Sang+Yup+Lee
-
 Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 39567
Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 39567 -
 Efficiently Producing Fatty Acids and Biofuels from Glucose
Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production.
The newly developed strain, created by Distinguished Professor Sang Yup Lee and his team, showed the highest efficiency in producing fatty acids and biodiesels ever reported. It will be expected to serve as a new platform to sustainably produce a wide array of fatty acid-based products from glucose and other carbon substrates.
Fossil fuels, which have long been energy resources for our daily lives, are now facing serious challenges: depletion of their reserves and their role in global warming. The production of sustainable bio-based renewable energy has emerged as an essential alternative and many studies to replace fossil fuels are underway. One of the representative examples is biodiesel. Currently, it is mainly being produced through the transesterification of vegetable oils or animal fats.
The research team engineered oleaginous microorganisms, Rhodococcus opacus, to produce fatty acids and their derivatives that can be used as biodiesel from glucose, one of the most abundant and cheap sugars derived from non-edible biomass.
Professor Lee’s team has already engineered Escherichia coli to produce short-chain hydrocarbons, which can be used as gasoline (published in Nature as the cover paper in 2013). However, the production efficiency of the short-chain hydrocarbons using E. coli (0.58 g/L) fell short of the levels required for commercialization.
To overcome these issues, the team employed oil-accumulating Rhodococcus opacus as a host strain in this study. First, the team optimized the cultivation conditions of Rhodococcus opacus to maximize the accumulation of oil (triacylglycerol), which serves as a precursor for the biosynthesis of fatty acids and their derivatives. Then, they systematically analyzed the metabolism of the strain and redesigned it to enable higher levels of fatty acids and two kinds of fatty acid-derived biodiesels (fatty acid ethyl esters and long-chain hydrocarbons) to be produced.
They found that the resulting strains produced 50.2, 21.3, and 5.2 g/L of fatty acids, fatty acid ethyl esters, and long-chain hydrocarbons, respectively. These are all the highest concentrations ever reported by microbial fermentations. It is expected that these strains can contribute to the future industrialization of microbial-based biodiesel production.
“This technology creates fatty acids and biodiesel with high efficiency by utilizing lignocellulose, one of the most abundant resources on the Earth, without depending on fossil fuels and vegetable or animal oils. This will provide new opportunities for oil and petroleum industries, which have long relied on fossil fuels, to turn to sustainable and eco-friendly biotechnologies,” said Professor Lee.
This paper titled “Engineering of an oleaginous bacterium for the production of fatty acids and fuels” was published in Nature Chemical Biology on June 17.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557).
(Figure: Metabolic engineering for the production of free fatty acids (FFAs), fatty acid ethyl esters (FAEEs), and long-chain hydrocarbons (LCHCs) in Rhodococcus opacus PD630. Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production.)
# # #
Source:
Hye Mi Kim, Tong Un Chae, So Young Choi, Won Jun Kim and Sang Yup Lee. Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nature Chemical Biology ( https://www.nature.com/nchembio/ ) DOI: 10.1038/s41589-019-0295-5
Profile
Dr. Sang Yup Lee
leesy@kaist.ac.kr
Distinguished Professor at the Department of Chemical and Biomolecular Engineering
KAIST
2019.06.19 View 51341
Efficiently Producing Fatty Acids and Biofuels from Glucose
Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production.
The newly developed strain, created by Distinguished Professor Sang Yup Lee and his team, showed the highest efficiency in producing fatty acids and biodiesels ever reported. It will be expected to serve as a new platform to sustainably produce a wide array of fatty acid-based products from glucose and other carbon substrates.
Fossil fuels, which have long been energy resources for our daily lives, are now facing serious challenges: depletion of their reserves and their role in global warming. The production of sustainable bio-based renewable energy has emerged as an essential alternative and many studies to replace fossil fuels are underway. One of the representative examples is biodiesel. Currently, it is mainly being produced through the transesterification of vegetable oils or animal fats.
The research team engineered oleaginous microorganisms, Rhodococcus opacus, to produce fatty acids and their derivatives that can be used as biodiesel from glucose, one of the most abundant and cheap sugars derived from non-edible biomass.
Professor Lee’s team has already engineered Escherichia coli to produce short-chain hydrocarbons, which can be used as gasoline (published in Nature as the cover paper in 2013). However, the production efficiency of the short-chain hydrocarbons using E. coli (0.58 g/L) fell short of the levels required for commercialization.
To overcome these issues, the team employed oil-accumulating Rhodococcus opacus as a host strain in this study. First, the team optimized the cultivation conditions of Rhodococcus opacus to maximize the accumulation of oil (triacylglycerol), which serves as a precursor for the biosynthesis of fatty acids and their derivatives. Then, they systematically analyzed the metabolism of the strain and redesigned it to enable higher levels of fatty acids and two kinds of fatty acid-derived biodiesels (fatty acid ethyl esters and long-chain hydrocarbons) to be produced.
They found that the resulting strains produced 50.2, 21.3, and 5.2 g/L of fatty acids, fatty acid ethyl esters, and long-chain hydrocarbons, respectively. These are all the highest concentrations ever reported by microbial fermentations. It is expected that these strains can contribute to the future industrialization of microbial-based biodiesel production.
“This technology creates fatty acids and biodiesel with high efficiency by utilizing lignocellulose, one of the most abundant resources on the Earth, without depending on fossil fuels and vegetable or animal oils. This will provide new opportunities for oil and petroleum industries, which have long relied on fossil fuels, to turn to sustainable and eco-friendly biotechnologies,” said Professor Lee.
This paper titled “Engineering of an oleaginous bacterium for the production of fatty acids and fuels” was published in Nature Chemical Biology on June 17.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557).
(Figure: Metabolic engineering for the production of free fatty acids (FFAs), fatty acid ethyl esters (FAEEs), and long-chain hydrocarbons (LCHCs) in Rhodococcus opacus PD630. Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production.)
# # #
Source:
Hye Mi Kim, Tong Un Chae, So Young Choi, Won Jun Kim and Sang Yup Lee. Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nature Chemical Biology ( https://www.nature.com/nchembio/ ) DOI: 10.1038/s41589-019-0295-5
Profile
Dr. Sang Yup Lee
leesy@kaist.ac.kr
Distinguished Professor at the Department of Chemical and Biomolecular Engineering
KAIST
2019.06.19 View 51341 -
 Engineered Microbial Production of Grape Flavoring
(Image 1: Engineered bacteria that produce grape flavoring.)
Researchers report a microbial method for producing an artificial grape flavor. Methyl anthranilate (MANT) is a common grape flavoring and odorant compound currently produced through a petroleum-based process that uses large volumes of toxic acid catalysts.
Professor Sang-Yup Lee’s team at the Department of Chemical and Biomolecular Engineering demonstrated production of MANT, a naturally occurring compound, via engineered bacteria. The authors engineered strains of Escherichia coli and Corynebacetrium glutamicum to produce MANT through a plant-based engineered metabolic pathway.
The authors tuned the bacterial metabolic pathway by optimizing the levels of AAMT1, the key enzyme in the process. To maximize production of MANT, the authors tested six strategies, including increasing the supply of a precursor compound and enhancing the availability of a co-substrate. The most productive strategy proved to be a two-phase extractive culture, in which MANT was extracted into a solvent. This strategy produced MANT on the scale of 4.47 to 5.74 grams per liter, a significant amount, considering that engineered microbes produce most natural products at a scale of milligrams or micrograms per liter.
According to the authors, the results suggest that MANT and other related molecules produced through industrial processes can be produced at scale by engineered microbes in a manner that would allow them to be marketed as natural one, instead of artificial one.
This study, featured at the Proceeding of the National Academy of Sciences of the USA on May 13, was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT.
