cancer
-
 KAIST Presents a Breakthrough in Overcoming Drug Resistance in Cancer – Hope for Treating Intractable Diseases like Diabetes
<(From the left) Prof. Hyun Uk Kim, Ph.D candiate Hae Deok Jung, Ph.D candidate Jina Lim, Prof.Yoosik Kim from the Department of Chemical and Biomolecular Engineering>
One of the biggest obstacles in cancer treatment is drug resistance in cancer cells. Conventional efforts have focused on identifying new drug targets to eliminate these resistant cells, but such approaches can often lead to even stronger resistance. Now, researchers at KAIST have developed a computational framework to predict key metabolic genes that can re-sensitize resistant cancer cells to treatment. This technique holds promise not only for a variety of cancer therapies but also for treating metabolic diseases such as diabetes.
On the 7th of July, KAIST (President Kwang Hyung Lee) announced that a research team led by Professors Hyun Uk Kim and Yoosik Kim from the Department of Chemical and Biomolecular Engineering had developed a computational framework that predicts metabolic gene targets to re-sensitize drug-resistant breast cancer cells. This was achieved using a metabolic network model capable of simulating human metabolism.
Focusing on metabolic alterations—key characteristics in the formation of drug resistance—the researchers developed a metabolism-based approach to identify gene targets that could enhance drug responsiveness by regulating the metabolism of drug-resistant breast cancer cells.
< Computational framework that can identify metabolic gene targets to revert the metabolic state of the drug-resistant cells to that of the drug-sensitive parental cells>
The team first constructed cell-specific metabolic network models by integrating proteomic data obtained from two different types of drug-resistant MCF7 breast cancer cell lines: one resistant to doxorubicin and the other to paclitaxel. They then performed gene knockout simulations* on all of the metabolic genes and analyzed the results.
*Gene knockout simulation: A computational method to predict changes in a biological network by virtually removing specific genes.
As a result, they discovered that suppressing certain genes could make previously resistant cancer cells responsive to anticancer drugs again. Specifically, they identified GOT1 as a target in doxorubicin-resistant cells, GPI in paclitaxel-resistant cells, and SLC1A5 as a common target for both drugs.
The predictions were experimentally validated by suppressing proteins encoded by these genes, which led to the re-sensitization of the drug-resistant cancer cells.
Furthermore, consistent re-sensitization effects were also observed when the same proteins were inhibited in other types of breast cancer cells that had developed resistance to the same drugs.
Professor Yoosik Kim remarked, “Cellular metabolism plays a crucial role in various intractable diseases including infectious and degenerative conditions. This new technology, which predicts metabolic regulation switches, can serve as a foundational tool not only for treating drug-resistant breast cancer but also for a wide range of diseases that currently lack effective therapies.”
Professor Hyun Uk Kim, who led the study, emphasized, “The significance of this research lies in our ability to accurately predict key metabolic genes that can make resistant cancer cells responsive to treatment again—using only computer simulations and minimal experimental data. This framework can be widely applied to discover new therapeutic targets in various cancers and metabolic diseases.”
The study, in which Ph.D. candidates JinA Lim and Hae Deok Jung from KAIST participated as co-first authors, was published online on June 25 in Proceedings of the National Academy of Sciences (PNAS), a leading multidisciplinary journal that covers top-tier research in life sciences, physics, engineering, and social sciences.
※ Title: Genome-scale knockout simulation and clustering analysis of drug-resistant breast cancer cells reveal drug sensitization targets ※ DOI: https://doi.org/10.1073/pnas.2425384122 ※ Authors: JinA Lim (KAIST, co-first author), Hae Deok Jung (KAIST, co-first author), Han Suk Ryu (Seoul National University Hospital, corresponding author), Yoosik Kim (KAIST, corresponding author), Hyun Uk Kim (KAIST, corresponding author), and five others.
This research was supported by the Ministry of Science and ICT through the National Research Foundation of Korea, and the Electronics and Telecommunications Research Institute (ETRI).
2025.07.08 View 472
KAIST Presents a Breakthrough in Overcoming Drug Resistance in Cancer – Hope for Treating Intractable Diseases like Diabetes
<(From the left) Prof. Hyun Uk Kim, Ph.D candiate Hae Deok Jung, Ph.D candidate Jina Lim, Prof.Yoosik Kim from the Department of Chemical and Biomolecular Engineering>
One of the biggest obstacles in cancer treatment is drug resistance in cancer cells. Conventional efforts have focused on identifying new drug targets to eliminate these resistant cells, but such approaches can often lead to even stronger resistance. Now, researchers at KAIST have developed a computational framework to predict key metabolic genes that can re-sensitize resistant cancer cells to treatment. This technique holds promise not only for a variety of cancer therapies but also for treating metabolic diseases such as diabetes.
On the 7th of July, KAIST (President Kwang Hyung Lee) announced that a research team led by Professors Hyun Uk Kim and Yoosik Kim from the Department of Chemical and Biomolecular Engineering had developed a computational framework that predicts metabolic gene targets to re-sensitize drug-resistant breast cancer cells. This was achieved using a metabolic network model capable of simulating human metabolism.
Focusing on metabolic alterations—key characteristics in the formation of drug resistance—the researchers developed a metabolism-based approach to identify gene targets that could enhance drug responsiveness by regulating the metabolism of drug-resistant breast cancer cells.
< Computational framework that can identify metabolic gene targets to revert the metabolic state of the drug-resistant cells to that of the drug-sensitive parental cells>
The team first constructed cell-specific metabolic network models by integrating proteomic data obtained from two different types of drug-resistant MCF7 breast cancer cell lines: one resistant to doxorubicin and the other to paclitaxel. They then performed gene knockout simulations* on all of the metabolic genes and analyzed the results.
*Gene knockout simulation: A computational method to predict changes in a biological network by virtually removing specific genes.
As a result, they discovered that suppressing certain genes could make previously resistant cancer cells responsive to anticancer drugs again. Specifically, they identified GOT1 as a target in doxorubicin-resistant cells, GPI in paclitaxel-resistant cells, and SLC1A5 as a common target for both drugs.
The predictions were experimentally validated by suppressing proteins encoded by these genes, which led to the re-sensitization of the drug-resistant cancer cells.
Furthermore, consistent re-sensitization effects were also observed when the same proteins were inhibited in other types of breast cancer cells that had developed resistance to the same drugs.
Professor Yoosik Kim remarked, “Cellular metabolism plays a crucial role in various intractable diseases including infectious and degenerative conditions. This new technology, which predicts metabolic regulation switches, can serve as a foundational tool not only for treating drug-resistant breast cancer but also for a wide range of diseases that currently lack effective therapies.”
Professor Hyun Uk Kim, who led the study, emphasized, “The significance of this research lies in our ability to accurately predict key metabolic genes that can make resistant cancer cells responsive to treatment again—using only computer simulations and minimal experimental data. This framework can be widely applied to discover new therapeutic targets in various cancers and metabolic diseases.”
The study, in which Ph.D. candidates JinA Lim and Hae Deok Jung from KAIST participated as co-first authors, was published online on June 25 in Proceedings of the National Academy of Sciences (PNAS), a leading multidisciplinary journal that covers top-tier research in life sciences, physics, engineering, and social sciences.
※ Title: Genome-scale knockout simulation and clustering analysis of drug-resistant breast cancer cells reveal drug sensitization targets ※ DOI: https://doi.org/10.1073/pnas.2425384122 ※ Authors: JinA Lim (KAIST, co-first author), Hae Deok Jung (KAIST, co-first author), Han Suk Ryu (Seoul National University Hospital, corresponding author), Yoosik Kim (KAIST, corresponding author), Hyun Uk Kim (KAIST, corresponding author), and five others.
This research was supported by the Ministry of Science and ICT through the National Research Foundation of Korea, and the Electronics and Telecommunications Research Institute (ETRI).
2025.07.08 View 472 -
 KAIST Develops Virtual Staining Technology for 3D Histopathology
Moving beyond traditional methods of observing thinly sliced and stained cancer tissues, a collaborative international research team led by KAIST has successfully developed a groundbreaking technology. This innovation uses advanced optical techniques combined with an artificial intelligence-based deep learning algorithm to create realistic, virtually stained 3D images of cancer tissue without the need for serial sectioning nor staining. This breakthrough is anticipated to pave the way for next-generation non-invasive pathological diagnosis.
< Photo 1. (From left) Juyeon Park (Ph.D. Candidate, Department of Physics), Professor YongKeun Park (Department of Physics) (Top left) Professor Su-Jin Shin (Gangnam Severance Hospital), Professor Tae Hyun Hwang (Vanderbilt University School of Medicine) >
KAIST (President Kwang Hyung Lee) announced on the 26th that a research team led by Professor YongKeun Park of the Department of Physics, in collaboration with Professor Su-Jin Shin's team at Yonsei University Gangnam Severance Hospital, Professor Tae Hyun Hwang's team at Mayo Clinic, and Tomocube's AI research team, has developed an innovative technology capable of vividly displaying the 3D structure of cancer tissues without separate staining.
For over 200 years, conventional pathology has relied on observing cancer tissues under a microscope, a method that only shows specific cross-sections of the 3D cancer tissue. This has limited the ability to understand the three-dimensional connections and spatial arrangements between cells.
To overcome this, the research team utilized holotomography (HT), an advanced optical technology, to measure the 3D refractive index information of tissues. They then integrated an AI-based deep learning algorithm to successfully generate virtual H&E* images.* H&E (Hematoxylin & Eosin): The most widely used staining method for observing pathological tissues. Hematoxylin stains cell nuclei blue, and eosin stains cytoplasm pink.
The research team quantitatively demonstrated that the images generated by this technology are highly similar to actual stained tissue images. Furthermore, the technology exhibited consistent performance across various organs and tissues, proving its versatility and reliability as a next-generation pathological analysis tool.
< Figure 1. Comparison of conventional 3D tissue pathology procedure and the 3D virtual H&E staining technology proposed in this study. The traditional method requires preparing and staining dozens of tissue slides, while the proposed technology can reduce the number of slides by up to 10 times and quickly generate H&E images without the staining process. >
Moreover, by validating the feasibility of this technology through joint research with hospitals and research institutions in Korea and the United States, utilizing Tomocube's holotomography equipment, the team demonstrated its potential for full-scale adoption in real-world pathological research settings.
