Biophysics
-
 KAIST Captures Protein Reaction in Just Six Milliseconds
Understanding biomolecular processes - such as protein-protein interactions and enzyme-substrate reactions that occur on the microseconds to millisecond time scale is essential for comprehending life processes and advancing drug development. KAIST researchers have developed a method for freezing and analyzing biochemical reaction dynamics within a span of just a few milliseconds, marking a significant step forward in better understanding complex biological reactions.
< Photo. (From left) Professor Jin Young Kang and Haerang Hwang of the Integrated Master's and Doctoral Program of the Department of Chemistry, along with Professor Wonhee Lee of the Department of Physics >
KAIST (represented by President Kwang Hyung Lee) announced on the 24th of March that a joint research team led by Professor Jin Young Kang from the Department of Chemistry and Professor Wonhee Lee from the Department of Physics has developed a parylene-based thin-film microfluidic mixing-and-spraying device for ultra-fast biochemical reaction studies.
*Parylene: A key material for microfluidic devices used to observe protein dynamics at ultra-high speeds. It can be fabricated into a few micrometer-thick films, which can be used in making a spray nozzle for microfluidic devices.
This research overcomes the limitations of the existing time-resolved cryo-electron microscopy (TRCEM) method by reducing sample consumption to one-third of the conventional amount while improving the minimum time resolution—down to just six milliseconds (6 ms).
TRCEM is a technique that rapidly freezes protein complexes during intermediate reaction stages under cryogenic conditions, which allows researchers to analyze their structures. This approach has gained significant attention recently for its ability to capture transient biochemical events.
< Figure 1. Time-resolved cryo-EM (TRCEM) technique using microfluidic channels. In order to capture the intermediate structure of biomolecules during a biochemical reaction over time, biomolecules and reaction substrates are mixed in a microfluidic channel, and then sprayed on a grid after a certain reaction time and frozen in liquid ethane to prepare a cryo-EM sample. This can then be analyzed by cryo-EM to observe the structural changes of proteins over time. >
Transient intermediate structures of protein complexes could not be captured by traditional cryo-electron microscopy due to their extremely short lifespans. Although several TRCEM techniques have been developed to address this issue, previous methods were hindered by large sample consumption and limited time resolution. To overcome these challenges, the KAIST team developed a new mixing-and-spraying device using ultra-thin parylene films. The integrated design of the device further enhanced the precision and reproducibility of experiments.
< Figure 2. TRCEM grid fabrication setup using a parylene-based thin-film microfluidic device and actual appearance of the device. You can see that a thin-film parylene channel is inserted into the injection nozzle. The integration of the reaction channel and the injection nozzle allowed the residence time in the device to be reduced to at least 0.5 ms. >
“This research makes TRCEM more practical and paves the way for diverse applications of the parylene thin-film device in structural biology, drug development, enzyme reaction studies, and biosensor research.” Professor Jin Young Kang explained, emphasizing the significance of the study.
Professor Wonhee Lee added, “The team aims to continue this research, focusing on improvement of the technique to achieve higher time resolution with minimal sample consumption.”
< Figure 3. Comparison of the spraying patterns of the parylene mixing-jet device and the conventional mixing-jet device and the filament length in the resulting RecA-ssDNA filament formation reaction. It was shown that the thin film spray nozzle structure affects the uniformity and accuracy of the final reaction time. >
The research findings, with Haerang Hwang (a graduate student in the integrated master's and Ph.D. program in the Department of Chemistry) as the first author, were published online on January 28, 2025, in the international journal Advanced Functional Materials. (Paper Title: “Integrated Parylene-Based Thin-Film Microfluidic Device for Time-Resolved Cryo-Electron Microscopy”, DOI: doi.org/10.1002/adfm.202418224)
This research was supported by the National Research Foundation of Korea (NRF), the Samsung Future Technology Development Program, and the CELINE consortium.
2025.03.24 View 114
KAIST Captures Protein Reaction in Just Six Milliseconds
Understanding biomolecular processes - such as protein-protein interactions and enzyme-substrate reactions that occur on the microseconds to millisecond time scale is essential for comprehending life processes and advancing drug development. KAIST researchers have developed a method for freezing and analyzing biochemical reaction dynamics within a span of just a few milliseconds, marking a significant step forward in better understanding complex biological reactions.