(Image 2. Overview of the strategies applied for the microbial production of grape flavoring.)
2019.05.15 View 57244
Engineered Microbial Production of Grape Flavoring
(Image 1: Engineered bacteria that produce grape flavoring.)
Researchers report a microbial method for producing an artificial grape flavor. Methyl anthranilate (MANT) is a common grape flavoring and odorant compound currently produced through a petroleum-based process that uses large volumes of toxic acid catalysts.
Professor Sang-Yup Lee’s team at the Department of Chemical and Biomolecular Engineering demonstrated production of MANT, a naturally occurring compound, via engineered bacteria. The authors engineered strains of Escherichia coli and Corynebacetrium glutamicum to produce MANT through a plant-based engineered metabolic pathway.
The authors tuned the bacterial metabolic pathway by optimizing the levels of AAMT1, the key enzyme in the process. To maximize production of MANT, the authors tested six strategies, including increasing the supply of a precursor compound and enhancing the availability of a co-substrate. The most productive strategy proved to be a two-phase extractive culture, in which MANT was extracted into a solvent. This strategy produced MANT on the scale of 4.47 to 5.74 grams per liter, a significant amount, considering that engineered microbes produce most natural products at a scale of milligrams or micrograms per liter.
According to the authors, the results suggest that MANT and other related molecules produced through industrial processes can be produced at scale by engineered microbes in a manner that would allow them to be marketed as natural one, instead of artificial one.
This study, featured at the Proceeding of the National Academy of Sciences of the USA on May 13, was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT.
(Image 2. Overview of the strategies applied for the microbial production of grape flavoring.)
2019.05.15 View 57244 -
 A Comprehensive Metabolic Map for Bio-Based Chemicals Production
A KAIST research team completed a metabolic map that charts all available strategies and pathways of chemical reactions that lead to the production of various industrial bio-based chemicals.
The team was led by Distinguished Professor Sang Yup Lee, who has produced high-quality metabolic engineering and systems engineering research for decades, and made the hallmark chemicals map after seven years of studies.
The team presented a very detailed analysis on metabolic engineering for the production of a wide range of industrial chemicals, fuels, and materials. Surveying the current trends in the bio-based production of chemicals in industrial biotechnology, the team thoroughly examined the current status of industrial chemicals produced using biological and/or chemical reactions.
This comprehensive map is expected to serve as a blueprint for the visual and intuitive inspection of biological and/or chemical reactions for the production of interest from renewable resources. The team also compiled an accompanying poster to visually present the synthetic pathways of chemicals in the context of their microbial metabolism.
As metabolic engineering has become increasing powerful in addressing limited fossil resources, climate change, and other environmental issues, the number of microbially produced chemicals using biomass as a carbon source has increased substantially. The sustainable production of industrial chemicals and materials has been explored with micro-organisms as cell factories and renewable nonfood biomass as raw materials for alternative petroleum. The engineering of these micro-organism has increasingly become more efficient and effective with the help of metabolic engineering – a practice of engineering using the metabolism of living organisms to produce a desired metabolite.
With the establishment of systems metabolic engineering – the integration of metabolic engineering with tools and strategies from systems biology, synthetic biology and evolutionary engineering – the speed at which micro-organisms are being engineered has reached an unparalleled pace.
In order to evaluate the current state at which metabolically engineered micro-organisms can produce a large portfolio of industrial chemicals, the team conducted an extensive review of the literature and mapped them out on a poster. This resulting poster, termed the bio-based chemicals map, presents synthetic pathways for industrial chemicals, which consist of biological and/or chemical reactions.
Industrial chemicals and their production routes are presented in the context of central carbon metabolic pathways as these key metabolites serve as precursors for the chemicals to be produced. The resulting biochemical map allows the detection and analysis of optimal synthetic pathways for a given industrial chemical. In addition to the poster, the authors have compiled a list of chemicals that have successfully been produced using micro-organisms and a list of the corresponding companies producing them commercially. This thorough review of the literature and the accompanying analytical summary will be an important resource for researchers interested in the production of chemicals from renewable biomass sources.
Metabolically engineered micro-organisms have already made a huge contribution toward the sustainable production of chemicals using renewable resources. Professor Lee said he wanted a detailed survey of the current state and capacity of bio-based chemicals production.
“We are so excited that this review and poster will expand further discussion on the production of important chemicals through engineered micro-organisms and also combined biological and chemical means in a more sustainable manner,” he explained.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biofineries from the Ministry of Science and ICT through the National Research Foundation of Korea.
For further information, Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST ( leesy@kaist.ac.kr , Tel: +82-42-350-3930)
Figure: Bio-based chemicals production through biological and chemical routes. This metabolic map describes representative chemicals that can be produced either by biological and/or chemical means. Red arrows represent chemical routes and blue arrows represent biological routes. Intermediate metabolites in the metabolism of a living organism can serve as a platform toward the production of industrially relevant chemicals. A more comprehensive map presented by the team can be found as a poster in the review.
2019.01.15 View 7929
A Comprehensive Metabolic Map for Bio-Based Chemicals Production
A KAIST research team completed a metabolic map that charts all available strategies and pathways of chemical reactions that lead to the production of various industrial bio-based chemicals.
The team was led by Distinguished Professor Sang Yup Lee, who has produced high-quality metabolic engineering and systems engineering research for decades, and made the hallmark chemicals map after seven years of studies.
The team presented a very detailed analysis on metabolic engineering for the production of a wide range of industrial chemicals, fuels, and materials. Surveying the current trends in the bio-based production of chemicals in industrial biotechnology, the team thoroughly examined the current status of industrial chemicals produced using biological and/or chemical reactions.
This comprehensive map is expected to serve as a blueprint for the visual and intuitive inspection of biological and/or chemical reactions for the production of interest from renewable resources. The team also compiled an accompanying poster to visually present the synthetic pathways of chemicals in the context of their microbial metabolism.
As metabolic engineering has become increasing powerful in addressing limited fossil resources, climate change, and other environmental issues, the number of microbially produced chemicals using biomass as a carbon source has increased substantially. The sustainable production of industrial chemicals and materials has been explored with micro-organisms as cell factories and renewable nonfood biomass as raw materials for alternative petroleum. The engineering of these micro-organism has increasingly become more efficient and effective with the help of metabolic engineering – a practice of engineering using the metabolism of living organisms to produce a desired metabolite.
With the establishment of systems metabolic engineering – the integration of metabolic engineering with tools and strategies from systems biology, synthetic biology and evolutionary engineering – the speed at which micro-organisms are being engineered has reached an unparalleled pace.
In order to evaluate the current state at which metabolically engineered micro-organisms can produce a large portfolio of industrial chemicals, the team conducted an extensive review of the literature and mapped them out on a poster. This resulting poster, termed the bio-based chemicals map, presents synthetic pathways for industrial chemicals, which consist of biological and/or chemical reactions.
Industrial chemicals and their production routes are presented in the context of central carbon metabolic pathways as these key metabolites serve as precursors for the chemicals to be produced. The resulting biochemical map allows the detection and analysis of optimal synthetic pathways for a given industrial chemical. In addition to the poster, the authors have compiled a list of chemicals that have successfully been produced using micro-organisms and a list of the corresponding companies producing them commercially. This thorough review of the literature and the accompanying analytical summary will be an important resource for researchers interested in the production of chemicals from renewable biomass sources.
Metabolically engineered micro-organisms have already made a huge contribution toward the sustainable production of chemicals using renewable resources. Professor Lee said he wanted a detailed survey of the current state and capacity of bio-based chemicals production.
“We are so excited that this review and poster will expand further discussion on the production of important chemicals through engineered micro-organisms and also combined biological and chemical means in a more sustainable manner,” he explained.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biofineries from the Ministry of Science and ICT through the National Research Foundation of Korea.