Professor YongKeun Park stated, "This research marks a major advancement by transitioning pathological analysis from conventional 2D methods to comprehensive 3D imaging. It will greatly enhance biomedical research and clinical diagnostics, particularly in understanding cancer tumor boundaries and the intricate spatial arrangements of cells within tumor microenvironments."
< Figure 2. Results of AI-based 3D virtual H&E staining and quantitative analysis of pathological tissue. The virtually stained images enabled 3D reconstruction of key pathological features such as cell nuclei and glandular lumens. Based on this, various quantitative indicators, including cell nuclear distribution, volume, and surface area, could be extracted. >
This research, with Juyeon Park, a student of the Integrated Master’s and Ph.D. Program at KAIST, as the first author, was published online in the prestigious journal Nature Communications on May 22.
(Paper title: Revealing 3D microanatomical structures of unlabeled thick cancer tissues using holotomography and virtual H&E staining.
[https://doi.org/10.1038/s41467-025-59820-0]
This study was supported by the Leader Researcher Program of the National Research Foundation of Korea, the Global Industry Technology Cooperation Center Project of the Korea Institute for Advancement of Technology, and the Korea Health Industry Development Institute.
2025.05.26 View 4051
KAIST Develops Virtual Staining Technology for 3D Histopathology
Moving beyond traditional methods of observing thinly sliced and stained cancer tissues, a collaborative international research team led by KAIST has successfully developed a groundbreaking technology. This innovation uses advanced optical techniques combined with an artificial intelligence-based deep learning algorithm to create realistic, virtually stained 3D images of cancer tissue without the need for serial sectioning nor staining. This breakthrough is anticipated to pave the way for next-generation non-invasive pathological diagnosis.
< Photo 1. (From left) Juyeon Park (Ph.D. Candidate, Department of Physics), Professor YongKeun Park (Department of Physics) (Top left) Professor Su-Jin Shin (Gangnam Severance Hospital), Professor Tae Hyun Hwang (Vanderbilt University School of Medicine) >
KAIST (President Kwang Hyung Lee) announced on the 26th that a research team led by Professor YongKeun Park of the Department of Physics, in collaboration with Professor Su-Jin Shin's team at Yonsei University Gangnam Severance Hospital, Professor Tae Hyun Hwang's team at Mayo Clinic, and Tomocube's AI research team, has developed an innovative technology capable of vividly displaying the 3D structure of cancer tissues without separate staining.
For over 200 years, conventional pathology has relied on observing cancer tissues under a microscope, a method that only shows specific cross-sections of the 3D cancer tissue. This has limited the ability to understand the three-dimensional connections and spatial arrangements between cells.
To overcome this, the research team utilized holotomography (HT), an advanced optical technology, to measure the 3D refractive index information of tissues. They then integrated an AI-based deep learning algorithm to successfully generate virtual H&E* images.* H&E (Hematoxylin & Eosin): The most widely used staining method for observing pathological tissues. Hematoxylin stains cell nuclei blue, and eosin stains cytoplasm pink.
The research team quantitatively demonstrated that the images generated by this technology are highly similar to actual stained tissue images. Furthermore, the technology exhibited consistent performance across various organs and tissues, proving its versatility and reliability as a next-generation pathological analysis tool.
< Figure 1. Comparison of conventional 3D tissue pathology procedure and the 3D virtual H&E staining technology proposed in this study. The traditional method requires preparing and staining dozens of tissue slides, while the proposed technology can reduce the number of slides by up to 10 times and quickly generate H&E images without the staining process. >
Moreover, by validating the feasibility of this technology through joint research with hospitals and research institutions in Korea and the United States, utilizing Tomocube's holotomography equipment, the team demonstrated its potential for full-scale adoption in real-world pathological research settings.
Professor YongKeun Park stated, "This research marks a major advancement by transitioning pathological analysis from conventional 2D methods to comprehensive 3D imaging. It will greatly enhance biomedical research and clinical diagnostics, particularly in understanding cancer tumor boundaries and the intricate spatial arrangements of cells within tumor microenvironments."
< Figure 2. Results of AI-based 3D virtual H&E staining and quantitative analysis of pathological tissue. The virtually stained images enabled 3D reconstruction of key pathological features such as cell nuclei and glandular lumens. Based on this, various quantitative indicators, including cell nuclear distribution, volume, and surface area, could be extracted. >
This research, with Juyeon Park, a student of the Integrated Master’s and Ph.D. Program at KAIST, as the first author, was published online in the prestigious journal Nature Communications on May 22.
(Paper title: Revealing 3D microanatomical structures of unlabeled thick cancer tissues using holotomography and virtual H&E staining.
[https://doi.org/10.1038/s41467-025-59820-0]
This study was supported by the Leader Researcher Program of the National Research Foundation of Korea, the Global Industry Technology Cooperation Center Project of the Korea Institute for Advancement of Technology, and the Korea Health Industry Development Institute.
2025.05.26 View 4051 -
 KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 28611
KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 28611 -
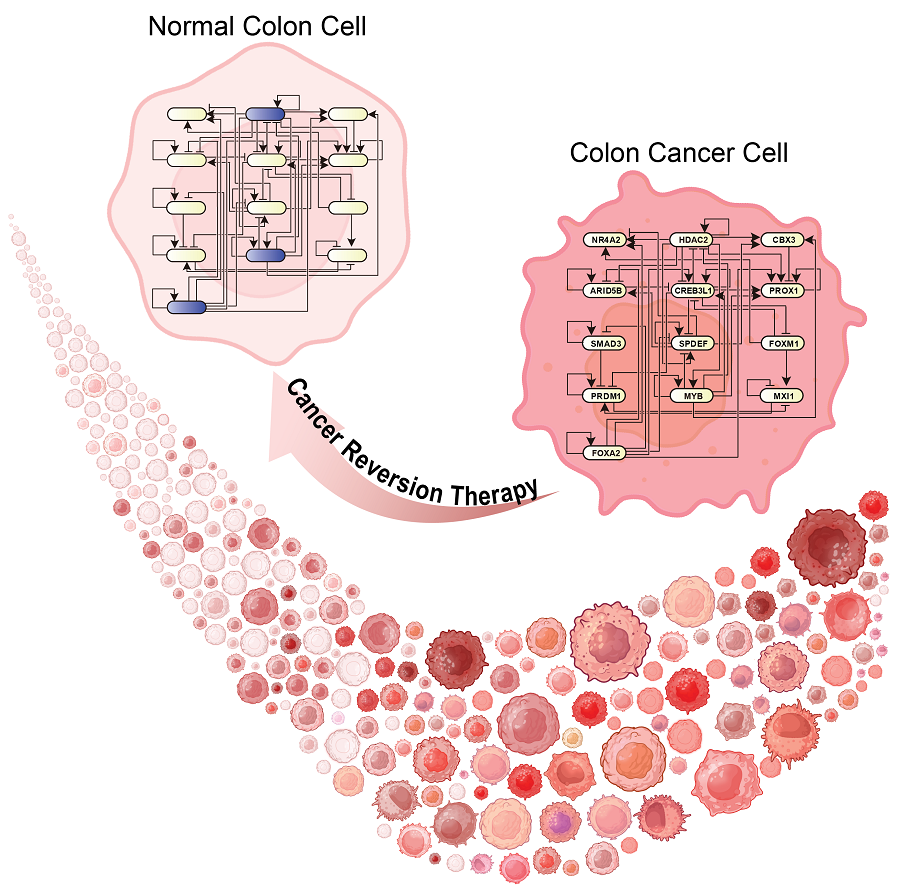 KAIST Develops Foundational Technology to Revert Cancer Cells to Normal Cells
Despite the development of numerous cancer treatment technologies, the common goal of current cancer therapies is to eliminate cancer cells. This approach, however, faces fundamental limitations, including cancer cells developing resistance and returning, as well as severe side effects from the destruction of healthy cells.
< (From top left) Bio and Brain Engineering PhD candidates Juhee Kim, Jeong-Ryeol Gong, Chun-Kyung Lee, and Hoon-Min Kim posed for a group photo with Professor Kwang-Hyun Cho >
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of December that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering has developed a groundbreaking technology that can treat colon cancer by converting cancer cells into a state resembling normal colon cells without killing them, thus avoiding side effects.
The research team focused on the observation that during the oncogenesis process, normal cells regress along their differentiation trajectory. Building on this insight, they developed a technology to create a digital twin of the gene network associated with the differentiation trajectory of normal cells.
< Figure 1. Technology for creating a digital twin of a gene network from single-cell transcriptome data of a normal cell differentiation trajectory. Professor Kwang-Hyun Cho's research team developed a digital twin creation technology that precisely observes the dynamics of gene regulatory relationships during the process of normal cells differentiating along a differentiation trajectory and analyzes the relationships among key genes to build a mathematical model that can be simulated (A-F). In addition, they developed a technology to discover key regulatory factors that control the differentiation trajectory of normal cells by simulating and analyzing this digital twin. >
< Figure 2. Digital twin simulation simulating the differentiation trajectory of normal colon cells. The dynamics of single-cell transcriptome data for the differentiation trajectory of normal colon cells were analyzed (A) and a digital twin of the gene network was developed representing the regulatory relationships of key genes in this differentiation trajectory (B). The simulation results of the digital twin confirm that it readily reproduces the dynamics of single-cell transcriptome data (C, D). >
Through simulation analysis, the team systematically identified master molecular switches that induce normal cell differentiation. When these switches were applied to colon cancer cells, the cancer cells reverted to a normal-like state, a result confirmed through molecular and cellular experiments as well as animal studies.
< Figure 3. Discovery of top-level key control factors that induce differentiation of normal colon cells. By applying control factor discovery technology to the digital twin model, three genes, HDAC2, FOXA2, and MYB, were discovered as key control factors that induce differentiation of normal colon cells (A, B). The results of simulation analysis of the regulatory effects of the discovered control factors through the digital twin confirmed that they could induce complete differentiation of colon cells (C). >
< Figure 4. Verification of the effect of the key control factors discovered using colon cancer cells and animal experiments on the reversibility of colon cancer. The key control factors of the normal colon cell differentiation trajectory discovered through digital twin simulation analysis were applied to actual colon cancer cells and colon cancer mouse animal models to experimentally verify the effect of cancer reversibility. The key control factors significantly reduced the proliferation of three colon cancer cell lines (A), and this was confirmed in the same way in animal models (B-D). >
This research demonstrates that cancer cell reversion can be systematically achieved by analyzing and utilizing the digital twin of the cancer cell gene network, rather than relying on serendipitous discoveries. The findings hold significant promise for developing reversible cancer therapies that can be applied to various types of cancer.