< Photo. (From left) Professor Jin Young Kang and Haerang Hwang of the Integrated Master's and Doctoral Program of the Department of Chemistry, along with Professor Wonhee Lee of the Department of Physics >
KAIST (represented by President Kwang Hyung Lee) announced on the 24th of March that a joint research team led by Professor Jin Young Kang from the Department of Chemistry and Professor Wonhee Lee from the Department of Physics has developed a parylene-based thin-film microfluidic mixing-and-spraying device for ultra-fast biochemical reaction studies.
*Parylene: A key material for microfluidic devices used to observe protein dynamics at ultra-high speeds. It can be fabricated into a few micrometer-thick films, which can be used in making a spray nozzle for microfluidic devices.
This research overcomes the limitations of the existing time-resolved cryo-electron microscopy (TRCEM) method by reducing sample consumption to one-third of the conventional amount while improving the minimum time resolution—down to just six milliseconds (6 ms).
TRCEM is a technique that rapidly freezes protein complexes during intermediate reaction stages under cryogenic conditions, which allows researchers to analyze their structures. This approach has gained significant attention recently for its ability to capture transient biochemical events.
< Figure 1. Time-resolved cryo-EM (TRCEM) technique using microfluidic channels. In order to capture the intermediate structure of biomolecules during a biochemical reaction over time, biomolecules and reaction substrates are mixed in a microfluidic channel, and then sprayed on a grid after a certain reaction time and frozen in liquid ethane to prepare a cryo-EM sample. This can then be analyzed by cryo-EM to observe the structural changes of proteins over time. >
Transient intermediate structures of protein complexes could not be captured by traditional cryo-electron microscopy due to their extremely short lifespans. Although several TRCEM techniques have been developed to address this issue, previous methods were hindered by large sample consumption and limited time resolution. To overcome these challenges, the KAIST team developed a new mixing-and-spraying device using ultra-thin parylene films. The integrated design of the device further enhanced the precision and reproducibility of experiments.
< Figure 2. TRCEM grid fabrication setup using a parylene-based thin-film microfluidic device and actual appearance of the device. You can see that a thin-film parylene channel is inserted into the injection nozzle. The integration of the reaction channel and the injection nozzle allowed the residence time in the device to be reduced to at least 0.5 ms. >
“This research makes TRCEM more practical and paves the way for diverse applications of the parylene thin-film device in structural biology, drug development, enzyme reaction studies, and biosensor research.” Professor Jin Young Kang explained, emphasizing the significance of the study.
Professor Wonhee Lee added, “The team aims to continue this research, focusing on improvement of the technique to achieve higher time resolution with minimal sample consumption.”
< Figure 3. Comparison of the spraying patterns of the parylene mixing-jet device and the conventional mixing-jet device and the filament length in the resulting RecA-ssDNA filament formation reaction. It was shown that the thin film spray nozzle structure affects the uniformity and accuracy of the final reaction time. >
The research findings, with Haerang Hwang (a graduate student in the integrated master's and Ph.D. program in the Department of Chemistry) as the first author, were published online on January 28, 2025, in the international journal Advanced Functional Materials. (Paper Title: “Integrated Parylene-Based Thin-Film Microfluidic Device for Time-Resolved Cryo-Electron Microscopy”, DOI: doi.org/10.1002/adfm.202418224)
This research was supported by the National Research Foundation of Korea (NRF), the Samsung Future Technology Development Program, and the CELINE consortium.