For further information, Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST ( leesy@kaist.ac.kr , Tel: +82-42-350-3930)
Figure: Bio-based chemicals production through biological and chemical routes. This metabolic map describes representative chemicals that can be produced either by biological and/or chemical means. Red arrows represent chemical routes and blue arrows represent biological routes. Intermediate metabolites in the metabolism of a living organism can serve as a platform toward the production of industrially relevant chemicals. A more comprehensive map presented by the team can be found as a poster in the review.
2019.01.15 View 7929 -
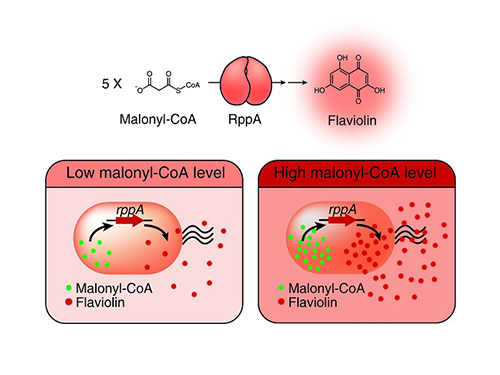 A Novel Biosensor to Advance Diverse High-Level Production of Microbial Cell Factories
A research group at KAIST presented a novel biosensor which can produce diverse, high-level microbial cell factories. The biosensor monitors the concentration of products and even intermediates when new strains are being developed. This strategy provides a new platform for manufacturing diverse natural products from renewable resources. The team succeeded in creating four natural products of high-level pharmaceutical importance with this strategy.
Malonyl-CoA is a major building block for many value-added chemicals including diverse natural products with pharmaceutical importance. However, due to the low availability of malonyl-CoA in bacteria, many malonyl-CoA-derived natural products have been produced by chemical synthesis or extraction from natural resources that are harmful to the environment and are unsustainable. For the sustainable biological production of malonyl-CoA-derived natural products, increasing the intracellular malonyl-CoA pool is necessary. To this end, the development of a robust and efficient malonyl-CoA biosensor was required to monitor the concentration of intracellular malonyl-CoA abundance as new strains are developed.
Metabolic engineering researchers at KAIST addressed this issue. This research reports the development of a simple and robust malonyl-CoA biosensor by repurposing a type III polyketide synthase (also known as RppA), which produces flaviolin, a colorimetric indicator of malonyl-CoA. Subsequently, the RppA biosensor was used for the rapid and efficient colorimetric screening of gene manipulation targets enabling enhanced malonyl-CoA abundance. The screened beneficial gene targets were employed for the high-level production of four representative natural products derived from malonyl-CoA. Compared with the previous strategies, which were expensive and time-consuming, the new biosensor could be easily applied to industrially relevant bacteria including Escherichia coli, Pseudomonas putida, and Corynebacterium glutamicum to enable a one-step process.
The study employs synthetic small regulatory RNA (sRNA) technology to rapidly and efficiently reduce endogenous target gene expression for improved malonyl-CoA production. The researchers constructed an E. coli genome-scale synthetic sRNA library targeting 1,858 genes covering all major metabolic genes in E. coli. This library was employed with the RppA biosensor to screen for gene targets which are believed to be beneficial for enhancing malonyl-CoA accumulation upon their expression knockdown.
From this colorimetric screening, 14 gene targets were selected, all of which were successful at significantly increasing the production of four natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin). Although specific examples are demonstrated in E. coli as a host, the researchers showed that the biosensor is also functional in P. putida and C. glutamicum, industrially important representative gram-negative and gram-positive bacteria, respectively. The malonyl-CoA biosensor developed in this research will serve as an efficient platform for the rapid development of strains capable of producing natural products crucial for the pharmaceutical, chemical, cosmetics, and food industries.
An important aspect of this work is that the high-performance strains constructed in this research were developed rapidly and easily by utilizing the simple approach of colorimetric screening, without involving extensive metabolic engineering approaches. 6-Methylsalicylic acid (an antibiotic) could be produced to the highest titer reported for E. coli, and the microbial production of aloesone (a precursor of aloesin, an anti-inflammatory agent/whitening agent) was achieved for the first time.
“A sustainable process for producing diverse natural products using renewable resources is of great interest. This study represents the development of a robust and efficient malonyl-CoA biosensor generally applicable to a wide range of industrially important bacteria. The capability of this biosensor for screening a large library was demonstrated to show that the rapid and efficient construction of high-performance strains is feasible. This research will be useful for further accelerating the development process of strains capable of producing valuable chemicals to industrially relevant levels,” said Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, who led the research.
This study entitled “Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria,” was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on October 02.
PhD students Dongsoo Yang and Won Jun Kim, MS student Shin Hee Ha, research staff Mun Hee Lee, Research Professor Seung Min Yoo, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Dr. Jong Hyun Choi of the Applied Microbiology Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB) participated in this research.
Figure: Type III polyketide synthase (RppA) as a malonyl-CoA biosensor. RppA converts five molecules of malonyl-CoA into one molecule of red-colored flaviolin. This schematic diagram shows the overall conceptualization of the malonyl-CoA biosensor by indicating that higher malonyl-CoA abundance leads to higher production and secretion of flaviolin, resulting in a deeper red color of the culture. This system was employed for the enhanced production of four representative natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin) from engineered E. coli strains.
2018.10.11 View 10702
A Novel Biosensor to Advance Diverse High-Level Production of Microbial Cell Factories
A research group at KAIST presented a novel biosensor which can produce diverse, high-level microbial cell factories. The biosensor monitors the concentration of products and even intermediates when new strains are being developed. This strategy provides a new platform for manufacturing diverse natural products from renewable resources. The team succeeded in creating four natural products of high-level pharmaceutical importance with this strategy.
Malonyl-CoA is a major building block for many value-added chemicals including diverse natural products with pharmaceutical importance. However, due to the low availability of malonyl-CoA in bacteria, many malonyl-CoA-derived natural products have been produced by chemical synthesis or extraction from natural resources that are harmful to the environment and are unsustainable. For the sustainable biological production of malonyl-CoA-derived natural products, increasing the intracellular malonyl-CoA pool is necessary. To this end, the development of a robust and efficient malonyl-CoA biosensor was required to monitor the concentration of intracellular malonyl-CoA abundance as new strains are developed.
Metabolic engineering researchers at KAIST addressed this issue. This research reports the development of a simple and robust malonyl-CoA biosensor by repurposing a type III polyketide synthase (also known as RppA), which produces flaviolin, a colorimetric indicator of malonyl-CoA. Subsequently, the RppA biosensor was used for the rapid and efficient colorimetric screening of gene manipulation targets enabling enhanced malonyl-CoA abundance. The screened beneficial gene targets were employed for the high-level production of four representative natural products derived from malonyl-CoA. Compared with the previous strategies, which were expensive and time-consuming, the new biosensor could be easily applied to industrially relevant bacteria including Escherichia coli, Pseudomonas putida, and Corynebacterium glutamicum to enable a one-step process.
The study employs synthetic small regulatory RNA (sRNA) technology to rapidly and efficiently reduce endogenous target gene expression for improved malonyl-CoA production. The researchers constructed an E. coli genome-scale synthetic sRNA library targeting 1,858 genes covering all major metabolic genes in E. coli. This library was employed with the RppA biosensor to screen for gene targets which are believed to be beneficial for enhancing malonyl-CoA accumulation upon their expression knockdown.
From this colorimetric screening, 14 gene targets were selected, all of which were successful at significantly increasing the production of four natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin). Although specific examples are demonstrated in E. coli as a host, the researchers showed that the biosensor is also functional in P. putida and C. glutamicum, industrially important representative gram-negative and gram-positive bacteria, respectively. The malonyl-CoA biosensor developed in this research will serve as an efficient platform for the rapid development of strains capable of producing natural products crucial for the pharmaceutical, chemical, cosmetics, and food industries.