< Figure 5. The change in overall gene expression was confirmed through the regulation of the identified key regulatory factors, which converted the state of colon cancer cells to that of normal colon cells. The transcriptomes of colon cancer tissues and normal colon tissues from more than 400 colon cancer patients were compared with the transcriptomes of colon cancer cell lines and reversible colon cancer cell lines, respectively. The comparison results confirmed that the regulation of the identified key regulatory factors converted all three colon cancer cell lines to a state similar to the transcriptome expression of normal colon tissues. >
Professor Kwang-Hyun Cho remarked, "The fact that cancer cells can be converted back to normal cells is an astonishing phenomenon. This study proves that such reversion can be systematically induced."
He further emphasized, "This research introduces the novel concept of reversible cancer therapy by reverting cancer cells to normal cells. It also develops foundational technology for identifying targets for cancer reversion through the systematic analysis of normal cell differentiation trajectories."
This research included contributions from Jeong-Ryeol Gong, Chun-Kyung Lee, Hoon-Min Kim, Juhee Kim, and Jaeog Jeon, and was published in the online edition of the international journal Advanced Science by Wiley on December 11. (Title: “Control of Cellular Differentiation Trajectories for Cancer Reversion”) DOI: https://doi.org/10.1002/advs.202402132
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed a source technology to systematically discover key control factors that can induce reversibility of colon cancer cells through a systems biology approach and a digital twin simulation analysis of the differentiation trajectory of normal colon cells, and verified the effects of reversion on actual colon cancer through molecular cell experiments and animal experiments. >
The study was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Researcher Program and Basic Research Laboratory Program. The research findings have been transferred to BioRevert Inc., where they will be used for the development of practical cancer reversion therapies.
2024.12.23 View 104331
KAIST Develops Foundational Technology to Revert Cancer Cells to Normal Cells
Despite the development of numerous cancer treatment technologies, the common goal of current cancer therapies is to eliminate cancer cells. This approach, however, faces fundamental limitations, including cancer cells developing resistance and returning, as well as severe side effects from the destruction of healthy cells.
< (From top left) Bio and Brain Engineering PhD candidates Juhee Kim, Jeong-Ryeol Gong, Chun-Kyung Lee, and Hoon-Min Kim posed for a group photo with Professor Kwang-Hyun Cho >
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of December that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering has developed a groundbreaking technology that can treat colon cancer by converting cancer cells into a state resembling normal colon cells without killing them, thus avoiding side effects.
The research team focused on the observation that during the oncogenesis process, normal cells regress along their differentiation trajectory. Building on this insight, they developed a technology to create a digital twin of the gene network associated with the differentiation trajectory of normal cells.
< Figure 1. Technology for creating a digital twin of a gene network from single-cell transcriptome data of a normal cell differentiation trajectory. Professor Kwang-Hyun Cho's research team developed a digital twin creation technology that precisely observes the dynamics of gene regulatory relationships during the process of normal cells differentiating along a differentiation trajectory and analyzes the relationships among key genes to build a mathematical model that can be simulated (A-F). In addition, they developed a technology to discover key regulatory factors that control the differentiation trajectory of normal cells by simulating and analyzing this digital twin. >
< Figure 2. Digital twin simulation simulating the differentiation trajectory of normal colon cells. The dynamics of single-cell transcriptome data for the differentiation trajectory of normal colon cells were analyzed (A) and a digital twin of the gene network was developed representing the regulatory relationships of key genes in this differentiation trajectory (B). The simulation results of the digital twin confirm that it readily reproduces the dynamics of single-cell transcriptome data (C, D). >
Through simulation analysis, the team systematically identified master molecular switches that induce normal cell differentiation. When these switches were applied to colon cancer cells, the cancer cells reverted to a normal-like state, a result confirmed through molecular and cellular experiments as well as animal studies.
< Figure 3. Discovery of top-level key control factors that induce differentiation of normal colon cells. By applying control factor discovery technology to the digital twin model, three genes, HDAC2, FOXA2, and MYB, were discovered as key control factors that induce differentiation of normal colon cells (A, B). The results of simulation analysis of the regulatory effects of the discovered control factors through the digital twin confirmed that they could induce complete differentiation of colon cells (C). >
< Figure 4. Verification of the effect of the key control factors discovered using colon cancer cells and animal experiments on the reversibility of colon cancer. The key control factors of the normal colon cell differentiation trajectory discovered through digital twin simulation analysis were applied to actual colon cancer cells and colon cancer mouse animal models to experimentally verify the effect of cancer reversibility. The key control factors significantly reduced the proliferation of three colon cancer cell lines (A), and this was confirmed in the same way in animal models (B-D). >
This research demonstrates that cancer cell reversion can be systematically achieved by analyzing and utilizing the digital twin of the cancer cell gene network, rather than relying on serendipitous discoveries. The findings hold significant promise for developing reversible cancer therapies that can be applied to various types of cancer.
< Figure 5. The change in overall gene expression was confirmed through the regulation of the identified key regulatory factors, which converted the state of colon cancer cells to that of normal colon cells. The transcriptomes of colon cancer tissues and normal colon tissues from more than 400 colon cancer patients were compared with the transcriptomes of colon cancer cell lines and reversible colon cancer cell lines, respectively. The comparison results confirmed that the regulation of the identified key regulatory factors converted all three colon cancer cell lines to a state similar to the transcriptome expression of normal colon tissues. >
Professor Kwang-Hyun Cho remarked, "The fact that cancer cells can be converted back to normal cells is an astonishing phenomenon. This study proves that such reversion can be systematically induced."
He further emphasized, "This research introduces the novel concept of reversible cancer therapy by reverting cancer cells to normal cells. It also develops foundational technology for identifying targets for cancer reversion through the systematic analysis of normal cell differentiation trajectories."
This research included contributions from Jeong-Ryeol Gong, Chun-Kyung Lee, Hoon-Min Kim, Juhee Kim, and Jaeog Jeon, and was published in the online edition of the international journal Advanced Science by Wiley on December 11. (Title: “Control of Cellular Differentiation Trajectories for Cancer Reversion”) DOI: https://doi.org/10.1002/advs.202402132
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed a source technology to systematically discover key control factors that can induce reversibility of colon cancer cells through a systems biology approach and a digital twin simulation analysis of the differentiation trajectory of normal colon cells, and verified the effects of reversion on actual colon cancer through molecular cell experiments and animal experiments. >
The study was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Researcher Program and Basic Research Laboratory Program. The research findings have been transferred to BioRevert Inc., where they will be used for the development of practical cancer reversion therapies.
2024.12.23 View 104331 -
 A KAIST-SNUH Team Devises a Way to Make Mathematical Predictions to find Metabolites Related to Somatic Mutations in Cancers
Cancer is characterized by abnormal metabolic processes different from those of normal cells. Therefore, cancer metabolism has been extensively studied to develop effective diagnosis and treatment strategies. Notable achievements of cancer metabolism studies include the discovery of oncometabolites* and the approval of anticancer drugs by the U.S. Food and Drug Administration (FDA) that target enzymes associated with oncometabolites. Approved anticancer drugs such as ‘Tibsovo (active ingredient: ivosidenib)’ and ‘Idhifa (active ingredient: enasidenib)’ are both used for the treatment of acute myeloid leukemia. Despite such achievements, studying cancer metabolism, especially oncometabolites, remains challenging due to time-consuming and expensive methodologies such as metabolomics. Thus, the number of confirmed oncometabolites is very small although a relatively large number of cancer-associated gene mutations have been well studied.
*Oncometabolite: A metabolite that shows pro-oncogenic function when abnormally accumulated in cancer cells. An oncometabolite is often generated as a result of gene mutations, and this accumulation promotes the growth and survival of cancer cells. Representative oncometabolites include 2-hydroxyglutarate, succinate, and fumarate.
On March 18th, a KAIST research team led by Professor Hyun Uk Kim from the Department of Chemical and Biomolecular Engineering developed a computational workflow that systematically predicts metabolites and metabolic pathways associated with somatic mutations in cancer through collaboration with research teams under Prof Youngil Koh, Prof. Hongseok Yun, and Prof. Chang Wook Jeong from Seoul National University Hospital.
The research teams have successfully reconstructed patient-specific genome-scale metabolic models (GEMs)* for 1,043 cancer patients across 24 cancer types by integrating publicly available cancer patients’ transcriptome data (i.e., from international cancer genome consortiums such as PCAWG and TCGA) into a generic human GEM. The resulting patient-specific GEMs make it possible to predict each patient’s metabolic phenotypes.
*Genome-scale metabolic model (GEM): A computational model that mathematically describes all of the biochemical reactions that take place inside a cell. It allows for the prediction of the cell’s metabolic phenotypes under various genetic and/or environmental conditions.
< Figure 1. Schematic diagram of a computational methodology for predicting metabolites and metabolic pathways associated with cancer somatic mutations. of a computational methodology for predicting metabolites and metabolic pathways associated with cancer somatic mutations. >
The team developed a four-step computational workflow using the patient-specific GEMs from 1,043 cancer patients and somatic mutation data obtained from the corresponding cancer patients. This workflow begins with the calculation of the flux-sum value of each metabolite by simulating the patient-specific GEMs. The flux-sum value quantifies the intracellular importance of a metabolite. Next, the workflow identifies metabolites that appear to be significantly associated with specific gene mutations through a statistical analysis of the predicted flux-sum data and the mutation data. Finally, the workflow selects altered metabolic pathways that significantly contribute to the biosynthesis of the predicted oncometabolite candidates, ultimately generating metabolite-gene-pathway sets as an output.
The two co-first authors, Dr. GaRyoung Lee (currently a postdoctoral fellow at the Dana-Farber Cancer Institute and Harvard Medical School) and Dr. Sang Mi Lee (currently a postdoctoral fellow at Harvard Medical School) said, “The computational workflow developed can systematically predict how genetic mutations affect cellular metabolism through metabolic pathways. Importantly, it can easily be applied to different types of cancer based on the mutation and transcriptome data of cancer patient cohorts.”
Prof. Kim said, “The computational workflow and its resulting prediction outcomes will serve as the groundwork for identifying novel oncometabolites and for facilitating the development of various treatment and diagnosis strategies”.
