicate with each other almost simultaneously even with very short distance signaling. The research was reported at Nature Chemical Biology.
Cells often communicate using signaling molecules, which can travel only a short distance. Nevertheless, the cells can also communicate over large distances to spur collective action. The team revealed a cell communication mechanism that quickly forms a network of local interactions to spur collective action, even in large communities.
The research team used an engineered transcriptional circuit of combined positive and negative feedback loops in E. coli, which can periodically release two types of signaling molecules: activator and repressor. As the signaling molecules travel over a short distance, cells can only talk to their nearest neighbors. However, cell communities synchronize oscillatory gene expression in spatially extended systems as long as the transcriptional circuit contains a positive feedback loop for the activator.
Professor Kim said that analyzing and understanding such high-dimensional dynamics was extremely difficult. He explained, “That’s why we used high-dimensional partial differential equation to describe the system based on the interactions among various types of molecules.” Surprisingly, the mathematical model accurately simulates the synthesis of the signaling molecules in the cell and their spatial diffusion throughout the chamber and their effect on neighboring cells.
The team simplified the high-dimensional system into a one-dimensional orbit, noting that the system repeats periodically. This allowed them to discover that cells can make one voice when they lowered their own voice and listened to the others. “It turns out the positive feedback loop reduces the distance between moving points and finally makes them move all together. That’s why you clap louder when you hear applause from nearby neighbors and everyone eventually claps together at almost the same time,” said Professor Kim.
Professor Kim added, “Math is a powerful as it simplifies complex thing so that we can find an essential underlying property. This finding would not have been possible without the simplification of complex systems using mathematics."
The National Institutes of Health, the National Science Foundation, the Robert A. Welch Foundation, the Hamill Foundation, the National Research Foundation of Korea, and the T.J. Park Science Fellowship of POSCO supported the research.
(Figure: Complex molecular interactions among microbial consortia is simplified as interactions among points on a limit cycle (right).)
2019.10.15 View 29923 -
 Professor Jae Kyoung Kim Receives the 2017 HSFP Award
The Human Frontier Science Program (HSFP), one of the most competitive research grants in life sciences, has funded researchers worldwide across and beyond the field since 1990. Each year, the program selects a handful of recipients who push the envelope of basic research in biology to bring breakthroughs from novel approaches. Among its 7,000 recipients thus far, 26 scientists have received the Nobel Prize. For that reason, HSFP grants are often referred to as “Nobel Prize Grants.”
Professor Jae Kyoung Kim of the Mathematical Sciences Department at KAIST and his international collaborators, Professor Robert Havekes from the University of Groningen, the Netherlands, Professor Sara Aton from the University of Michigan in Ann Arbor, the United States, and Professor Matias Zurbriggen from the University of Düsseldorf, Germany, won the Young Investigator Grants of the 2017 HSFP.
The 30 winning teams of the 2017 competition (in 9 Young Investigator Grants and 21 Program Grants) went through a rigorous year-long review process from a total of 1,073 applications submitted from more than 60 countries around the world. Each winning team will receive financial support averaging 110,000-125,000 USD per year for three years.
Although Professor Kim was trained as a mathematician, he has extended his research focus into biological sciences and attempted to solve some of the most difficult problems in biology by employing mathematical theories and applications including nonlinear dynamics, stochastic process, singular perturbation, and parameter estimation.
The project that won the Young Investigator Grants was a study on how a molecular circadian clock may affect sleep-regulated neurophysiology in mammals. Physiological and metabolic processes such as sleep, blood pressure, and hormone secretion exhibit circadian rhythms in mammals. Professor Kim used mathematical modeling and analysis to explain that the mammalian circadian clock is a hierarchical system, in which the master clock in the superchiasmatic nucleus, a tiny region in the brain that controls circadian rhythms, functions as a pacemaker and synchronizer of peripheral clocks to generate coherent systematic rhythms throughout the body.
Professor Kim said, “The mechanisms of our neuronal and hormonal activities regulating many of our bodily functions over a 24-hour cycle are not yet fully known. We go to sleep every night, but do not really know how it affects our brain functions. I hope my experience in mathematics, along with insights from biologists, can find meaningful answers to some of today’s puzzling problems in biological sciences, for example, revealing the complexities of our brains and showing how they work.”
“In the meantime, I hope collaborations between the fields of mathematics and biology, as yet a rare phenomenon in the Korean scientific community, will become more popular in the near future.”