An important aspect of this work is that the high-performance strains constructed in this research were developed rapidly and easily by utilizing the simple approach of colorimetric screening, without involving extensive metabolic engineering approaches. 6-Methylsalicylic acid (an antibiotic) could be produced to the highest titer reported for E. coli, and the microbial production of aloesone (a precursor of aloesin, an anti-inflammatory agent/whitening agent) was achieved for the first time.
“A sustainable process for producing diverse natural products using renewable resources is of great interest. This study represents the development of a robust and efficient malonyl-CoA biosensor generally applicable to a wide range of industrially important bacteria. The capability of this biosensor for screening a large library was demonstrated to show that the rapid and efficient construction of high-performance strains is feasible. This research will be useful for further accelerating the development process of strains capable of producing valuable chemicals to industrially relevant levels,” said Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, who led the research.
This study entitled “Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria,” was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on October 02.
PhD students Dongsoo Yang and Won Jun Kim, MS student Shin Hee Ha, research staff Mun Hee Lee, Research Professor Seung Min Yoo, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Dr. Jong Hyun Choi of the Applied Microbiology Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB) participated in this research.
Figure: Type III polyketide synthase (RppA) as a malonyl-CoA biosensor. RppA converts five molecules of malonyl-CoA into one molecule of red-colored flaviolin. This schematic diagram shows the overall conceptualization of the malonyl-CoA biosensor by indicating that higher malonyl-CoA abundance leads to higher production and secretion of flaviolin, resulting in a deeper red color of the culture. This system was employed for the enhanced production of four representative natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin) from engineered E. coli strains.
2018.10.11 View 10702 -
 Engineered E. coli Using Formic Acid and CO2 As a C1-Refinery Platform Strain
(Figure: Formic acid and CO2 assimilation pathways consisting of the reconstructed THF cycle and reverse glycine cleavage reaction. This schematic diagram shows the formic acid and CO2 assimilation procedure through the pathway. Plasmids used in this study and the genetic engineering performed in this study are illustrated.)
A research group at KAIST has developed an engineered E. coli strain that converts formic acid and CO2 to pyruvate and produces cellular energy from formic acid through reconstructed one-carbon pathways. The strategy described in this study provides a new platform for producing value-added chemicals from one-carbon sources.
Formic acid is a carboxylic acid composed of one carbon. Formic acid was produced from CO2 by the chemical method. Recently, the C1 Gas Refinery R&D Center has successfully developed a biological process that produces formic acid from carbon monoxide for the first time. Formic acid is in a liquid state when at room temperature and atmospheric pressure. In addition, it is chemically stable and less toxic, thus, easy to store and transport. Therefore, it can be used as an alternative carbon source in the microbial fermentation process. In order to produce value-added chemicals using formic acid, a metabolic pathway that converts formic acid into cellular molecules composed of multiple carbons is required. However, a metabolic pathway that can efficiently convert formic acid into cellular molecules has not been developed. This acted as an obstacle for the production of value-added chemicals using formic acid
A research group of Ph.D. student Junho Bang and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering addressed this issue. This study, entitled “Assimilation of Formic Acid and CO2 by Engineered Escherichia coli Equipped with Reconstructed One-Carbon Assimilation Pathways”, has been published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on September 18.
There has been increasing interest in utilizing formic acid as an alternative carbon source for the production of value-added chemicals. This research reports the development of an engineered E. coli strain that can convert formic acid and CO2 to pyruvate and produce cellular energy from formic acid through the reconstructed one-carbon pathways.
The metabolic pathway that efficiently converts formic acid and CO2 into pyruvate was constructed by the combined use of the tetrahydrofolate cycle and reverse glycine cleavage reaction. The tetrahydrofolate cycle was reconstructed by utilizing Methylobacterium extorquens formate-THF ligase, methenyl-THF cyclohydrolase, and methylene-THF dehydrogenase. The glycine cleavage reaction was reversed by knocking out the repressor gene (gcvR) and overexpressing the gcvTHP genes that encode enzymes related with the glycine cleavage reaction. Formic acid and CO2 conversion to pyruvate was increased via metabolic engineering of the E. coli strain equipped with the one-carbon assimilation pathway.
In addition, in order to reduce glucose consumption and increase formic acid consumption, Candida boidnii formate dehydrogenase was additionally introduced to construct a cellular energy producing pathway from formic acid. This reduces glucose consumption and increases formic acid consumption.
The reconstructed one-carbon pathways can supply cellular molecules and cellular energies from the formic acid and CO2. Thus, the engineered E. coli strain equipped with the formic acid and CO2 assimilation pathway and cellular energy producing pathway from formic acid showed cell growth from formic acid and CO2 without glucose. Cell growth was monitored and 13C isotope analysis was performed to confirm E. coli growth from the formic acid and CO2. It was found that the engineered E. coli strain sustained cell growth from the formic acid and CO2 without glucose.
Professor Lee said, “To construct the C1-refinery system, a platform strain that can convert one-carbon materials to higher carbon materials needs to be developed. In this report, a one-carbon pathway that can efficiently convert formic acid and CO2 to pyruvate was developed and a cellular energy producing pathway from formic acid was introduced. This resulted in an engineered E. coli strain that can efficiently utilize formic acid as a carbon source while glucose consumption was reduced. The reconstructed one-carbon pathways in this research will be useful for the construction of the C1-refinery system.”
This work was supported by the C1 Gas Refinery Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2016M3D3A1A01913250). For further information: Sang Yup Lee, Distinguished Professor of Chemical and Biomolecular Engineering, KAIST (leesy@kaist.ac.kr, Tel: +82-42-350-3930)
2018.09.18 View 7452
Engineered E. coli Using Formic Acid and CO2 As a C1-Refinery Platform Strain
(Figure: Formic acid and CO2 assimilation pathways consisting of the reconstructed THF cycle and reverse glycine cleavage reaction. This schematic diagram shows the formic acid and CO2 assimilation procedure through the pathway. Plasmids used in this study and the genetic engineering performed in this study are illustrated.)
A research group at KAIST has developed an engineered E. coli strain that converts formic acid and CO2 to pyruvate and produces cellular energy from formic acid through reconstructed one-carbon pathways. The strategy described in this study provides a new platform for producing value-added chemicals from one-carbon sources.
Formic acid is a carboxylic acid composed of one carbon. Formic acid was produced from CO2 by the chemical method. Recently, the C1 Gas Refinery R&D Center has successfully developed a biological process that produces formic acid from carbon monoxide for the first time. Formic acid is in a liquid state when at room temperature and atmospheric pressure. In addition, it is chemically stable and less toxic, thus, easy to store and transport. Therefore, it can be used as an alternative carbon source in the microbial fermentation process. In order to produce value-added chemicals using formic acid, a metabolic pathway that converts formic acid into cellular molecules composed of multiple carbons is required. However, a metabolic pathway that can efficiently convert formic acid into cellular molecules has not been developed. This acted as an obstacle for the production of value-added chemicals using formic acid
A research group of Ph.D. student Junho Bang and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering addressed this issue. This study, entitled “Assimilation of Formic Acid and CO2 by Engineered Escherichia coli Equipped with Reconstructed One-Carbon Assimilation Pathways”, has been published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on September 18.
There has been increasing interest in utilizing formic acid as an alternative carbon source for the production of value-added chemicals. This research reports the development of an engineered E. coli strain that can convert formic acid and CO2 to pyruvate and produce cellular energy from formic acid through the reconstructed one-carbon pathways.