This study, which was supported by the National Research Foundation of Korea, has been published online in Genome Biology, a representative journal in the field of biotechnology and genetics, under the title "Prediction of metabolites associated with somatic mutations in cancers by using genome‑scale metabolic models and mutation data".
2024.03.18 View 8006
A KAIST-SNUH Team Devises a Way to Make Mathematical Predictions to find Metabolites Related to Somatic Mutations in Cancers
Cancer is characterized by abnormal metabolic processes different from those of normal cells. Therefore, cancer metabolism has been extensively studied to develop effective diagnosis and treatment strategies. Notable achievements of cancer metabolism studies include the discovery of oncometabolites* and the approval of anticancer drugs by the U.S. Food and Drug Administration (FDA) that target enzymes associated with oncometabolites. Approved anticancer drugs such as ‘Tibsovo (active ingredient: ivosidenib)’ and ‘Idhifa (active ingredient: enasidenib)’ are both used for the treatment of acute myeloid leukemia. Despite such achievements, studying cancer metabolism, especially oncometabolites, remains challenging due to time-consuming and expensive methodologies such as metabolomics. Thus, the number of confirmed oncometabolites is very small although a relatively large number of cancer-associated gene mutations have been well studied.
*Oncometabolite: A metabolite that shows pro-oncogenic function when abnormally accumulated in cancer cells. An oncometabolite is often generated as a result of gene mutations, and this accumulation promotes the growth and survival of cancer cells. Representative oncometabolites include 2-hydroxyglutarate, succinate, and fumarate.
On March 18th, a KAIST research team led by Professor Hyun Uk Kim from the Department of Chemical and Biomolecular Engineering developed a computational workflow that systematically predicts metabolites and metabolic pathways associated with somatic mutations in cancer through collaboration with research teams under Prof Youngil Koh, Prof. Hongseok Yun, and Prof. Chang Wook Jeong from Seoul National University Hospital.
The research teams have successfully reconstructed patient-specific genome-scale metabolic models (GEMs)* for 1,043 cancer patients across 24 cancer types by integrating publicly available cancer patients’ transcriptome data (i.e., from international cancer genome consortiums such as PCAWG and TCGA) into a generic human GEM. The resulting patient-specific GEMs make it possible to predict each patient’s metabolic phenotypes.
*Genome-scale metabolic model (GEM): A computational model that mathematically describes all of the biochemical reactions that take place inside a cell. It allows for the prediction of the cell’s metabolic phenotypes under various genetic and/or environmental conditions.
< Figure 1. Schematic diagram of a computational methodology for predicting metabolites and metabolic pathways associated with cancer somatic mutations. of a computational methodology for predicting metabolites and metabolic pathways associated with cancer somatic mutations. >
The team developed a four-step computational workflow using the patient-specific GEMs from 1,043 cancer patients and somatic mutation data obtained from the corresponding cancer patients. This workflow begins with the calculation of the flux-sum value of each metabolite by simulating the patient-specific GEMs. The flux-sum value quantifies the intracellular importance of a metabolite. Next, the workflow identifies metabolites that appear to be significantly associated with specific gene mutations through a statistical analysis of the predicted flux-sum data and the mutation data. Finally, the workflow selects altered metabolic pathways that significantly contribute to the biosynthesis of the predicted oncometabolite candidates, ultimately generating metabolite-gene-pathway sets as an output.
The two co-first authors, Dr. GaRyoung Lee (currently a postdoctoral fellow at the Dana-Farber Cancer Institute and Harvard Medical School) and Dr. Sang Mi Lee (currently a postdoctoral fellow at Harvard Medical School) said, “The computational workflow developed can systematically predict how genetic mutations affect cellular metabolism through metabolic pathways. Importantly, it can easily be applied to different types of cancer based on the mutation and transcriptome data of cancer patient cohorts.”
Prof. Kim said, “The computational workflow and its resulting prediction outcomes will serve as the groundwork for identifying novel oncometabolites and for facilitating the development of various treatment and diagnosis strategies”.
This study, which was supported by the National Research Foundation of Korea, has been published online in Genome Biology, a representative journal in the field of biotechnology and genetics, under the title "Prediction of metabolites associated with somatic mutations in cancers by using genome‑scale metabolic models and mutation data".
2024.03.18 View 8006 -
 Genome Sequencing Unveils Mutational Impacts of Radiation on Mammalian Cells
Recent release of the waste water from Japan's Fukushima nuclear disaster stirred apprehension regarding the health implications of radiation exposure. Classified as a Group 1 carcinogen, ionizing radiation has long been associated with various cancers and genetic disorders, as evidenced by survivors and descendants of atomic bombings and the Chernobyl disaster. Despite much smaller amount, we remain consistently exposed to low levels of radiation in everyday life and medical procedures.
Radiation, whether in the form of high-energy particles or electromagnetic waves, is conventionally known to break our cellular DNA, leading to cancer and genetic disorders. Yet, our understanding of the quantitative and qualitative mutational impacts of ionizing radiation has been incomplete.
On the 14th, Professor Young Seok Ju and his research team from KAIST, in collaboration with Dr. Tae Gen Son from the Dongnam Institute of Radiological and Medical Science, and Professors Kyung Su Kim and Ji Hyun Chang from Seoul National University, unveiled a breakthrough. Their study, led by joint first authors Drs. Jeonghwan Youk, Hyun Woo Kwon, Joonoh Lim, Eunji Kim and Tae-Woo Kim, titled "Quantitative and qualitative mutational impact of ionizing radiation on normal cells," was published in Cell Genomics.
Employing meticulous techniques, the research team comprehensively analyzed the whole-genome sequences of cells pre- and post-radiation exposure, pinpointing radiation-induced DNA mutations. Experiments involving cells from different organs of humans and mice exposed to varying radiation doses revealed mutation patterns correlating with exposure levels. (Figure 1)
Notably, exposure to 1 Gray (Gy) of radiation resulted in on average 14 mutations in every post-exposure cell. (Figure 2) Unlike other carcinogens, radiation-induced mutations primarily comprised short base deletions and a set of structural variations including inversions, translocations, and various complex genomic rearrangements. (Figure 3) Interestingly, experiments subjecting cells to low radiation dose rate over 100 days demonstrated that mutation quantities, under equivalent total radiation doses, mirrored those of high-dose exposure.
"Through this study, we have clearly elucidated the effects of radiation on cells at the molecular level," said Prof. Ju at KAIST. "Now we understand better how radiation changes the DNA of our cells," he added.
Dr. Son from the Dongnam Institute of Radiological and Medical Science stated, "Based on this study, we will continue to research the effects of very low and very high doses of radiation on the human body," and further remarked, "We will advance the development of safe and effective radiation therapy techniques."
Professors Kim and Chang from Seoul National University College of Medicine expressed their views, saying, "Through this study, we believe we now have a tool to accurately understand the impact of radiation on human DNA," and added, "We hope that many subsequent studies will emerge using the research methodologies employed in this study."
This research represents a significant leap forward in radiation studies, made possible through collaborative efforts and interdisciplinary approaches. This pioneering research engaged scholars from diverse backgrounds, spanning from the Genetic Engineering Research Institute at Seoul National University, the Cambridge Stem Cell Institute in the UK, the Institute for Molecular Biotechnology in Austria (IMBA), and the Genome Insight Inc. (a KAIST spin-off start-up). This study was supported by various institutions including the National Research Foundation of Korea, Dongnam Institute of Radiological and Medical Science (supported by Ministry of Science and ICT, the government of South Korea), the Suh Kyungbae Foundation, the Human Frontier Science Program (HFSP), and the Korea University Anam Hospital Korea Foundation for the Advancement of Science and Creativity, the Ministry of Science and ICT, and the National R&D Program.
2024.02.15 View 9869
Genome Sequencing Unveils Mutational Impacts of Radiation on Mammalian Cells
Recent release of the waste water from Japan's Fukushima nuclear disaster stirred apprehension regarding the health implications of radiation exposure. Classified as a Group 1 carcinogen, ionizing radiation has long been associated with various cancers and genetic disorders, as evidenced by survivors and descendants of atomic bombings and the Chernobyl disaster. Despite much smaller amount, we remain consistently exposed to low levels of radiation in everyday life and medical procedures.
Radiation, whether in the form of high-energy particles or electromagnetic waves, is conventionally known to break our cellular DNA, leading to cancer and genetic disorders. Yet, our understanding of the quantitative and qualitative mutational impacts of ionizing radiation has been incomplete.
On the 14th, Professor Young Seok Ju and his research team from KAIST, in collaboration with Dr. Tae Gen Son from the Dongnam Institute of Radiological and Medical Science, and Professors Kyung Su Kim and Ji Hyun Chang from Seoul National University, unveiled a breakthrough. Their study, led by joint first authors Drs. Jeonghwan Youk, Hyun Woo Kwon, Joonoh Lim, Eunji Kim and Tae-Woo Kim, titled "Quantitative and qualitative mutational impact of ionizing radiation on normal cells," was published in Cell Genomics.
Employing meticulous techniques, the research team comprehensively analyzed the whole-genome sequences of cells pre- and post-radiation exposure, pinpointing radiation-induced DNA mutations. Experiments involving cells from different organs of humans and mice exposed to varying radiation doses revealed mutation patterns correlating with exposure levels. (Figure 1)
Notably, exposure to 1 Gray (Gy) of radiation resulted in on average 14 mutations in every post-exposure cell. (Figure 2) Unlike other carcinogens, radiation-induced mutations primarily comprised short base deletions and a set of structural variations including inversions, translocations, and various complex genomic rearrangements. (Figure 3) Interestingly, experiments subjecting cells to low radiation dose rate over 100 days demonstrated that mutation quantities, under equivalent total radiation doses, mirrored those of high-dose exposure.
"Through this study, we have clearly elucidated the effects of radiation on cells at the molecular level," said Prof. Ju at KAIST. "Now we understand better how radiation changes the DNA of our cells," he added.
Dr. Son from the Dongnam Institute of Radiological and Medical Science stated, "Based on this study, we will continue to research the effects of very low and very high doses of radiation on the human body," and further remarked, "We will advance the development of safe and effective radiation therapy techniques."
Professors Kim and Chang from Seoul National University College of Medicine expressed their views, saying, "Through this study, we believe we now have a tool to accurately understand the impact of radiation on human DNA," and added, "We hope that many subsequent studies will emerge using the research methodologies employed in this study."