2025.03.24 View 114 -
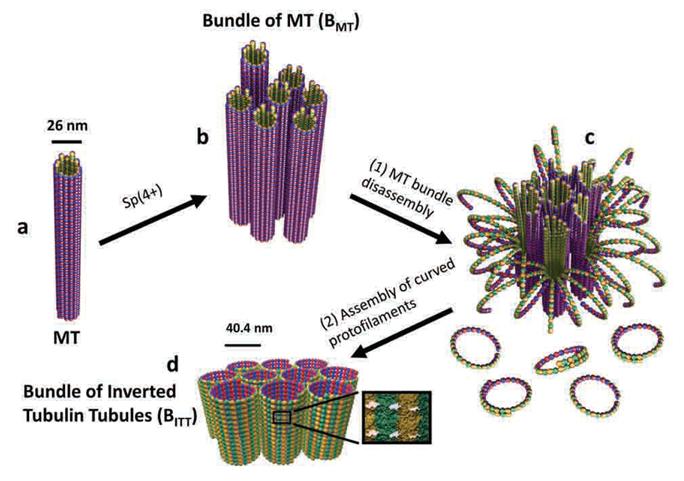 A Molecular Switch Controlling Self-Assembly of Protein Nanotubes Discovered
International collaborative research among South Korea, United States, and Israel research institutionsThe key to the treatment of cancer and brain disease mechanism The molecular switch that controls the self-assembly structure of the protein nanotubes, which plays crucial role in cell division and intracellular transport of materials, has been discovered. KAIST Bio and Brain Engineering Department’s Professor Myeong-Cheol Choi and Professor Chae-Yeon Song conducted the research, in collaboration with the University of California in Santa Barbara, U.S., and Hebrew University in Israel. The findings of the research were published in Nature Materials on the 19th. Microtubules are tube shaped and composed of protein that plays a key role in cell division, cytoskeleton, and intercellular material transport and is only 25nm in diameter (1/100,000 thickness of a human hair). Conventionally, cancer treatment focused on disrupting the formation of microtubules to suppress the division of cancer cells. In addition Alzheimer’s is known to be caused by the diminishing of structural integrity of microtubules responsible for intercellular material transport which leads to failure in signal transfer. The research team utilized synchrotron x-ray scattering and transmission electron microscope to analyze the self assemble structure of protein nanotubes to subnanometer accuracy. As a result, the microtubules were found to assemble into 25nm thickness tubules by stacking protein blocks 4 x 5 x 8nm in dimension. In the process, the research team discovered the molecular switch that controls the shape of these protein blocks. In addition the research team was successful in creating a new protein tube structure. Professor Choi commented that they were successful in introducing a new paradigm that suggests the possibility of controlling the complex biological functions of human’s biological system with the simple use of physical principles. He commented further that it is anticipated that the findings will allow for the application of bio nanotubes in engineering and that this is a small step in finding the mechanism behind cancer treatment and neural diseases.
2014.02.03 View 10582
A Molecular Switch Controlling Self-Assembly of Protein Nanotubes Discovered
International collaborative research among South Korea, United States, and Israel research institutionsThe key to the treatment of cancer and brain disease mechanism The molecular switch that controls the self-assembly structure of the protein nanotubes, which plays crucial role in cell division and intracellular transport of materials, has been discovered. KAIST Bio and Brain Engineering Department’s Professor Myeong-Cheol Choi and Professor Chae-Yeon Song conducted the research, in collaboration with the University of California in Santa Barbara, U.S., and Hebrew University in Israel. The findings of the research were published in Nature Materials on the 19th. Microtubules are tube shaped and composed of protein that plays a key role in cell division, cytoskeleton, and intercellular material transport and is only 25nm in diameter (1/100,000 thickness of a human hair). Conventionally, cancer treatment focused on disrupting the formation of microtubules to suppress the division of cancer cells. In addition Alzheimer’s is known to be caused by the diminishing of structural integrity of microtubules responsible for intercellular material transport which leads to failure in signal transfer. The research team utilized synchrotron x-ray scattering and transmission electron microscope to analyze the self assemble structure of protein nanotubes to subnanometer accuracy. As a result, the microtubules were found to assemble into 25nm thickness tubules by stacking protein blocks 4 x 5 x 8nm in dimension. In the process, the research team discovered the molecular switch that controls the shape of these protein blocks. In addition the research team was successful in creating a new protein tube structure. Professor Choi commented that they were successful in introducing a new paradigm that suggests the possibility of controlling the complex biological functions of human’s biological system with the simple use of physical principles. He commented further that it is anticipated that the findings will allow for the application of bio nanotubes in engineering and that this is a small step in finding the mechanism behind cancer treatment and neural diseases.