Professor Kim received his doctoral degree in Applied and Interdisciplinary Mathematics in 2013 from the University of Michigan and joined KAIST in 2015. He has published numerous articles in reputable science journals such as Science, Molecular Cell, Proceedings of the National Academy of Sciences, and Nature Communications.
Both the Program Grants and Young Investigator Grants support international teams with members from at least two countries for innovative and creative research. This year, the Program Grants were awarded to research topics ranging from the evolution of counting and the role of extracellular vesicles in breast cancer bone metastasis to the examination of obesity from a mechanobiological point of view.
The Young Investigator Grants are limited to teams that established their independent research within the last five years and received their doctoral degrees within the last decade. Besides Professor Kim’s study, such topics as the use of infrasound for navigation by seabirds and protein formation in photochemistry and photophysics were awarded in 2017.
Full lists of the 2017 HFSP winners are available at: http://www.hfsp.org/awardees/newly-awarded.
About the Human Frontier Science Program (HFSP):
The HFSP is a research funding program implemented by the International Human Frontier Science Program (HFSPO) based in Strasbourg, France. It promotes intercontinental collaboration and training in cutting-edge, interdisciplinary research specializing in life sciences. Founded in 1989, the HFSPO consists of the European Union and 14 other countries including the G7 nations and South Korea.
2017.03.21 View 12154
Professor Jae Kyoung Kim Receives the 2017 HSFP Award
The Human Frontier Science Program (HSFP), one of the most competitive research grants in life sciences, has funded researchers worldwide across and beyond the field since 1990. Each year, the program selects a handful of recipients who push the envelope of basic research in biology to bring breakthroughs from novel approaches. Among its 7,000 recipients thus far, 26 scientists have received the Nobel Prize. For that reason, HSFP grants are often referred to as “Nobel Prize Grants.”
Professor Jae Kyoung Kim of the Mathematical Sciences Department at KAIST and his international collaborators, Professor Robert Havekes from the University of Groningen, the Netherlands, Professor Sara Aton from the University of Michigan in Ann Arbor, the United States, and Professor Matias Zurbriggen from the University of Düsseldorf, Germany, won the Young Investigator Grants of the 2017 HSFP.
The 30 winning teams of the 2017 competition (in 9 Young Investigator Grants and 21 Program Grants) went through a rigorous year-long review process from a total of 1,073 applications submitted from more than 60 countries around the world. Each winning team will receive financial support averaging 110,000-125,000 USD per year for three years.
Although Professor Kim was trained as a mathematician, he has extended his research focus into biological sciences and attempted to solve some of the most difficult problems in biology by employing mathematical theories and applications including nonlinear dynamics, stochastic process, singular perturbation, and parameter estimation.
The project that won the Young Investigator Grants was a study on how a molecular circadian clock may affect sleep-regulated neurophysiology in mammals. Physiological and metabolic processes such as sleep, blood pressure, and hormone secretion exhibit circadian rhythms in mammals. Professor Kim used mathematical modeling and analysis to explain that the mammalian circadian clock is a hierarchical system, in which the master clock in the superchiasmatic nucleus, a tiny region in the brain that controls circadian rhythms, functions as a pacemaker and synchronizer of peripheral clocks to generate coherent systematic rhythms throughout the body.
Professor Kim said, “The mechanisms of our neuronal and hormonal activities regulating many of our bodily functions over a 24-hour cycle are not yet fully known. We go to sleep every night, but do not really know how it affects our brain functions. I hope my experience in mathematics, along with insights from biologists, can find meaningful answers to some of today’s puzzling problems in biological sciences, for example, revealing the complexities of our brains and showing how they work.”
“In the meantime, I hope collaborations between the fields of mathematics and biology, as yet a rare phenomenon in the Korean scientific community, will become more popular in the near future.”
Professor Kim received his doctoral degree in Applied and Interdisciplinary Mathematics in 2013 from the University of Michigan and joined KAIST in 2015. He has published numerous articles in reputable science journals such as Science, Molecular Cell, Proceedings of the National Academy of Sciences, and Nature Communications.
Both the Program Grants and Young Investigator Grants support international teams with members from at least two countries for innovative and creative research. This year, the Program Grants were awarded to research topics ranging from the evolution of counting and the role of extracellular vesicles in breast cancer bone metastasis to the examination of obesity from a mechanobiological point of view.