This research represents a significant leap forward in radiation studies, made possible through collaborative efforts and interdisciplinary approaches. This pioneering research engaged scholars from diverse backgrounds, spanning from the Genetic Engineering Research Institute at Seoul National University, the Cambridge Stem Cell Institute in the UK, the Institute for Molecular Biotechnology in Austria (IMBA), and the Genome Insight Inc. (a KAIST spin-off start-up). This study was supported by various institutions including the National Research Foundation of Korea, Dongnam Institute of Radiological and Medical Science (supported by Ministry of Science and ICT, the government of South Korea), the Suh Kyungbae Foundation, the Human Frontier Science Program (HFSP), and the Korea University Anam Hospital Korea Foundation for the Advancement of Science and Creativity, the Ministry of Science and ICT, and the National R&D Program.
2024.02.15 View 9869 -
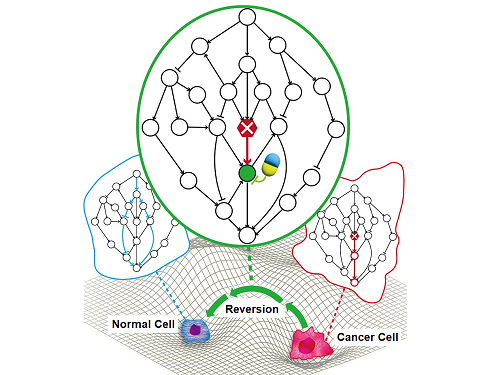 A KAIST Research Team Identifies a Cancer Reversion Mechanism
Despite decades of intensive cancer research by numerous biomedical scientists, cancer still holds its place as the number one cause of death in Korea. The fundamental reason behind the limitations of current cancer treatment methods is the fact that they all aim to completely destroy cancer cells, which eventually allows the cancer cells to acquire immunity. In other words, recurrences and side-effects caused by the destruction of healthy cells are inevitable. To this end, some have suggested anticancer treatment methods based on cancer reversion, which can revert cancer cells back to normal or near-normal cells under certain conditions. However, the practical development of this idea has not yet been attempted.
On June 8, a KAIST research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering reported to have successfully identified the fundamental principle of a process that can revert cancer cells back to normal cells without killing the cells.
Professor Cho’s team focused on the fact that unlike normal cells, which react according to external stimuli, cancer cells tend to ignore such stimuli and only undergo uncontrolled cell division. Through computer simulation analysis, the team discovered that the input-output (I/O) relationships that were distorted by genetic mutations could be reverted back to normal I/O relationships under certain conditions. The team then demonstrated through molecular cell experiments that such I/O relationship recovery also occurred in real cancer cells.
The results of this study, written by Dr. Jae Il Joo and Dr. Hwa-Jeong Park, were published in Wiley’s Advanced Science online on June 2 under the title, "Normalizing input-output relationships of cancer networks for reversion therapy."
< Image 1. Input-output (I/O) relationships in gene regulatory networks >
Professor Kwang-Hyun Cho's research team classified genes into four types by simulation-analyzing the effect of gene mutations on the I/O relationship of gene regulatory networks. (Figure A-J) In addition, by analyzing 18 genes of the cancer-related gene regulatory network, it was confirmed that when mutations occur in more than half of the genes constituting each network, reversibility is possible through appropriate control. (Figure K)
Professor Cho’s team uncovered that the reason the distorted I/O relationships of cancer cells could be reverted back to normal ones was the robustness and redundancy of intracellular gene control networks that developed over the course of evolution. In addition, they found that some genes were more promising as targets for cancer reversion than others, and showed through molecular cell experiments that controlling such genes could revert the distorted I/O relationships of cancer cells back to normal ones.
< Image 2. Simulation results of restoration of bladder cancer gene regulation network and I/O relationship of bladder cancer cells. >
The research team classified the effects of gene mutations on the I/O relationship in the bladder cancer gene regulation network by simulation analysis and classified them into 4 types. (Figure A) Through this, it was found that the distorted input-output relationship between bladder cancer cell lines KU-1919 and HCT-1197 could be restored to normal. (Figure B)
< Image 3. Analysis of survival of bladder cancer patients according to reversible gene mutation and I/O recovery experiment of bladder cancer cells. >
As predicted through network simulation analysis, Professor Kwang-Hyun Cho's research team confirmed through molecular cell experiments that the response to TGF-b was normally restored when AKT and MAP3K1 were inhibited in the bladder cancer cell line KU-1919. (Figure A-G) In addition, it was confirmed that there is a difference in the survival rate of bladder cancer patients depending on the presence or absence of a reversible gene mutation. (Figure H)
The results of this research show that the reversion of real cancer cells does not happen by chance, and that it is possible to systematically explore targets that can induce this phenomenon, thereby creating the potential for the development of innovative anticancer drugs that can control such target genes.
< Image 4. Cancer cell reversibility principle >
The research team analyzed the reversibility, redundancy, and robustness of various networks and found that there was a positive correlation between them. From this, it was found that reversibility was additionally inherent in the process of evolution in which the gene regulatory network acquired redundancy and consistency.
Professor Cho said, “By uncovering the fundamental principles of a new cancer reversion treatment strategy that may overcome the unresolved limitations of existing chemotherapy, we have increased the possibility of developing new and innovative drugs that can improve both the prognosis and quality of life of cancer patients.”
< Image 5. Conceptual diagram of research results >
The research team identified the fundamental control principle of cancer cell reversibility through systems biology research. When the I/O relationship of the intracellular gene regulatory network is distorted by mutation, the distorted I/O relationship can be restored to a normal state by identifying and adjusting the reversible gene target based on the redundancy of the molecular circuit inherent in the complex network.
After Professor Cho’s team first suggested the concept of reversion treatment, they published their results for reverting colorectal cancer in January 2020, and in January 2022 they successfully re-programmed malignant breast cancer cells back into hormone-treatable ones. In January 2023, the team successfully removed the metastasis ability from lung cancer cells and reverted them back to a state that allowed improved drug reactivity. However, these results were case studies of specific types of cancer and did not reveal what common principle allowed cancer reversion across all cancer types, making this the first revelation of the general principle of cancer reversion and its evolutionary origins.
This research was funded by the Ministry of Science and ICT of the Republic of Korea and the National Research Foundation of Korea.
2023.06.20 View 12883
A KAIST Research Team Identifies a Cancer Reversion Mechanism
Despite decades of intensive cancer research by numerous biomedical scientists, cancer still holds its place as the number one cause of death in Korea. The fundamental reason behind the limitations of current cancer treatment methods is the fact that they all aim to completely destroy cancer cells, which eventually allows the cancer cells to acquire immunity. In other words, recurrences and side-effects caused by the destruction of healthy cells are inevitable. To this end, some have suggested anticancer treatment methods based on cancer reversion, which can revert cancer cells back to normal or near-normal cells under certain conditions. However, the practical development of this idea has not yet been attempted.
On June 8, a KAIST research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering reported to have successfully identified the fundamental principle of a process that can revert cancer cells back to normal cells without killing the cells.
Professor Cho’s team focused on the fact that unlike normal cells, which react according to external stimuli, cancer cells tend to ignore such stimuli and only undergo uncontrolled cell division. Through computer simulation analysis, the team discovered that the input-output (I/O) relationships that were distorted by genetic mutations could be reverted back to normal I/O relationships under certain conditions. The team then demonstrated through molecular cell experiments that such I/O relationship recovery also occurred in real cancer cells.
The results of this study, written by Dr. Jae Il Joo and Dr. Hwa-Jeong Park, were published in Wiley’s Advanced Science online on June 2 under the title, "Normalizing input-output relationships of cancer networks for reversion therapy."
< Image 1. Input-output (I/O) relationships in gene regulatory networks >
Professor Kwang-Hyun Cho's research team classified genes into four types by simulation-analyzing the effect of gene mutations on the I/O relationship of gene regulatory networks. (Figure A-J) In addition, by analyzing 18 genes of the cancer-related gene regulatory network, it was confirmed that when mutations occur in more than half of the genes constituting each network, reversibility is possible through appropriate control. (Figure K)
Professor Cho’s team uncovered that the reason the distorted I/O relationships of cancer cells could be reverted back to normal ones was the robustness and redundancy of intracellular gene control networks that developed over the course of evolution. In addition, they found that some genes were more promising as targets for cancer reversion than others, and showed through molecular cell experiments that controlling such genes could revert the distorted I/O relationships of cancer cells back to normal ones.
< Image 2. Simulation results of restoration of bladder cancer gene regulation network and I/O relationship of bladder cancer cells. >
The research team classified the effects of gene mutations on the I/O relationship in the bladder cancer gene regulation network by simulation analysis and classified them into 4 types. (Figure A) Through this, it was found that the distorted input-output relationship between bladder cancer cell lines KU-1919 and HCT-1197 could be restored to normal. (Figure B)
< Image 3. Analysis of survival of bladder cancer patients according to reversible gene mutation and I/O recovery experiment of bladder cancer cells. >
As predicted through network simulation analysis, Professor Kwang-Hyun Cho's research team confirmed through molecular cell experiments that the response to TGF-b was normally restored when AKT and MAP3K1 were inhibited in the bladder cancer cell line KU-1919. (Figure A-G) In addition, it was confirmed that there is a difference in the survival rate of bladder cancer patients depending on the presence or absence of a reversible gene mutation. (Figure H)
The results of this research show that the reversion of real cancer cells does not happen by chance, and that it is possible to systematically explore targets that can induce this phenomenon, thereby creating the potential for the development of innovative anticancer drugs that can control such target genes.
< Image 4. Cancer cell reversibility principle >
The research team analyzed the reversibility, redundancy, and robustness of various networks and found that there was a positive correlation between them. From this, it was found that reversibility was additionally inherent in the process of evolution in which the gene regulatory network acquired redundancy and consistency.
Professor Cho said, “By uncovering the fundamental principles of a new cancer reversion treatment strategy that may overcome the unresolved limitations of existing chemotherapy, we have increased the possibility of developing new and innovative drugs that can improve both the prognosis and quality of life of cancer patients.”
< Image 5. Conceptual diagram of research results >
The research team identified the fundamental control principle of cancer cell reversibility through systems biology research. When the I/O relationship of the intracellular gene regulatory network is distorted by mutation, the distorted I/O relationship can be restored to a normal state by identifying and adjusting the reversible gene target based on the redundancy of the molecular circuit inherent in the complex network.