2014.02.03 View 10582 -
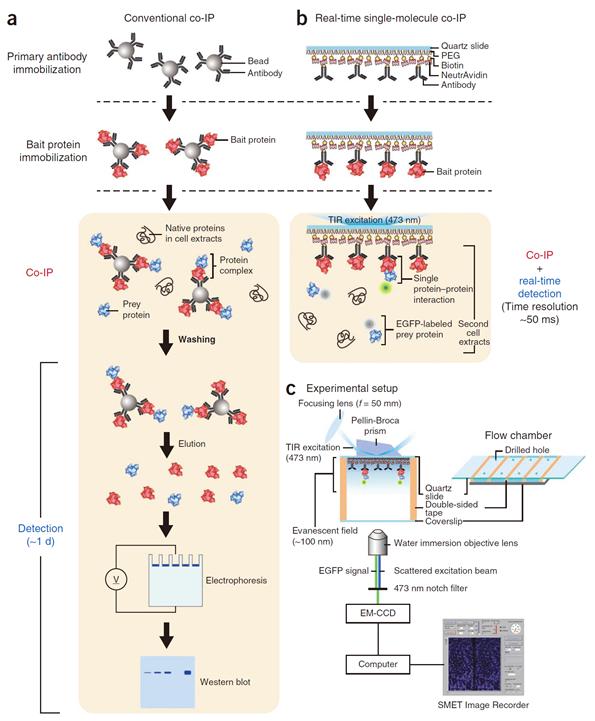 Success in Measuring Protein Interaction at the Molecular Level
Professor Tae Young Yoon
- Live observation of two protein interaction in molecular level successful- The limit in measurement and time resolution of immunoprecipitation technique improved by a hundred thousand fold
KAIST Department of Physics Professor Tae Young Yoon’s research team has successfully observed the interaction of two proteins live on molecular level and the findings were published in the October edition of Nature Protocols.
Professor Yoon’s research team developed a fluorescent microscope that can observe a single molecule. The team grafted the immunoprecipitation technique, traditionally used in protein interaction analysis, to the microscope to develop a “live molecular level immunoprecipitation technique”. The team successfully and accurately measured the reaction between two proteins by repeated momentary interactions in the unit of tens of milliseconds.
The existing immunoprecipitation technique required at least one day to detect interaction between two proteins. There were limitations in detecting momentary or weak interactions. Also, quantitative analysis of the results was difficult since the image was measured by protein-band strength. The technique could not be used for live observation.
The team aimed to drastically improve the existing technique and to develop accurate method of measurement on molecular level. The newly developed technology can enable observation of protein interaction within one hour. Also, the interaction can be measured live, thus the protein interaction phenomenon can be measured in depth.
Moreover, every programme used in the experiment was developed and distributed by the research team so source energy is secured and created the foundation for global infra.
Professor Tae Young Yoon said, “The newly developed technology does not require additional protein expression or purification. Hence, a very small sample of protein is enough to accurately analyse protein interaction on a kinetic level.” He continued, “Even cancerous protein from the tissue of a cancer patient can be analysed. Thus a platform for customised anti-cancer medicine in the future has been prepared, as well.”
Figure 1. Mimetic diagram comparing the existing immunoprecipitation technique and the newly developed live molecular level immunoprecipitation technique
2013.12.11 View 8125
Success in Measuring Protein Interaction at the Molecular Level
Professor Tae Young Yoon
- Live observation of two protein interaction in molecular level successful- The limit in measurement and time resolution of immunoprecipitation technique improved by a hundred thousand fold
KAIST Department of Physics Professor Tae Young Yoon’s research team has successfully observed the interaction of two proteins live on molecular level and the findings were published in the October edition of Nature Protocols.
Professor Yoon’s research team developed a fluorescent microscope that can observe a single molecule. The team grafted the immunoprecipitation technique, traditionally used in protein interaction analysis, to the microscope to develop a “live molecular level immunoprecipitation technique”. The team successfully and accurately measured the reaction between two proteins by repeated momentary interactions in the unit of tens of milliseconds.
The existing immunoprecipitation technique required at least one day to detect interaction between two proteins. There were limitations in detecting momentary or weak interactions. Also, quantitative analysis of the results was difficult since the image was measured by protein-band strength. The technique could not be used for live observation.