The metabolic pathway that efficiently converts formic acid and CO2 into pyruvate was constructed by the combined use of the tetrahydrofolate cycle and reverse glycine cleavage reaction. The tetrahydrofolate cycle was reconstructed by utilizing Methylobacterium extorquens formate-THF ligase, methenyl-THF cyclohydrolase, and methylene-THF dehydrogenase. The glycine cleavage reaction was reversed by knocking out the repressor gene (gcvR) and overexpressing the gcvTHP genes that encode enzymes related with the glycine cleavage reaction. Formic acid and CO2 conversion to pyruvate was increased via metabolic engineering of the E. coli strain equipped with the one-carbon assimilation pathway.
In addition, in order to reduce glucose consumption and increase formic acid consumption, Candida boidnii formate dehydrogenase was additionally introduced to construct a cellular energy producing pathway from formic acid. This reduces glucose consumption and increases formic acid consumption.
The reconstructed one-carbon pathways can supply cellular molecules and cellular energies from the formic acid and CO2. Thus, the engineered E. coli strain equipped with the formic acid and CO2 assimilation pathway and cellular energy producing pathway from formic acid showed cell growth from formic acid and CO2 without glucose. Cell growth was monitored and 13C isotope analysis was performed to confirm E. coli growth from the formic acid and CO2. It was found that the engineered E. coli strain sustained cell growth from the formic acid and CO2 without glucose.
Professor Lee said, “To construct the C1-refinery system, a platform strain that can convert one-carbon materials to higher carbon materials needs to be developed. In this report, a one-carbon pathway that can efficiently convert formic acid and CO2 to pyruvate was developed and a cellular energy producing pathway from formic acid was introduced. This resulted in an engineered E. coli strain that can efficiently utilize formic acid as a carbon source while glucose consumption was reduced. The reconstructed one-carbon pathways in this research will be useful for the construction of the C1-refinery system.”
This work was supported by the C1 Gas Refinery Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2016M3D3A1A01913250). For further information: Sang Yup Lee, Distinguished Professor of Chemical and Biomolecular Engineering, KAIST (leesy@kaist.ac.kr, Tel: +82-42-350-3930)
2018.09.18 View 7452 -
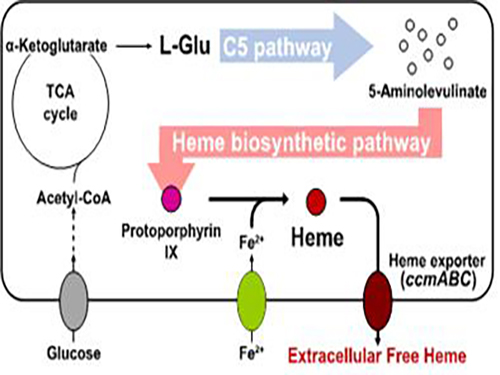 Metabolic Engineering of E. coli for the Secretory Production of Free Haem
Researchers of KAIST have defined a novel strategy for the secretory production of free haem using engineered Escherichia coli (E. coli) strains. They utilized the C5 pathway, the optimized downstream pathways, and the haem exporter to construct a recombinant micro-organism producing extracellular haem using fed-batch fermentation. This is the first report to extracellularly produce haem using engineered E. coli.
This strategy will expedite the efficient production of free haem to serve as a bioavailable iron-supplying agent and an important prosthetic group of multiple hemoproteins for medical uses. This study, led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, was published in Nature Catalysis on Aug. 28.
Haem, an organometallic compound complexed with a ferrous ion, is an essential molecule delivering oxygen in the blood of many animals. It is also a key component of electron transport chains responsible for the respiration of aerobic organisms including diverse bacteria. It is now being widely applied as a bioavailable iron-supplying agent in the healthcare and dietary supplement industries. The demand for haem and the need for the efficient production of this compound continue to grow.
Many previous researchers have attempted to produce free haem using engineered E. coli. However, none of the studies was successful in producing free haem extracellularly, requiring an additional step to extract the accumulated haem from cells for subsequent uses. The secretion of haem in the form of haem peptides or proteins also requires an extraction step to isolate the free haem from the secreted products. Thus, the secretory production of free haem is an important task for the economical production of haem that is suitable for human consumption.
Although some researchers could produce intracellular haem using recombinant E. coli strains, its final titer was extremely low, resulting from the use of sub-optimal metabolic pathways. Furthermore, the addition of the precursors L-glycine and succinate was deemed undesirable for massive industrial production. Thus, it is necessary to construct an optimized haem biosynthetic pathway to enable the efficient production of haem and examine the consequent secretion of free haem.
To address this issue, the KAIST team used multiple strategies to produce extracellular free haem by enhancing its biosynthesis in E. coli. First, the capacities of the C4 and C5 pathways to produce aminolevulinate (ALA) without feeding precursors were examined. After confirming the superior performance of the C5 pathway over the C4 pathway, the metabolic genes of the C5 pathway and downstream pathways for haem biosynthesis were overexpressed. Then, the metabolic pathways were optimized by adjusting the expression levels of the relevant genes and disrupting the putative haem degradation enzyme encoded by the yfeX gene.
Consequently, the resulting engineered strain secreted a significant amount of haem to the medium. Subsequent optimization of the cultivation conditions and the supplementation of nitrogen sources further increased both the titer of the total free haem and the amount of free haem secreted to the medium. Finally, the overexpression of the ccmABC genes encoding the haem exporter further enhanced the production and secretion of haem, producing the highest titer of haem both intracellularly and extracellularly from glucose.
Professor Lee said, “The eco-friendly and sustainable chemical industry is a key global agenda every nation faces. We are conducting research to bio-synthesize high concentrations, high yields, and high productivity in natural products. This novel technology will serve as an opportunity to advance the biochemical industry moving forward.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea. Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea ( leesy@kaist.ac.kr+82-42-350-3930).
2018.08.28 View 6320
Metabolic Engineering of E. coli for the Secretory Production of Free Haem
Researchers of KAIST have defined a novel strategy for the secretory production of free haem using engineered Escherichia coli (E. coli) strains. They utilized the C5 pathway, the optimized downstream pathways, and the haem exporter to construct a recombinant micro-organism producing extracellular haem using fed-batch fermentation. This is the first report to extracellularly produce haem using engineered E. coli.
This strategy will expedite the efficient production of free haem to serve as a bioavailable iron-supplying agent and an important prosthetic group of multiple hemoproteins for medical uses. This study, led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, was published in Nature Catalysis on Aug. 28.
Haem, an organometallic compound complexed with a ferrous ion, is an essential molecule delivering oxygen in the blood of many animals. It is also a key component of electron transport chains responsible for the respiration of aerobic organisms including diverse bacteria. It is now being widely applied as a bioavailable iron-supplying agent in the healthcare and dietary supplement industries. The demand for haem and the need for the efficient production of this compound continue to grow.
Many previous researchers have attempted to produce free haem using engineered E. coli. However, none of the studies was successful in producing free haem extracellularly, requiring an additional step to extract the accumulated haem from cells for subsequent uses. The secretion of haem in the form of haem peptides or proteins also requires an extraction step to isolate the free haem from the secreted products. Thus, the secretory production of free haem is an important task for the economical production of haem that is suitable for human consumption.
Although some researchers could produce intracellular haem using recombinant E. coli strains, its final titer was extremely low, resulting from the use of sub-optimal metabolic pathways. Furthermore, the addition of the precursors L-glycine and succinate was deemed undesirable for massive industrial production. Thus, it is necessary to construct an optimized haem biosynthetic pathway to enable the efficient production of haem and examine the consequent secretion of free haem.