The Young Investigator Grants are limited to teams that established their independent research within the last five years and received their doctoral degrees within the last decade. Besides Professor Kim’s study, such topics as the use of infrasound for navigation by seabirds and protein formation in photochemistry and photophysics were awarded in 2017.
Full lists of the 2017 HFSP winners are available at: http://www.hfsp.org/awardees/newly-awarded.
About the Human Frontier Science Program (HFSP):
The HFSP is a research funding program implemented by the International Human Frontier Science Program (HFSPO) based in Strasbourg, France. It promotes intercontinental collaboration and training in cutting-edge, interdisciplinary research specializing in life sciences. Founded in 1989, the HFSPO consists of the European Union and 14 other countries including the G7 nations and South Korea.
2017.03.21 View 12154 -
 KAIST's Mathematician Reveals the Mechanism for Sustaining Biological Rhythms
Our bodies have a variety of biological clocks that follow rhythms or oscillations with periods ranging from seconds to days. For example, our hearts beat every second, and cells divide periodically. The circadian clock located in the hypothalamus generates twenty-four hour rhythms, timing our sleep and hormone release. How do these biological clocks or circuits generate and sustain the stable rhythms that are essential to life?
Jae Kyoung Kim, who is an assistant professor in the Department of Mathematical Sciences at KAIST, has predicted how these biological circuits generate rhythms and control their robustness, utilizing mathematical modeling based on differential equations and stochastic parameter sampling. Based on his prediction, using synthetic biology, a research team headed by Matthew Bennett of Rice University constructed a novel biological circuit that spans two genetically engineered strains of bacteria, one serves as an activator and the other as a repressor to regulate gene expression across multiple cell types, and found that the circuit generates surprisingly robust rhythms under various conditions.
The results of the research conducted in collaboration with KAIST (Korea Institute of Science and Technology), Rice University, and the University of Houston were published in Science (August 28, 2015 issue). The title of the paper is "Emergent Genetic Oscillations in a Synthetic Microbial Consortium" .
The top-down research approach, which focuses on identifying the components of biological circuits, limits our understanding of the mechanisms in which the circuits generate rhythms. Synthetic biology, a rapidly growing field at the interface of biosciences and engineering, however, uses a bottom-up approach.
Synthetic biologists can create complex circuits out of simpler components, and some of these new genetic circuits are capable of fluctuation to regulate gene production. In the same way that electrical engineers understand how an electrical circuit works as they construct batteries, resistors, and wires, synthetic biologists can understand better about biological circuits if they put them together using genes and proteins. However, due to the complexity of biological systems, both experiments and mathematical modeling need to be applied hand in hand to design these biological circuits and understand their function.
In this research, an interdisciplinary approach proved that a synthetic intercellular singling circuit generates robust rhythms to create a cooperative microbial system. Specifically, Kim's mathematical analysis suggested, and experiments confirmed, that the presence of negative feedback loops in addition to a core transcriptional negative feedback loop can explain the robustness of rhythms in this system. This result provides important clues about the fundamental mechanism of robust rhythm generation in biological systems.
Furthermore, rather than constructing the entire circuit inside a single bacterial strain, the circuit was split among two strains of Escherichia coli bacterium. When the strains were grown together, the bacteria exchanged information, completing the circuit. Thus, this research also shows how, by regulating individual cells within the system, complex biological systems can be controlled, which in turn influences each other (e.g., the gut microbiome in humans).
###
Ye Chen, a graduate student in Bennett's laboratory at Rice University, and Jae Kyoung Kim, an assistant professor at KAIST and a former postdoctoral fellow at Ohio State University, are the lead authors of the paper. The co-authors are Rice graduate student Andrew Hirning and Krešimir Josic?, a professor of mathematics at the University of Houston. Bennett is the Assistant Professor of the Biochemistry and Cell Biology Department at Rice University.
About the researcher: While Jae Kyoung Kim is a mathematician, he has also solved various biological puzzles in collaboration with various experimental laboratories of Matthew Bennett at Rice University, David Virshup at Duke and the National University of Singapore, Carla Finkielstein at Virginia Polytechnic Institute and State University, Choo-Gon Lee at the Florida State University, Seung-Hee Yoo at the Medical School of the University of Texas, Toru Takumi at RIKEN Brain Science Institute, and Travis Wager at Pfizer Inc. He has used non-linear dynamics and stochastic analysis to understand the function of biochemical networks in biological systems. In particular, he is interested in mechanisms generating and regulating biological rhythms.