The team aimed to drastically improve the existing technique and to develop accurate method of measurement on molecular level. The newly developed technology can enable observation of protein interaction within one hour. Also, the interaction can be measured live, thus the protein interaction phenomenon can be measured in depth.
Moreover, every programme used in the experiment was developed and distributed by the research team so source energy is secured and created the foundation for global infra.
Professor Tae Young Yoon said, “The newly developed technology does not require additional protein expression or purification. Hence, a very small sample of protein is enough to accurately analyse protein interaction on a kinetic level.” He continued, “Even cancerous protein from the tissue of a cancer patient can be analysed. Thus a platform for customised anti-cancer medicine in the future has been prepared, as well.”
Figure 1. Mimetic diagram comparing the existing immunoprecipitation technique and the newly developed live molecular level immunoprecipitation technique
2013.12.11 View 8125 -
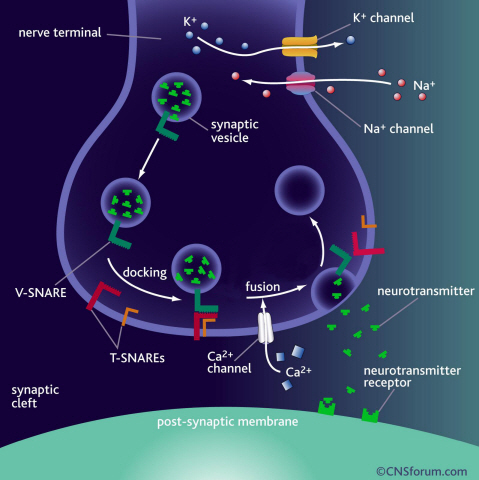 Neurotransmitter protein structure and operation principle identified
Professor Tae-Young Yoon
- Real-time measurement of structural change of bio-membrane fusion protein
- A new clue to degenerative brain diseases research
KAIST Physics Department’s Professor Tae-Young Yoon has successfully identified the hidden structure and operation mechanism of the SNARE protein, which has a central role in transporting neurotransmitters between neurons, using magnetic nanotweezers. SNARE protein’s cell membrane fusion function is closely related to degenerative brain diseases or neurological disorders such as Alzheimer’s. Hence, this research may provide a clue to the disease’s prevention and treatment.
Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. The SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters. The academia speculated the SNARE protein would regulate the exchange of neurotransmitters, but its precise function and structure has been unknown. Professor Yoon’s research team developed an experimental technique using nanotweezers to measure physical changes to nanometer level by pulling and releasing each protein with force of 1 pN (piconewton). The research identified the existence of hidden SNARE protein"s intermediate structure. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been identified.
Professor Yoon’s research team developed an experimental technique using magnetic nanotweezers to measure physical changes of proteins to nanometer level by pulling and releasing each protein with force of 1 pN. The research identified the existence of hidden SNARE protein"s intermediate structure and its formation. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been discovered.
Professor Yoon said, “Ground breaking research results have been produced. A simple experimental technique of applying the smallest possible forces to proteins (with tweezers) to see their hidden structure and formation process can produce the same result as real observation has been developed.” He continued, “This technique will be very important in researching biological object with physical experimental technique. It will be a vital foundation to consilient research of different academia in the future.”
This research was a joint project of Physics Department’s Professor Tae-Young Yoon, KAIST, and Biomedical Engineering Institute’s Professor Yeon-Kyun Shin at KIST. KAIST Physics Department’s Professor Yong-Hoon Cho, Ph.D. candidate Do-Yong Lee and KIAS Computational Sciences Department’s Professor Chang-Bong Hyun participated. The research was published on Nature Communications on April 16th.
a) Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. A SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters.
b) A schematic diagram using magnetic nanotweezers to measure protein structure changes on molecular level. The nanotweezers exert an exquisite pull and release of each protein with a force of 1 pN to measure physical changes to nanometer level in real-time to observe the hidden intermediate structure and operation principles of bio-membrane fusion protein.
2013.05.25 View 9401
Neurotransmitter protein structure and operation principle identified
Professor Tae-Young Yoon
- Real-time measurement of structural change of bio-membrane fusion protein
- A new clue to degenerative brain diseases research
KAIST Physics Department’s Professor Tae-Young Yoon has successfully identified the hidden structure and operation mechanism of the SNARE protein, which has a central role in transporting neurotransmitters between neurons, using magnetic nanotweezers. SNARE protein’s cell membrane fusion function is closely related to degenerative brain diseases or neurological disorders such as Alzheimer’s. Hence, this research may provide a clue to the disease’s prevention and treatment.
Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. The SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters. The academia speculated the SNARE protein would regulate the exchange of neurotransmitters, but its precise function and structure has been unknown. Professor Yoon’s research team developed an experimental technique using nanotweezers to measure physical changes to nanometer level by pulling and releasing each protein with force of 1 pN (piconewton). The research identified the existence of hidden SNARE protein"s intermediate structure. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been identified.
Professor Yoon’s research team developed an experimental technique using magnetic nanotweezers to measure physical changes of proteins to nanometer level by pulling and releasing each protein with force of 1 pN. The research identified the existence of hidden SNARE protein"s intermediate structure and its formation. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been discovered.
Professor Yoon said, “Ground breaking research results have been produced. A simple experimental technique of applying the smallest possible forces to proteins (with tweezers) to see their hidden structure and formation process can produce the same result as real observation has been developed.” He continued, “This technique will be very important in researching biological object with physical experimental technique. It will be a vital foundation to consilient research of different academia in the future.”
This research was a joint project of Physics Department’s Professor Tae-Young Yoon, KAIST, and Biomedical Engineering Institute’s Professor Yeon-Kyun Shin at KIST. KAIST Physics Department’s Professor Yong-Hoon Cho, Ph.D. candidate Do-Yong Lee and KIAS Computational Sciences Department’s Professor Chang-Bong Hyun participated. The research was published on Nature Communications on April 16th.
a) Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. A SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters.
b) A schematic diagram using magnetic nanotweezers to measure protein structure changes on molecular level. The nanotweezers exert an exquisite pull and release of each protein with a force of 1 pN to measure physical changes to nanometer level in real-time to observe the hidden intermediate structure and operation principles of bio-membrane fusion protein.
2013.05.25 View 9401 -
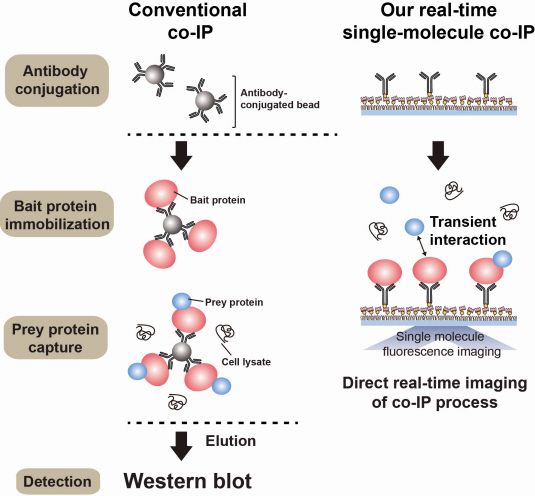 The new era of personalized cancer diagnosis and treatment
Professor Tae-Young Yoon
- Succeeded in observing carcinogenic protein at the molecular level
- “Paved the way to customized cancer treatment through accurate analysis of carcinogenic protein”
The joint KAIST research team of Professor Tae Young Yoon of the Department of Physics and Professor Won Do Huh of the Department of Biological Sciences have developed the technology to monitor characteristics of carcinogenic protein in cancer tissue – for the first time in the world.
The technology makes it possible to analyse the mechanism of cancer development through a small amount of carcinogenic protein from a cancer patient. Therefore, a personalised approach to diagnosis and treatment using the knowledge of the specific mechanism of cancer development in the patient may be possible in the future.
Until recently, modern medicine could only speculate on the cause of cancer through statistics. Although developed countries, such as the United States, are known to use a large sequencing technology that analyses the patient’s DNA, identification of the interactions between proteins responsible for causing cancer remained an unanswered question for a long time in medicine.
Firstly, Professor Yoon’s research team has developed a fluorescent microscope that can observe even a single molecule. Then, the “Immunoprecipitation method”, a technology to extract a specific protein exploiting the high affinity between antigens and antibodies was developed. Using this technology and the microscope, “Real-Time Single Molecule co-Immunoprecipitation Method” was created. In this way, the team succeeded in observing the interactions between carcinogenic and other proteins at a molecular level, in real time.