After Professor Cho’s team first suggested the concept of reversion treatment, they published their results for reverting colorectal cancer in January 2020, and in January 2022 they successfully re-programmed malignant breast cancer cells back into hormone-treatable ones. In January 2023, the team successfully removed the metastasis ability from lung cancer cells and reverted them back to a state that allowed improved drug reactivity. However, these results were case studies of specific types of cancer and did not reveal what common principle allowed cancer reversion across all cancer types, making this the first revelation of the general principle of cancer reversion and its evolutionary origins.
This research was funded by the Ministry of Science and ICT of the Republic of Korea and the National Research Foundation of Korea.
2023.06.20 View 12883 -
 'Jumping Genes' Found to Alter Human Colon Genomes, Offering Insights into Aging and Tumorigenesis
The Korea Advanced Institute of Science and Technology (KAIST) and their collaborators have conducted a groundbreaking study targeting 'jumping genes' in the entire genomes of the human large intestine. Published in Nature on May 18 2023, the research unveils the surprising activity of 'Long interspersed nuclear element-1 (L1),' a type of jumping gene previously thought to be mostly dormant in human genomes. The study shows that L1 genes can become activated and disrupt genomic functions throughout an individual's lifetime, particularly in the colorectal epithelium.
(Paper Title: Widespread somatic L1 retrotransposition in normal colorectal epithelium, https://www.nature.com/articles/s41586-023-06046-z)
With approximately 500,000 L1 jumping genes, accounting for 17% of the human genome, they have long been recognized for their contribution to the evolution of the human species by introducing 'disruptive innovation' to genome sequences. Until now, it was believed that most L1 elements had lost their ability to jump in normal tissues of modern humans. However, this study reveals that some L1 jumping genes can be widely activated in normal cells, leading to the accumulation of genomic mutations over an individual's lifetime. The rate of L1 jumping and resulting genomic changes vary among different cell types, with a notable concentration observed in aged colon epithelial cells. The study illustrates that every colonic epithelial cell experiences an L1 jumping event by the age of 40 on average.
The research, led by co-first authors Chang Hyun Nam (a graduate student at KAIST) and Dr. Jeonghwan Youk (former graduate student at KAIST and assistant clinical professor at Seoul National University Hospital), involved the analysis of whole-genome sequences from 899 single cells obtained from skin (fibroblasts), blood, and colon epithelial tissues collected from 28 individuals. The study uncovers the activation of L1 jumping genes in normal cells, resulting in the gradual accumulation of genomic mutations over time. Additionally, the team explored epigenomic (DNA methylation) sequences to understand the mechanism behind L1 jumping gene activation. They found that cells with activated L1 jumping genes exhibit epigenetic instability, suggesting the critical role of epigenetic changes in regulating L1 jumping gene activity. Most of these epigenomic instabilities were found to arise during the early stages of embryogenesis. The study provides valuable insights into the aging process and the development of diseases in human colorectal tissues.
"This study illustrates that genomic damage in normal cells is acquired not only through exposure to carcinogens but also through the activity of endogenous components whose impact was previously unclear. Genomes of apparently healthy aged cells, particularly in the colorectal epithelium, become mosaic due to the activity of L1 jumping genes," said Prof. Young Seok Ju at KAIST.
"We emphasize the essential and ongoing collaboration among researchers in clinical medicine and basic medical sciences," said Prof. Min Jung Kim of the Department of Surgery at Seoul National University Hospital. "This case highlights the critical role of systematically collected human tissues from clinical settings in unraveling the complex process of disease development in humans."
"I am delighted that the research team's advancements in single-cell genome technology have come to fruition. We will persistently strive to lead in single-cell genome technology," said Prof. Hyun Woo Kwon of the Department of Nuclear Medicine at Korea University School of Medicine.
The research team received support from the Research Leader Program and the Young Researcher Program of the National Research Foundation of Korea, a grant from the MD-PhD/Medical Scientist Training Program through the Korea Health Industry Development Institute, and the Suh Kyungbae Foundation.
< Figure 1. Experimental design of the study >
< Figure 2. Schematic diagram illustrating factors influencing the soL1R landscape. >
Genetic composition of rc-L1s is inherited from the parents. The methylation landscape of rc-L1 promoters is predominantly determined by global DNA demethylation, followed by remethylation processes in the developmental stages. Then, when an rc-L1 is promoter demethylated in a specific cell lineage, the source expresses L1 transcripts thus making possible the induction of soL1Rs.
2023.05.22 View 10369
'Jumping Genes' Found to Alter Human Colon Genomes, Offering Insights into Aging and Tumorigenesis
The Korea Advanced Institute of Science and Technology (KAIST) and their collaborators have conducted a groundbreaking study targeting 'jumping genes' in the entire genomes of the human large intestine. Published in Nature on May 18 2023, the research unveils the surprising activity of 'Long interspersed nuclear element-1 (L1),' a type of jumping gene previously thought to be mostly dormant in human genomes. The study shows that L1 genes can become activated and disrupt genomic functions throughout an individual's lifetime, particularly in the colorectal epithelium.
(Paper Title: Widespread somatic L1 retrotransposition in normal colorectal epithelium, https://www.nature.com/articles/s41586-023-06046-z)
With approximately 500,000 L1 jumping genes, accounting for 17% of the human genome, they have long been recognized for their contribution to the evolution of the human species by introducing 'disruptive innovation' to genome sequences. Until now, it was believed that most L1 elements had lost their ability to jump in normal tissues of modern humans. However, this study reveals that some L1 jumping genes can be widely activated in normal cells, leading to the accumulation of genomic mutations over an individual's lifetime. The rate of L1 jumping and resulting genomic changes vary among different cell types, with a notable concentration observed in aged colon epithelial cells. The study illustrates that every colonic epithelial cell experiences an L1 jumping event by the age of 40 on average.
The research, led by co-first authors Chang Hyun Nam (a graduate student at KAIST) and Dr. Jeonghwan Youk (former graduate student at KAIST and assistant clinical professor at Seoul National University Hospital), involved the analysis of whole-genome sequences from 899 single cells obtained from skin (fibroblasts), blood, and colon epithelial tissues collected from 28 individuals. The study uncovers the activation of L1 jumping genes in normal cells, resulting in the gradual accumulation of genomic mutations over time. Additionally, the team explored epigenomic (DNA methylation) sequences to understand the mechanism behind L1 jumping gene activation. They found that cells with activated L1 jumping genes exhibit epigenetic instability, suggesting the critical role of epigenetic changes in regulating L1 jumping gene activity. Most of these epigenomic instabilities were found to arise during the early stages of embryogenesis. The study provides valuable insights into the aging process and the development of diseases in human colorectal tissues.
"This study illustrates that genomic damage in normal cells is acquired not only through exposure to carcinogens but also through the activity of endogenous components whose impact was previously unclear. Genomes of apparently healthy aged cells, particularly in the colorectal epithelium, become mosaic due to the activity of L1 jumping genes," said Prof. Young Seok Ju at KAIST.
"We emphasize the essential and ongoing collaboration among researchers in clinical medicine and basic medical sciences," said Prof. Min Jung Kim of the Department of Surgery at Seoul National University Hospital. "This case highlights the critical role of systematically collected human tissues from clinical settings in unraveling the complex process of disease development in humans."
"I am delighted that the research team's advancements in single-cell genome technology have come to fruition. We will persistently strive to lead in single-cell genome technology," said Prof. Hyun Woo Kwon of the Department of Nuclear Medicine at Korea University School of Medicine.
The research team received support from the Research Leader Program and the Young Researcher Program of the National Research Foundation of Korea, a grant from the MD-PhD/Medical Scientist Training Program through the Korea Health Industry Development Institute, and the Suh Kyungbae Foundation.
< Figure 1. Experimental design of the study >
< Figure 2. Schematic diagram illustrating factors influencing the soL1R landscape. >
Genetic composition of rc-L1s is inherited from the parents. The methylation landscape of rc-L1 promoters is predominantly determined by global DNA demethylation, followed by remethylation processes in the developmental stages. Then, when an rc-L1 is promoter demethylated in a specific cell lineage, the source expresses L1 transcripts thus making possible the induction of soL1Rs.
2023.05.22 View 10369 -
 KAIST presents a fundamental technology to remove metastatic traits from lung cancer cells
KAIST (President Kwang Hyung Lee) announced on January 30th that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering succeeded in using systems biology research to change the properties of carcinogenic cells in the lungs and eliminate both drug resistance and their ability to proliferate out to other areas of the body.
As the incidences of cancer increase within aging populations, cancer has become the most lethal disease threatening healthy life. Fatality rates are especially high when early detection does not happen in time and metastasis has occurred in various organs. In order to resolve this problem, a series of attempts were made to remove or lower the ability of cancer cells to spread, but they resulted in cancer cells in the intermediate state becoming more unstable and even more malignant, which created serious treatment challenges.
Professor Kwang-Hyun Cho's research team simulated various cancer cell states in the Epithelial-to-Mesenchymal Transition (EMT) of lung cancer cells, between epithelial cells without metastatic ability and mesenchymal cells with metastatic ability. A mathematical model of molecular network was established, and key regulators that could reverse the state of invasive and drug resistant mesenchymal cells back to the epithelial state were discovered through computer simulation analysis and molecular cell experiments. In particular, this process succeeded in properly reverting the mesenchymal lung cancer cells to a state where they were sensitive to chemotherapy treatment while avoiding the unstable EMT hybrid cell state in the middle process, which had remained a difficult problem.
The results of this research, in which KAIST Ph.D. student Namhee Kim, Dr. Chae Young Hwang, Researcher Taeyoung Kim, and Ph.D. student Hyunjin Kim participated, were published as an online paper in the international journal “Cancer Research” published by the American Association for Cancer Research (AACR) on January 30th. (Paper title: A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states)
Cells in an EMT hybrid state, which are caused by incomplete transitions during the EMT process in cancer cells, have the characteristics of both epithelial cells and mesenchymal cells, and are known to have high drug resistance and metastatic potential by acquiring high stem cell capacity. In particular, EMT is further enhanced through factors such as transforming growth factor-beta (TGF-β) secreted from the tumor microenvironment (TME) and, as a result, various cell states with high plasticity appear. Due to the complexity of EMT, it has been very difficult to completely reverse the transitional process of the mesenchymal cancer cells to an epithelial cell state in which metastatic ability and drug resistance are eliminated while avoiding the EMT hybrid cell state with high metastatic ability and drug resistance.