To address this issue, the KAIST team used multiple strategies to produce extracellular free haem by enhancing its biosynthesis in E. coli. First, the capacities of the C4 and C5 pathways to produce aminolevulinate (ALA) without feeding precursors were examined. After confirming the superior performance of the C5 pathway over the C4 pathway, the metabolic genes of the C5 pathway and downstream pathways for haem biosynthesis were overexpressed. Then, the metabolic pathways were optimized by adjusting the expression levels of the relevant genes and disrupting the putative haem degradation enzyme encoded by the yfeX gene.
Consequently, the resulting engineered strain secreted a significant amount of haem to the medium. Subsequent optimization of the cultivation conditions and the supplementation of nitrogen sources further increased both the titer of the total free haem and the amount of free haem secreted to the medium. Finally, the overexpression of the ccmABC genes encoding the haem exporter further enhanced the production and secretion of haem, producing the highest titer of haem both intracellularly and extracellularly from glucose.
Professor Lee said, “The eco-friendly and sustainable chemical industry is a key global agenda every nation faces. We are conducting research to bio-synthesize high concentrations, high yields, and high productivity in natural products. This novel technology will serve as an opportunity to advance the biochemical industry moving forward.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea. Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea ( leesy@kaist.ac.kr+82-42-350-3930).
2018.08.28 View 6320 -
 Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi)
A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements.
The study analyzed the nanomaterial biosynthesis conditions using a Pourbaix diagram to predict the producibility and crystallinity. Researchers studied a Pourbaix diagram to predict the stable chemical species of each element for nanomaterial biosynthesis at varying levels of reduction potential (Eh) and pH. Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, resulting in the biosynthesis of various crystalline nanomaterials that were previously amorphous or not synthesized.
This strategy was extended to biosynthesize multi-element nanomaterials. Various single and multi-element nanomaterials biosynthesized in this research can potentially serve as new and novel nanomaterials for industrial applications such as catalysts, chemical sensors, biosensors, bioimaging, drug delivery, and cancer therapy.
A research group consisting of PhD candidate Yoojin Choi, Associate Professor Doh Chang Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST and Associate Professor Tae Jung Park of the Department of Chemistry at Chung-Ang University reported the synthesis. This study, entitled “Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials,” was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on May 21.
A recent successful biosynthesis of nanomaterials under mild conditions without requiring physical and chemical treatments has triggered the exploration of the full biosynthesis capacity of a biological system for producing a diverse range of nanomaterials as well as for understanding biosynthesis mechanisms for crystalline versus amorphous nanomaterials.
There has been increased interest in synthesizing various nanomaterials that have not yet been synthesized for various applications including semiconducting materials, enhanced solar cells, biomedical materials, and many others. This research reports the construction of a recombinant E. coli strain that co-expresses metallothionein, a metal binding protein, and phytochelatin synthase that synthesizes the metal-binding peptide phytochelatin for the biosynthesis of various nanomaterials. Subsequently, an E. coli strain was engineered to produce a diverse range of nanomaterials, including those never biosynthesized before, by using 35 individual elements from the periodic table and also by combining multi-elements.
Distinguished Professor Lee said, “An environmentally-friendly and sustainable process is of much interest for producing nanomaterials by not only chemical and physical methods but biological synthesis. Moreover, there has been much attention paid to producing diverse and novel nanomaterials for new industrial applications. This is the first report to predict the biosynthesis of various nanomaterials, by far the largest number of various single- and multi-elements nanomaterials. The strategies used for nanomaterial biosynthesis in this research will be useful for further diversifying the portfolio of nanomaterials that can be manufactured.”
Figure: The biosynthesis of diverse nanomaterials using recombinant E. coli. This schematic diagram shows the overall conceptualization of the biosynthesis of various single and multi-element nanomaterials using recombinant E. coli under incubation with corresponding elemental precursors. The 35 elements that were tested to biosynthesize nanomaterials are shown in black circles on the periodic table.
2018.05.23 View 12443
Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi)
A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements.
The study analyzed the nanomaterial biosynthesis conditions using a Pourbaix diagram to predict the producibility and crystallinity. Researchers studied a Pourbaix diagram to predict the stable chemical species of each element for nanomaterial biosynthesis at varying levels of reduction potential (Eh) and pH. Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, resulting in the biosynthesis of various crystalline nanomaterials that were previously amorphous or not synthesized.
This strategy was extended to biosynthesize multi-element nanomaterials. Various single and multi-element nanomaterials biosynthesized in this research can potentially serve as new and novel nanomaterials for industrial applications such as catalysts, chemical sensors, biosensors, bioimaging, drug delivery, and cancer therapy.
A research group consisting of PhD candidate Yoojin Choi, Associate Professor Doh Chang Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST and Associate Professor Tae Jung Park of the Department of Chemistry at Chung-Ang University reported the synthesis. This study, entitled “Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials,” was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on May 21.
A recent successful biosynthesis of nanomaterials under mild conditions without requiring physical and chemical treatments has triggered the exploration of the full biosynthesis capacity of a biological system for producing a diverse range of nanomaterials as well as for understanding biosynthesis mechanisms for crystalline versus amorphous nanomaterials.
There has been increased interest in synthesizing various nanomaterials that have not yet been synthesized for various applications including semiconducting materials, enhanced solar cells, biomedical materials, and many others. This research reports the construction of a recombinant E. coli strain that co-expresses metallothionein, a metal binding protein, and phytochelatin synthase that synthesizes the metal-binding peptide phytochelatin for the biosynthesis of various nanomaterials. Subsequently, an E. coli strain was engineered to produce a diverse range of nanomaterials, including those never biosynthesized before, by using 35 individual elements from the periodic table and also by combining multi-elements.
Distinguished Professor Lee said, “An environmentally-friendly and sustainable process is of much interest for producing nanomaterials by not only chemical and physical methods but biological synthesis. Moreover, there has been much attention paid to producing diverse and novel nanomaterials for new industrial applications. This is the first report to predict the biosynthesis of various nanomaterials, by far the largest number of various single- and multi-elements nanomaterials. The strategies used for nanomaterial biosynthesis in this research will be useful for further diversifying the portfolio of nanomaterials that can be manufactured.”
Figure: The biosynthesis of diverse nanomaterials using recombinant E. coli. This schematic diagram shows the overall conceptualization of the biosynthesis of various single and multi-element nanomaterials using recombinant E. coli under incubation with corresponding elemental precursors. The 35 elements that were tested to biosynthesize nanomaterials are shown in black circles on the periodic table.
2018.05.23 View 12443 -
 Cross-Generation Collaborative Labs Open
KAIST opened two cross-generation collaborative labs last month. This novel approach will pair up senior and junior faculty members for sustaining research and academic achievements even after the senior researcher retires.
This is one of the Vision 2031 innovation initiatives established to extend the spectrum of knowledge and research competitiveness. The selected labs will be funded for five years and the funding will be extended if necessary. KAIST will continue to select new labs every year.
A five-member selection committee including the Nobel Laureates Professor Klaus Von Klitzing at the Max-Planck Institute for Solid State Research and Dr. Kurt Wüthrich from ETH Zürich selected the first two labs with senior-junior pairs in March.
(Two renowned scholars' Cross-Generation Collaborative Labs which opened last month.
Distinguished Professor Lee's lab (above) andChair Professor Sung's lab)
Both labs are run by world-renowned scholars: the Systems Metabolic Engineering and Systems Healthcare Laboratory headed by Distinguished Professor Sang-Yup Lee in the Department of Chemical and Biomolecular Engineering and the Acousto-Microfluidics Research Center for Next-Generation Healthcare led by Chair Professor Hyung Jin Sung in the Department of Mechanical Engineering.
Distinguished Professor Lee will be teamed up with Professor Hyun Uk Kim, and their lab aims to mass produce new eco-friendly chemical materials as well as higher-value-added materials which will be used for medicine. The new platform technologies created in the lab are expected to provide information which will benefit human healthcare.