To validate the developed technology, the team investigated Ras, a carcinogenic protein; its mutation statistically is known to cause around 30% of cancers.
The experimental results confirmed that 30-50% of Ras protein was expressed in mouse tumour and human cancer cells. In normal cells, less than 5% of Ras protein was expressed. Thus, the experiment showed that unusual increase in activation of Ras protein induces cancer.
The increase in the ratio of active Ras protein can be inferred from existing research data but the measurement of specific numerical data has never been done before.
The team suggested a new molecular level diagnosis technique of identifying the progress of cancer in patients through measuring the percentage of activated carcinogenic protein in cancer tissue.
Professor Yoon Tae-young said, “This newly developed technology does not require a separate procedure of protein expression or refining, hence the existing proteins in real biological tissues or cancer cells can be observed directly.” He also said, “Since carcinogenic protein can be analyzed accurately, it has opened up the path to customized cancer treatment in the future.”
“Since the observation is possible on a molecular level, the technology confers the advantage that researchers can carry out various examinations on a small sample of the cancer patient.” He added, “The clinical trial will start in December 2012 and in a few years customized cancer diagnosis and treatment will be possible.”
Meanwhile, the research has been published in Nature Communications (February 19). Many researchers from various fields have participated, regardless of the differences in their speciality, and successfully produced interdisciplinary research. Professor Tae Young Yoon of the Department of Physics and Professors Dae Sik Lim and Won Do Huh of Biological Sciences at KAIST, and Professor Chang Bong Hyun of Computational Science of KIAS contributed to developing the technique.
Figure 1: Schematic diagram of observed interactions at the molecular level in real time using fluorescent microscope. The carcinogenic protein from a mouse tumour is fixed on the microchip, and its molecular characteristics are observed live.
Figure 2: Molecular interaction data using a molecular level fluorescent microscope. A signal in the form of spike is shown when two proteins combine. This is monitored live using an Electron Multiplying Charge Coupled Device (EMCCD). It shows signal results in bright dots.
An organism has an immune system as a defence mechanism to foreign intruders. The immune system is activated when unwanted pathogens or foreign protein are in the body. Antibodies form in recognition of the specific antigen to protect itself. Organisms evolved to form antibodies with high specificity to a certain antigen. Antibodies only react to its complementary antigens. The field of molecular biology uses the affinity between antigens and antibodies to extract specific proteins; a technology called immunoprecipitation. Even in a mixture of many proteins, the protein sought can be extracted using antibodies. Thus immunoprecipitation is widely used to detect pathogens or to extract specific proteins.
Technology co-IP is a well-known example that uses immunoprecipitation. The research on interactions between proteins uses co-IP in general. The basis of fixing the antigen on the antibody to extract antigen protein is the same as immunoprecipitation. Then, researchers inject and observe its reaction with the partner protein to observe the interactions and precipitate the antibodies. If the reaction occurs, the partner protein will be found with the antibodies in the precipitations. If not, then the partner protein will not be found. This shows that the two proteins interact.
However, the traditional co-IP can be used to infer the interactions between the two proteins although the information of the dynamics on how the reaction occurs is lost. To overcome these shortcomings, the Real-Time Single Molecule co-IP Method enables observation on individual protein level in real time. Therefore, the significance of the new technique is in making observation of interactions more direct and quantitative.
Additional Figure 1: Comparison between Conventional co-IP and Real-Time Single Molecule co-IP
2013.04.01 View 18940
The new era of personalized cancer diagnosis and treatment
Professor Tae-Young Yoon
- Succeeded in observing carcinogenic protein at the molecular level
- “Paved the way to customized cancer treatment through accurate analysis of carcinogenic protein”
The joint KAIST research team of Professor Tae Young Yoon of the Department of Physics and Professor Won Do Huh of the Department of Biological Sciences have developed the technology to monitor characteristics of carcinogenic protein in cancer tissue – for the first time in the world.
The technology makes it possible to analyse the mechanism of cancer development through a small amount of carcinogenic protein from a cancer patient. Therefore, a personalised approach to diagnosis and treatment using the knowledge of the specific mechanism of cancer development in the patient may be possible in the future.