Professor Kwang-Hyun Cho's research team established a mathematical model of the gene regulation network that governs the complex process of EMT, and then applied large-scale computer simulation analysis and complex system network control technology to identify and verify 'p53', 'SMAD4', and 'ERK1' and 'ERK 2' (collectively ERKs) through molecular cell experiments as the three key molecular targets that can transform lung cancer cells in the mesenchymal cell state, reversed back to an epithelial cell state that no longer demonstrates the ability to metastasize, while avoiding the EMT hybrid cell state.
In particular, by analyzing the molecular regulatory mechanism of the complex EMT process at the system level, the key pathways were identified that were linked to the positive feedback that plays an important role in completely returning cancer cells to an epithelial cell state in which metastatic ability and drug resistance are removed.
This discovery is significant in that it proved that mesenchymal cells can be reverted to the state of epithelial cells under conditions where TGF-β stimulation are present, like they are in the actual environment where cancer tissue forms in the human body.
Abnormal EMT in cancer cells leads to various malignant traits such as the migration and invasion of cancer cells, changes in responsiveness to chemotherapy treatment, enhanced stem cell function, and the dissemination of cancer. In particular, the acquisition of the metastatic ability of cancer cells is a key determinant factor for the prognosis of cancer patients. The EMT reversal technology in lung cancer cells developed in this research is a new anti-cancer treatment strategy that reprograms cancer cells to eliminate their high plasticity and metastatic potential and increase their responsiveness to chemotherapy.
Professor Kwang-Hyun Cho said, "By succeeding in reversing the state of lung cancer cells that acquired high metastatic traits and resistance to drugs and reverting them to a treatable epithelial cell state with renewed sensitivity to chemotherapy, the research findings propose a new strategy for treatments that can improve the prognosis of cancer patients.”
Professor Kwang-Hyun Cho's research team was the first to present the principle of reversal treatment to revert cancer cells to normal cells, following through with the announcement of the results of their study that reverted colon cancer cells to normal colon cells in January of 2020, and also presenting successful re-programming research where the most malignant basal type breast cancer cells turned into less-malignant luminal type breast cancer cells that were treatable with hormonal therapies in January of 2022. This latest research result is the third in the development of reversal technology where lung cancer cells that had acquired metastatic traits returned to a state in which their metastatic ability was removed and drug sensitivity was enhanced.
This research was carried out with support from the Ministry of Science and ICT and the National Research Foundation of Korea's Basic Research in Science & Engineering Program for Mid-Career Researchers.
< Figure 1. Construction of the mathematical model of the regulatory network to represent the EMT phenotype based on the interaction between various molecules related to EMT.
(A) Professor Kwang-Hyun Cho's research team investigated numerous literatures and databases related to complex EMT, and based on comparative analysis of cell line data showing epithelial and mesenchymal cell conditions, they extracted key signaling pathways related to EMT and built a mathematical model of regulatory network (B) By comparing the results of computer simulation analysis and the molecular cell experiments, it was verified how well the constructed mathematical model simulated the actual cellular phenomena. >
< Figure 2. Understanding of various EMT phenotypes through large-scale computer simulation analysis and complex system network control technology.
(A) Through computer simulation analysis and experiments, Professor Kwang-Hyun Cho's research team found that complete control of EMT is impossible with single-molecule control alone. In particular, through comparison of the relative stability of attractors, it was revealed that the cell state exhibiting EMT hybrid characteristics has unstable properties. (B), (C) Based on these results, Prof. Cho’s team identified two feedbacks (positive feedback consisting of Snail-miR-34 and ZEB1-miR-200) that play an important role in avoiding the EMT hybrid state that appeared in the TGF-β-ON state. It was found through computer simulation analysis that the two feedbacks restore relatively high stability when the excavated p53 and SMAD4 are regulated. In addition, molecular cell experiments demonstrated that the expression levels of E-cad and ZEB1, which are representative phenotypic markers of EMT, changed similarly to the expression profile in the epithelial cell state, despite the TGF-β-ON state. >
< Figure 3. Complex molecular network analysis and discovery of reprogramming molecular targets for intact elimination of EMT hybrid features.
(A) Controlling the expression of p53 and SMAD4 in lung cancer cell lines was expected to overcome drug resistance, but contrary to expectations, chemotherapy responsiveness was not restored. (B) Professor Kwang-Hyun Cho's research team additionally analyzed computer simulations, genome data, and experimental results and found that high expression levels of TWIST1 and EPCAM were related to drug resistance. (C) Prof. Cho’s team identified three key molecular targets: p53, SMAD4 and ERK1 & ERK2. (D), (E) Furthermore, they identified a key pathway that plays an important role in completely reversing into epithelial cells while avoiding EMT hybrid characteristics, and confirmed through network analysis and attractor analysis that high stability of the key pathway was restored when the proposed molecular target was controlled. >
< Figure 4. Verification through experiments with lung cancer cell lines.
When p53 was activated and SMAD4 and ERK1/2 were inhibited in lung cancer cell lines, (A), (B) E-cad protein expression increased and ZEB1 protein expression decreased, and (C) mesenchymal cell status including TWIST1 and EPCAM and gene expression of markers related to stem cell potential characteristics were completely inhibited. In addition, (D) it was confirmed that resistance to chemotherapy treatment was also overcome as the cell state was reversed by the regulated target. >
< Figure 5. A schematic representation of the research results.
Prof. Cho’s research team identified key molecular regulatory pathways to avoid high plasticity formed by abnormal EMT of cancer cells and reverse it to an epithelial cell state through systems biology research. From this analysis, a reprogramming molecular target that can reverse the state of mesenchymal cells with acquired invasiveness and drug resistance to the state of epithelial cells with restored drug responsiveness was discovered.
For lung cancer cells, when a drug that enhances the expression of p53, one of the molecular targets discovered, and inhibits the expression of SMAD4 and ERK1 & ERK2 is administered, the molecular network of genes in the state of mesenchymal cells is modified, eventually eliminating metastatic ability and it is reprogrammed to turn into epithelial cells without the resistance to chemotherapy treatments. >
2023.01.30 View 20327
KAIST presents a fundamental technology to remove metastatic traits from lung cancer cells
KAIST (President Kwang Hyung Lee) announced on January 30th that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering succeeded in using systems biology research to change the properties of carcinogenic cells in the lungs and eliminate both drug resistance and their ability to proliferate out to other areas of the body.
As the incidences of cancer increase within aging populations, cancer has become the most lethal disease threatening healthy life. Fatality rates are especially high when early detection does not happen in time and metastasis has occurred in various organs. In order to resolve this problem, a series of attempts were made to remove or lower the ability of cancer cells to spread, but they resulted in cancer cells in the intermediate state becoming more unstable and even more malignant, which created serious treatment challenges.
Professor Kwang-Hyun Cho's research team simulated various cancer cell states in the Epithelial-to-Mesenchymal Transition (EMT) of lung cancer cells, between epithelial cells without metastatic ability and mesenchymal cells with metastatic ability. A mathematical model of molecular network was established, and key regulators that could reverse the state of invasive and drug resistant mesenchymal cells back to the epithelial state were discovered through computer simulation analysis and molecular cell experiments. In particular, this process succeeded in properly reverting the mesenchymal lung cancer cells to a state where they were sensitive to chemotherapy treatment while avoiding the unstable EMT hybrid cell state in the middle process, which had remained a difficult problem.
The results of this research, in which KAIST Ph.D. student Namhee Kim, Dr. Chae Young Hwang, Researcher Taeyoung Kim, and Ph.D. student Hyunjin Kim participated, were published as an online paper in the international journal “Cancer Research” published by the American Association for Cancer Research (AACR) on January 30th. (Paper title: A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states)
Cells in an EMT hybrid state, which are caused by incomplete transitions during the EMT process in cancer cells, have the characteristics of both epithelial cells and mesenchymal cells, and are known to have high drug resistance and metastatic potential by acquiring high stem cell capacity. In particular, EMT is further enhanced through factors such as transforming growth factor-beta (TGF-β) secreted from the tumor microenvironment (TME) and, as a result, various cell states with high plasticity appear. Due to the complexity of EMT, it has been very difficult to completely reverse the transitional process of the mesenchymal cancer cells to an epithelial cell state in which metastatic ability and drug resistance are eliminated while avoiding the EMT hybrid cell state with high metastatic ability and drug resistance.
Professor Kwang-Hyun Cho's research team established a mathematical model of the gene regulation network that governs the complex process of EMT, and then applied large-scale computer simulation analysis and complex system network control technology to identify and verify 'p53', 'SMAD4', and 'ERK1' and 'ERK 2' (collectively ERKs) through molecular cell experiments as the three key molecular targets that can transform lung cancer cells in the mesenchymal cell state, reversed back to an epithelial cell state that no longer demonstrates the ability to metastasize, while avoiding the EMT hybrid cell state.
In particular, by analyzing the molecular regulatory mechanism of the complex EMT process at the system level, the key pathways were identified that were linked to the positive feedback that plays an important role in completely returning cancer cells to an epithelial cell state in which metastatic ability and drug resistance are removed.
This discovery is significant in that it proved that mesenchymal cells can be reverted to the state of epithelial cells under conditions where TGF-β stimulation are present, like they are in the actual environment where cancer tissue forms in the human body.
Abnormal EMT in cancer cells leads to various malignant traits such as the migration and invasion of cancer cells, changes in responsiveness to chemotherapy treatment, enhanced stem cell function, and the dissemination of cancer. In particular, the acquisition of the metastatic ability of cancer cells is a key determinant factor for the prognosis of cancer patients. The EMT reversal technology in lung cancer cells developed in this research is a new anti-cancer treatment strategy that reprograms cancer cells to eliminate their high plasticity and metastatic potential and increase their responsiveness to chemotherapy.
Professor Kwang-Hyun Cho said, "By succeeding in reversing the state of lung cancer cells that acquired high metastatic traits and resistance to drugs and reverting them to a treatable epithelial cell state with renewed sensitivity to chemotherapy, the research findings propose a new strategy for treatments that can improve the prognosis of cancer patients.”
Professor Kwang-Hyun Cho's research team was the first to present the principle of reversal treatment to revert cancer cells to normal cells, following through with the announcement of the results of their study that reverted colon cancer cells to normal colon cells in January of 2020, and also presenting successful re-programming research where the most malignant basal type breast cancer cells turned into less-malignant luminal type breast cancer cells that were treatable with hormonal therapies in January of 2022. This latest research result is the third in the development of reversal technology where lung cancer cells that had acquired metastatic traits returned to a state in which their metastatic ability was removed and drug sensitivity was enhanced.