Meanwhile, the Acousto-Microfluidics Research Center for Next-Generation Healthcare will team up with Professors Hyoungsoo Kim and Yeunwoo Cho under Chair Professor Sung. The lab will conduct research on controlling fluids and objects exquisitely on a micro-nano scale by using high-frequency acoustic waves. The lab plans to develop a next-generation healthcare platform for customized diagnoses as well as disease treatment.
KAIST President Sung-Chul Shin, who introduced this novel idea in his research innovation initiative, said that he hopes the Cross-Generation Collaborative Labs will contribute to honoring senior scholars’ research legacies and passing knowledge down to junior researchers in order to further develop their academic achievements. He said, “I sincerely hope the labs will make numerous research breakthroughs in the very near future.”
2018.05.03 View 13549
Cross-Generation Collaborative Labs Open
KAIST opened two cross-generation collaborative labs last month. This novel approach will pair up senior and junior faculty members for sustaining research and academic achievements even after the senior researcher retires.
This is one of the Vision 2031 innovation initiatives established to extend the spectrum of knowledge and research competitiveness. The selected labs will be funded for five years and the funding will be extended if necessary. KAIST will continue to select new labs every year.
A five-member selection committee including the Nobel Laureates Professor Klaus Von Klitzing at the Max-Planck Institute for Solid State Research and Dr. Kurt Wüthrich from ETH Zürich selected the first two labs with senior-junior pairs in March.
(Two renowned scholars' Cross-Generation Collaborative Labs which opened last month.
Distinguished Professor Lee's lab (above) andChair Professor Sung's lab)
Both labs are run by world-renowned scholars: the Systems Metabolic Engineering and Systems Healthcare Laboratory headed by Distinguished Professor Sang-Yup Lee in the Department of Chemical and Biomolecular Engineering and the Acousto-Microfluidics Research Center for Next-Generation Healthcare led by Chair Professor Hyung Jin Sung in the Department of Mechanical Engineering.
Distinguished Professor Lee will be teamed up with Professor Hyun Uk Kim, and their lab aims to mass produce new eco-friendly chemical materials as well as higher-value-added materials which will be used for medicine. The new platform technologies created in the lab are expected to provide information which will benefit human healthcare.
Meanwhile, the Acousto-Microfluidics Research Center for Next-Generation Healthcare will team up with Professors Hyoungsoo Kim and Yeunwoo Cho under Chair Professor Sung. The lab will conduct research on controlling fluids and objects exquisitely on a micro-nano scale by using high-frequency acoustic waves. The lab plans to develop a next-generation healthcare platform for customized diagnoses as well as disease treatment.
KAIST President Sung-Chul Shin, who introduced this novel idea in his research innovation initiative, said that he hopes the Cross-Generation Collaborative Labs will contribute to honoring senior scholars’ research legacies and passing knowledge down to junior researchers in order to further develop their academic achievements. He said, “I sincerely hope the labs will make numerous research breakthroughs in the very near future.”
2018.05.03 View 13549 -
 Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 13070
Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 13070 -
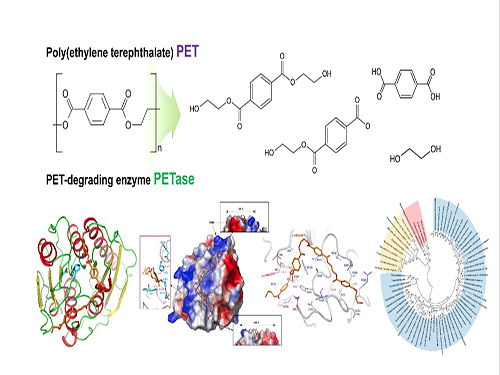 Structural Insight into the Molecular Mechanism of PET Degradation
A KAIST metabolic engineering research team has newly suggested a molecular mechanism showing superior degradability of poly ethylene terephthalate (PET).
This is the first report to simultaneously determine the 3D crystal structure of Ideonella sakaiensis PETase and develop the new variant with enhanced PET degradation.
Recently, diverse research projects are working to address the non-degradability of materials. A poly ethylene terephthalate (PET)-degrading bacterium called Ideonella sakaiensis was recently identified for the possible degradation and recycling of PET by Japanese team in Science journal (Yoshida et al., 2016). However, the detailed molecular mechanism of PET degradation has not been yet identified.
The team under Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and the team under Professor Kyung-Jin Kim of the Department of Biotechnology at Kyungpook National University conducted this research. The findings were published in Nature Communications on January 26.
This research predicts a special molecular mechanism based on the docking simulation between PETase and a PET alternative mimic substrate. Furthermore, they succeeded in constructing the variant for IsPETase with enhanced PET-degrading activity using structural-based protein engineering.
It is expected that the new approaches taken in this research can be background for further study of other enzymes capable of degrading not only PET but other plastics as well.
PET is very important source in our daily lives. However, PET after use causes tremendous contamination issues to our environment due to its non-biodegradability, which has been a major advantage of PET. Conventionally, PET is disposed of in landfills, using incineration, and sometimes recycling using chemical methods, which induces additional environmental pollution. Therefore, a new development for highly-efficient PET degrading enzymes is essential to degrade PET using bio-based eco-friendly methods.
Recently, a new bacterial species, Ideonella sakaiensis, which can use PET as a carbon source, was isolated. The PETase of I. sakaiensis (IsPETase) can degrade PET with relatively higher success than other PET-degrading enzymes. However, the detailed enzyme mechanism has not been elucidated, hindering further studies.
The research teams investigated how the substrate binds to the enzyme and which differences in enzyme structure result in significantly higher PET degrading activity compared with other cutinases and esterases, which make IsPETase highly attractive for industrial applications toward PET waste recycling.
Based on the 3D structure and related biochemical studies, they successfully predicted the reasons for extraordinary PET degrading activity of IsPETase and suggested other enzymes that can degrade PET with a newly-classified phylogenetic tree. The team proposed that 4 MHET moieties are the most properly matched substrates due to a cleft on structure even with the 10-20-mers for PET. This is meaningful in that it is the first docking simulation between PETase and PET, not its monomer.
Furthermore, they succeeded in developing a new variant with much higher PET-degrading activity using a crystal structure of this variant to show that the changed structure is better to accommodate PET substrates than wild type PETase, which will lead to developing further superior enzymes and constructing platforms for microbial plastic recycling.
Professor Lee said, “Environmental pollution from plastics remains one of the greatest challenges worldwide with the increasing consumption of plastics. We successfully constructed a new superior PET-degrading variant with the determination of a crystal structure of PETase and its degrading molecular mechanism. This novel technology will help further studies to engineer more superior enzymes with high efficiency in degrading. This will be the subject of our team’s ongoing research projects to address the global environmental pollution problem for next generation.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea. Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
(Figure: Structural insight into the molecular mechanism of poly(ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes. This schematic diagram shows the overall conceptualization for structural insight into the molecular mechanism of poly (ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes.)
2018.01.31 View 9911
Structural Insight into the Molecular Mechanism of PET Degradation
A KAIST metabolic engineering research team has newly suggested a molecular mechanism showing superior degradability of poly ethylene terephthalate (PET).
This is the first report to simultaneously determine the 3D crystal structure of Ideonella sakaiensis PETase and develop the new variant with enhanced PET degradation.
Recently, diverse research projects are working to address the non-degradability of materials. A poly ethylene terephthalate (PET)-degrading bacterium called Ideonella sakaiensis was recently identified for the possible degradation and recycling of PET by Japanese team in Science journal (Yoshida et al., 2016). However, the detailed molecular mechanism of PET degradation has not been yet identified.
The team under Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and the team under Professor Kyung-Jin Kim of the Department of Biotechnology at Kyungpook National University conducted this research. The findings were published in Nature Communications on January 26.