Until recently, modern medicine could only speculate on the cause of cancer through statistics. Although developed countries, such as the United States, are known to use a large sequencing technology that analyses the patient’s DNA, identification of the interactions between proteins responsible for causing cancer remained an unanswered question for a long time in medicine.
Firstly, Professor Yoon’s research team has developed a fluorescent microscope that can observe even a single molecule. Then, the “Immunoprecipitation method”, a technology to extract a specific protein exploiting the high affinity between antigens and antibodies was developed. Using this technology and the microscope, “Real-Time Single Molecule co-Immunoprecipitation Method” was created. In this way, the team succeeded in observing the interactions between carcinogenic and other proteins at a molecular level, in real time.
To validate the developed technology, the team investigated Ras, a carcinogenic protein; its mutation statistically is known to cause around 30% of cancers.
The experimental results confirmed that 30-50% of Ras protein was expressed in mouse tumour and human cancer cells. In normal cells, less than 5% of Ras protein was expressed. Thus, the experiment showed that unusual increase in activation of Ras protein induces cancer.
The increase in the ratio of active Ras protein can be inferred from existing research data but the measurement of specific numerical data has never been done before.
The team suggested a new molecular level diagnosis technique of identifying the progress of cancer in patients through measuring the percentage of activated carcinogenic protein in cancer tissue.
Professor Yoon Tae-young said, “This newly developed technology does not require a separate procedure of protein expression or refining, hence the existing proteins in real biological tissues or cancer cells can be observed directly.” He also said, “Since carcinogenic protein can be analyzed accurately, it has opened up the path to customized cancer treatment in the future.”
“Since the observation is possible on a molecular level, the technology confers the advantage that researchers can carry out various examinations on a small sample of the cancer patient.” He added, “The clinical trial will start in December 2012 and in a few years customized cancer diagnosis and treatment will be possible.”
Meanwhile, the research has been published in Nature Communications (February 19). Many researchers from various fields have participated, regardless of the differences in their speciality, and successfully produced interdisciplinary research. Professor Tae Young Yoon of the Department of Physics and Professors Dae Sik Lim and Won Do Huh of Biological Sciences at KAIST, and Professor Chang Bong Hyun of Computational Science of KIAS contributed to developing the technique.
Figure 1: Schematic diagram of observed interactions at the molecular level in real time using fluorescent microscope. The carcinogenic protein from a mouse tumour is fixed on the microchip, and its molecular characteristics are observed live.
Figure 2: Molecular interaction data using a molecular level fluorescent microscope. A signal in the form of spike is shown when two proteins combine. This is monitored live using an Electron Multiplying Charge Coupled Device (EMCCD). It shows signal results in bright dots.
An organism has an immune system as a defence mechanism to foreign intruders. The immune system is activated when unwanted pathogens or foreign protein are in the body. Antibodies form in recognition of the specific antigen to protect itself. Organisms evolved to form antibodies with high specificity to a certain antigen. Antibodies only react to its complementary antigens. The field of molecular biology uses the affinity between antigens and antibodies to extract specific proteins; a technology called immunoprecipitation. Even in a mixture of many proteins, the protein sought can be extracted using antibodies. Thus immunoprecipitation is widely used to detect pathogens or to extract specific proteins.
Technology co-IP is a well-known example that uses immunoprecipitation. The research on interactions between proteins uses co-IP in general. The basis of fixing the antigen on the antibody to extract antigen protein is the same as immunoprecipitation. Then, researchers inject and observe its reaction with the partner protein to observe the interactions and precipitate the antibodies. If the reaction occurs, the partner protein will be found with the antibodies in the precipitations. If not, then the partner protein will not be found. This shows that the two proteins interact.
However, the traditional co-IP can be used to infer the interactions between the two proteins although the information of the dynamics on how the reaction occurs is lost. To overcome these shortcomings, the Real-Time Single Molecule co-IP Method enables observation on individual protein level in real time. Therefore, the significance of the new technique is in making observation of interactions more direct and quantitative.
Additional Figure 1: Comparison between Conventional co-IP and Real-Time Single Molecule co-IP
2013.04.01 View 18940