Picture 1: This schematic image is the design of a biological circuit between two strains of bacteria and the part of differential equations used to understand the function of the biological circuit.
Picture 2: The core transcriptional negative feedback loop and additional negative feedback loop in the biological circuit (picture 1) generate robust rhythms. The snapshots correspond the red dots in the time series graph.
2015.08.31 View 9393
KAIST's Mathematician Reveals the Mechanism for Sustaining Biological Rhythms
Our bodies have a variety of biological clocks that follow rhythms or oscillations with periods ranging from seconds to days. For example, our hearts beat every second, and cells divide periodically. The circadian clock located in the hypothalamus generates twenty-four hour rhythms, timing our sleep and hormone release. How do these biological clocks or circuits generate and sustain the stable rhythms that are essential to life?
Jae Kyoung Kim, who is an assistant professor in the Department of Mathematical Sciences at KAIST, has predicted how these biological circuits generate rhythms and control their robustness, utilizing mathematical modeling based on differential equations and stochastic parameter sampling. Based on his prediction, using synthetic biology, a research team headed by Matthew Bennett of Rice University constructed a novel biological circuit that spans two genetically engineered strains of bacteria, one serves as an activator and the other as a repressor to regulate gene expression across multiple cell types, and found that the circuit generates surprisingly robust rhythms under various conditions.
The results of the research conducted in collaboration with KAIST (Korea Institute of Science and Technology), Rice University, and the University of Houston were published in Science (August 28, 2015 issue). The title of the paper is "Emergent Genetic Oscillations in a Synthetic Microbial Consortium" .
The top-down research approach, which focuses on identifying the components of biological circuits, limits our understanding of the mechanisms in which the circuits generate rhythms. Synthetic biology, a rapidly growing field at the interface of biosciences and engineering, however, uses a bottom-up approach.
Synthetic biologists can create complex circuits out of simpler components, and some of these new genetic circuits are capable of fluctuation to regulate gene production. In the same way that electrical engineers understand how an electrical circuit works as they construct batteries, resistors, and wires, synthetic biologists can understand better about biological circuits if they put them together using genes and proteins. However, due to the complexity of biological systems, both experiments and mathematical modeling need to be applied hand in hand to design these biological circuits and understand their function.
In this research, an interdisciplinary approach proved that a synthetic intercellular singling circuit generates robust rhythms to create a cooperative microbial system. Specifically, Kim's mathematical analysis suggested, and experiments confirmed, that the presence of negative feedback loops in addition to a core transcriptional negative feedback loop can explain the robustness of rhythms in this system. This result provides important clues about the fundamental mechanism of robust rhythm generation in biological systems.
Furthermore, rather than constructing the entire circuit inside a single bacterial strain, the circuit was split among two strains of Escherichia coli bacterium. When the strains were grown together, the bacteria exchanged information, completing the circuit. Thus, this research also shows how, by regulating individual cells within the system, complex biological systems can be controlled, which in turn influences each other (e.g., the gut microbiome in humans).
###
Ye Chen, a graduate student in Bennett's laboratory at Rice University, and Jae Kyoung Kim, an assistant professor at KAIST and a former postdoctoral fellow at Ohio State University, are the lead authors of the paper. The co-authors are Rice graduate student Andrew Hirning and Krešimir Josic?, a professor of mathematics at the University of Houston. Bennett is the Assistant Professor of the Biochemistry and Cell Biology Department at Rice University.
About the researcher: While Jae Kyoung Kim is a mathematician, he has also solved various biological puzzles in collaboration with various experimental laboratories of Matthew Bennett at Rice University, David Virshup at Duke and the National University of Singapore, Carla Finkielstein at Virginia Polytechnic Institute and State University, Choo-Gon Lee at the Florida State University, Seung-Hee Yoo at the Medical School of the University of Texas, Toru Takumi at RIKEN Brain Science Institute, and Travis Wager at Pfizer Inc. He has used non-linear dynamics and stochastic analysis to understand the function of biochemical networks in biological systems. In particular, he is interested in mechanisms generating and regulating biological rhythms.
Picture 1: This schematic image is the design of a biological circuit between two strains of bacteria and the part of differential equations used to understand the function of the biological circuit.
Picture 2: The core transcriptional negative feedback loop and additional negative feedback loop in the biological circuit (picture 1) generate robust rhythms. The snapshots correspond the red dots in the time series graph.
2015.08.31 View 9393