This research predicts a special molecular mechanism based on the docking simulation between PETase and a PET alternative mimic substrate. Furthermore, they succeeded in constructing the variant for IsPETase with enhanced PET-degrading activity using structural-based protein engineering.
It is expected that the new approaches taken in this research can be background for further study of other enzymes capable of degrading not only PET but other plastics as well.
PET is very important source in our daily lives. However, PET after use causes tremendous contamination issues to our environment due to its non-biodegradability, which has been a major advantage of PET. Conventionally, PET is disposed of in landfills, using incineration, and sometimes recycling using chemical methods, which induces additional environmental pollution. Therefore, a new development for highly-efficient PET degrading enzymes is essential to degrade PET using bio-based eco-friendly methods.
Recently, a new bacterial species, Ideonella sakaiensis, which can use PET as a carbon source, was isolated. The PETase of I. sakaiensis (IsPETase) can degrade PET with relatively higher success than other PET-degrading enzymes. However, the detailed enzyme mechanism has not been elucidated, hindering further studies.
The research teams investigated how the substrate binds to the enzyme and which differences in enzyme structure result in significantly higher PET degrading activity compared with other cutinases and esterases, which make IsPETase highly attractive for industrial applications toward PET waste recycling.
Based on the 3D structure and related biochemical studies, they successfully predicted the reasons for extraordinary PET degrading activity of IsPETase and suggested other enzymes that can degrade PET with a newly-classified phylogenetic tree. The team proposed that 4 MHET moieties are the most properly matched substrates due to a cleft on structure even with the 10-20-mers for PET. This is meaningful in that it is the first docking simulation between PETase and PET, not its monomer.
Furthermore, they succeeded in developing a new variant with much higher PET-degrading activity using a crystal structure of this variant to show that the changed structure is better to accommodate PET substrates than wild type PETase, which will lead to developing further superior enzymes and constructing platforms for microbial plastic recycling.
Professor Lee said, “Environmental pollution from plastics remains one of the greatest challenges worldwide with the increasing consumption of plastics. We successfully constructed a new superior PET-degrading variant with the determination of a crystal structure of PETase and its degrading molecular mechanism. This novel technology will help further studies to engineer more superior enzymes with high efficiency in degrading. This will be the subject of our team’s ongoing research projects to address the global environmental pollution problem for next generation.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea. Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
(Figure: Structural insight into the molecular mechanism of poly(ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes. This schematic diagram shows the overall conceptualization for structural insight into the molecular mechanism of poly (ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes.)
2018.01.31 View 9911 -
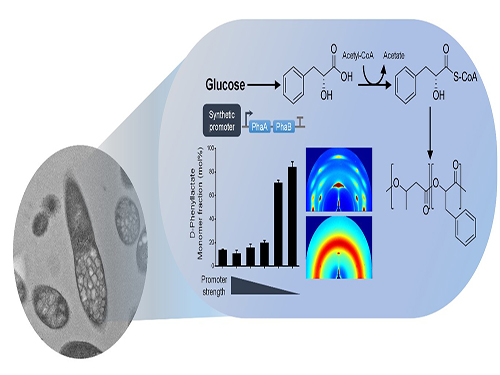 One-Step Production of Aromatic Polyesters by E. coli Strains
KAIST systems metabolic engineers defined a novel strategy for microbial aromatic polyesters production fused with synthetic biology from renewable biomass. The team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering produced aromatic polyesters from Escherichia coli (E. coli) strains by applying microbial fermentation, employing direct microbial fermentation from renewable feedstock carbohydrates.
This is the first report to determine a platform strain of engineered E. coli capable of producing environmentally friendly aromatic polyesters. This engineered E. coli strain, if desired, has the potential to be used as a platform strain capable of producing various high-valued aromatic polyesters from renewable biomass. This research was published in Nature Communications on January 8.
Conventionally, aromatic polyesters boast solid strength and heat stability so that there has been a great deal of interest in fermentative production of aromatic polyesters from renewable non-food biomass, but without success.
However, aromatic polyesters are only made by feeding the cells with corresponding aromatic monomers as substrates, and have not been produced by direct fermentation from renewable feedstock carbohydrates such as glucose.
To address this issue, the team prescribed the detailed procedure for aromatic polyester production through identifying CoA-transferase that activates phenylalkanoates into their corresponding CoA derivatives. In this process, researchers employed metabolic engineering of E. coli to produce phenylalkanoates from glucose based on genome-scale metabolic flux analysis. In particular, the KAIST team made a modulation of gene expression to produce various aromatic polyesters having different monomer fractions.
The research team successfully produced aromatic polyesters, a non-natural polymer using the strategy that combines systems metabolic engineering and synthetic biology. They succeeded in biosynthesis of various kinds of aromatic polyesters through the system, thus proving the technical excellence of the environmentally friendly biosynthetic system of this research. Furthermore, his team also proved the potential of expanding the range of aromatic polyesters from renewable resources, which is expected to play an important role in the bio-plastic industry.
Professor Lee said, “An eco-friendly and sustainable chemical industry is the key global agenda every nation faces. We are making a research focus to a biochemical industry free from petroleum dependence, and conducting diverse research activities to address the issue. This novel technology we are presenting will serve as an opportunity to advance the biochemical industry moving forward.”
This work was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) and also by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea.
Figure: Biosynthesis of aromatic polyesters by metabolically engineered E. coli.This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced aromatic polyesters from glucose.
2018.01.09 View 7991
One-Step Production of Aromatic Polyesters by E. coli Strains
KAIST systems metabolic engineers defined a novel strategy for microbial aromatic polyesters production fused with synthetic biology from renewable biomass. The team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering produced aromatic polyesters from Escherichia coli (E. coli) strains by applying microbial fermentation, employing direct microbial fermentation from renewable feedstock carbohydrates.
This is the first report to determine a platform strain of engineered E. coli capable of producing environmentally friendly aromatic polyesters. This engineered E. coli strain, if desired, has the potential to be used as a platform strain capable of producing various high-valued aromatic polyesters from renewable biomass. This research was published in Nature Communications on January 8.
Conventionally, aromatic polyesters boast solid strength and heat stability so that there has been a great deal of interest in fermentative production of aromatic polyesters from renewable non-food biomass, but without success.
However, aromatic polyesters are only made by feeding the cells with corresponding aromatic monomers as substrates, and have not been produced by direct fermentation from renewable feedstock carbohydrates such as glucose.
To address this issue, the team prescribed the detailed procedure for aromatic polyester production through identifying CoA-transferase that activates phenylalkanoates into their corresponding CoA derivatives. In this process, researchers employed metabolic engineering of E. coli to produce phenylalkanoates from glucose based on genome-scale metabolic flux analysis. In particular, the KAIST team made a modulation of gene expression to produce various aromatic polyesters having different monomer fractions.
The research team successfully produced aromatic polyesters, a non-natural polymer using the strategy that combines systems metabolic engineering and synthetic biology. They succeeded in biosynthesis of various kinds of aromatic polyesters through the system, thus proving the technical excellence of the environmentally friendly biosynthetic system of this research. Furthermore, his team also proved the potential of expanding the range of aromatic polyesters from renewable resources, which is expected to play an important role in the bio-plastic industry.
Professor Lee said, “An eco-friendly and sustainable chemical industry is the key global agenda every nation faces. We are making a research focus to a biochemical industry free from petroleum dependence, and conducting diverse research activities to address the issue. This novel technology we are presenting will serve as an opportunity to advance the biochemical industry moving forward.”
This work was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) and also by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea.
Figure: Biosynthesis of aromatic polyesters by metabolically engineered E. coli.This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced aromatic polyesters from glucose.
2018.01.09 View 7991