This research was carried out with support from the Ministry of Science and ICT and the National Research Foundation of Korea's Basic Research in Science & Engineering Program for Mid-Career Researchers.
< Figure 1. Construction of the mathematical model of the regulatory network to represent the EMT phenotype based on the interaction between various molecules related to EMT.
(A) Professor Kwang-Hyun Cho's research team investigated numerous literatures and databases related to complex EMT, and based on comparative analysis of cell line data showing epithelial and mesenchymal cell conditions, they extracted key signaling pathways related to EMT and built a mathematical model of regulatory network (B) By comparing the results of computer simulation analysis and the molecular cell experiments, it was verified how well the constructed mathematical model simulated the actual cellular phenomena. >
< Figure 2. Understanding of various EMT phenotypes through large-scale computer simulation analysis and complex system network control technology.
(A) Through computer simulation analysis and experiments, Professor Kwang-Hyun Cho's research team found that complete control of EMT is impossible with single-molecule control alone. In particular, through comparison of the relative stability of attractors, it was revealed that the cell state exhibiting EMT hybrid characteristics has unstable properties. (B), (C) Based on these results, Prof. Cho’s team identified two feedbacks (positive feedback consisting of Snail-miR-34 and ZEB1-miR-200) that play an important role in avoiding the EMT hybrid state that appeared in the TGF-β-ON state. It was found through computer simulation analysis that the two feedbacks restore relatively high stability when the excavated p53 and SMAD4 are regulated. In addition, molecular cell experiments demonstrated that the expression levels of E-cad and ZEB1, which are representative phenotypic markers of EMT, changed similarly to the expression profile in the epithelial cell state, despite the TGF-β-ON state. >
< Figure 3. Complex molecular network analysis and discovery of reprogramming molecular targets for intact elimination of EMT hybrid features.
(A) Controlling the expression of p53 and SMAD4 in lung cancer cell lines was expected to overcome drug resistance, but contrary to expectations, chemotherapy responsiveness was not restored. (B) Professor Kwang-Hyun Cho's research team additionally analyzed computer simulations, genome data, and experimental results and found that high expression levels of TWIST1 and EPCAM were related to drug resistance. (C) Prof. Cho’s team identified three key molecular targets: p53, SMAD4 and ERK1 & ERK2. (D), (E) Furthermore, they identified a key pathway that plays an important role in completely reversing into epithelial cells while avoiding EMT hybrid characteristics, and confirmed through network analysis and attractor analysis that high stability of the key pathway was restored when the proposed molecular target was controlled. >
< Figure 4. Verification through experiments with lung cancer cell lines.
When p53 was activated and SMAD4 and ERK1/2 were inhibited in lung cancer cell lines, (A), (B) E-cad protein expression increased and ZEB1 protein expression decreased, and (C) mesenchymal cell status including TWIST1 and EPCAM and gene expression of markers related to stem cell potential characteristics were completely inhibited. In addition, (D) it was confirmed that resistance to chemotherapy treatment was also overcome as the cell state was reversed by the regulated target. >
< Figure 5. A schematic representation of the research results.
Prof. Cho’s research team identified key molecular regulatory pathways to avoid high plasticity formed by abnormal EMT of cancer cells and reverse it to an epithelial cell state through systems biology research. From this analysis, a reprogramming molecular target that can reverse the state of mesenchymal cells with acquired invasiveness and drug resistance to the state of epithelial cells with restored drug responsiveness was discovered.
For lung cancer cells, when a drug that enhances the expression of p53, one of the molecular targets discovered, and inhibits the expression of SMAD4 and ERK1 & ERK2 is administered, the molecular network of genes in the state of mesenchymal cells is modified, eventually eliminating metastatic ability and it is reprogrammed to turn into epithelial cells without the resistance to chemotherapy treatments. >
2023.01.30 View 20327 -
 Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 12840
Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 12840 -
 Identification of How Chemotherapy Drug Works Could Deliver Personalized Cancer Treatment
The chemotherapy drug decitabine is commonly used to treat patients with blood cancers, but its response rate is somewhat low. Researchers have now identified why this is the case, opening the door to more personalized cancer therapies for those with these types of cancers, and perhaps further afield.
Researchers have identified the genetic and molecular mechanisms within cells that make the chemotherapy drug decitabine—used to treat patients with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) —work for some patients but not others. The findings should assist clinicians in developing more patient-specific treatment strategies.
The findings were published in the Proceedings of the National Academies of Science on March 30.
The chemotherapy drug decitabine, also known by its brand name Dacogen, works by modifying our DNA that in turn switches on genes that stop the cancer cells from growing and replicating. However, decitabine’s response rate is somewhat low (showing improvement in just 30-35% of patients), which leaves something of a mystery as to why it works well for some patients but not for others. To find out why this happens, researchers from the KAIST investigated the molecular mediators that are involved with regulating the effects of the drug.
Decitabine works to activate the production of endogenous retroviruses (ERVs), which in turn induces an immune response. ERVs are viruses that long ago inserted dormant copies of themselves into the human genome. Decitabine in essence, ‘reactivates’ these viral elements and produces double-stranded RNAs (dsRNAs) that the immune system views as a foreign body.
“However, the mechanisms involved in this process, in particular how production and transport of these ERV dsRNAs were regulated within the cell were understudied,” said corresponding author Yoosik Kim, professor in the Department of Chemical and Biomolecular Engineering at KAIST.
“So to explain why decitabine works in some patients but not others, we investigated what these molecular mechanisms were,” added Kim.
To do so, the researchers used image-based RNA interference (RNAi) screening. This is a relatively new technique in which specific sequences within a genome are knocked out of action or “downregulated.” Large-scale screening, which can be performed in cultured cells or within live organisms, works to investigate the function of different genes. The KAIST researchers collaborated with the Institut Pasteur Korea to analyze the effect of downregulating genes that recognize ERV dsRNAs and could be involved in the cellular response to decitabine.
From these initial screening results, they performed an even more detailed downregulation screening analysis. Through the screening, they were able to identify two particular gene sequences involved in the production of an RNA-binding protein called Staufen1 and the production of a strand of RNA that does not in turn produce any proteins called TINCR that play a key regulatory role in response to the drug. Staufen1 binds directly to dsRNAs and stabilizes them in concert with the TINCR.
If a patient is not producing sufficient Staufen1 and TINCR, then the dsRNA viral mimics quickly degrade before the immune system can spot them. And, crucially for cancer therapy, this means that patients with lower expression (activation) of these sequences will show inferior response to decitabine. Indeed, the researchers confirmed that MDS/AML patients with low Staufen1 and TINCR expression did not benefit from decitabine therapy.
“We can now isolate patients who will not benefit from the therapy and direct them to a different type of therapy,” said first author Yongsuk Ku. “This serves as an important step toward developing a patient-specific treatment cancer strategy.”
As the researchers used patient samples taken from bone marrow, the next step will be to try to develop a testing method that can identify the problem from just blood samples, which are much easier to acquire from patients.
The team plans to investigate if the analysis can be extended to patients with solid tumors in addition to those with blood cancers.
-Profile
Professor Yoosik Kim
https://qcbio.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering
KAIST
-Publication
Noncanonical immune response to the inhibition of DNA methylation by Staufen1 via stabilization of endogenous retrovirus RNAs, PNAS
2021.05.24 View 12523
Identification of How Chemotherapy Drug Works Could Deliver Personalized Cancer Treatment
The chemotherapy drug decitabine is commonly used to treat patients with blood cancers, but its response rate is somewhat low. Researchers have now identified why this is the case, opening the door to more personalized cancer therapies for those with these types of cancers, and perhaps further afield.
Researchers have identified the genetic and molecular mechanisms within cells that make the chemotherapy drug decitabine—used to treat patients with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) —work for some patients but not others. The findings should assist clinicians in developing more patient-specific treatment strategies.
The findings were published in the Proceedings of the National Academies of Science on March 30.
The chemotherapy drug decitabine, also known by its brand name Dacogen, works by modifying our DNA that in turn switches on genes that stop the cancer cells from growing and replicating. However, decitabine’s response rate is somewhat low (showing improvement in just 30-35% of patients), which leaves something of a mystery as to why it works well for some patients but not for others. To find out why this happens, researchers from the KAIST investigated the molecular mediators that are involved with regulating the effects of the drug.
Decitabine works to activate the production of endogenous retroviruses (ERVs), which in turn induces an immune response. ERVs are viruses that long ago inserted dormant copies of themselves into the human genome. Decitabine in essence, ‘reactivates’ these viral elements and produces double-stranded RNAs (dsRNAs) that the immune system views as a foreign body.
“However, the mechanisms involved in this process, in particular how production and transport of these ERV dsRNAs were regulated within the cell were understudied,” said corresponding author Yoosik Kim, professor in the Department of Chemical and Biomolecular Engineering at KAIST.
“So to explain why decitabine works in some patients but not others, we investigated what these molecular mechanisms were,” added Kim.
To do so, the researchers used image-based RNA interference (RNAi) screening. This is a relatively new technique in which specific sequences within a genome are knocked out of action or “downregulated.” Large-scale screening, which can be performed in cultured cells or within live organisms, works to investigate the function of different genes. The KAIST researchers collaborated with the Institut Pasteur Korea to analyze the effect of downregulating genes that recognize ERV dsRNAs and could be involved in the cellular response to decitabine.
From these initial screening results, they performed an even more detailed downregulation screening analysis. Through the screening, they were able to identify two particular gene sequences involved in the production of an RNA-binding protein called Staufen1 and the production of a strand of RNA that does not in turn produce any proteins called TINCR that play a key regulatory role in response to the drug. Staufen1 binds directly to dsRNAs and stabilizes them in concert with the TINCR.
If a patient is not producing sufficient Staufen1 and TINCR, then the dsRNA viral mimics quickly degrade before the immune system can spot them. And, crucially for cancer therapy, this means that patients with lower expression (activation) of these sequences will show inferior response to decitabine. Indeed, the researchers confirmed that MDS/AML patients with low Staufen1 and TINCR expression did not benefit from decitabine therapy.
“We can now isolate patients who will not benefit from the therapy and direct them to a different type of therapy,” said first author Yongsuk Ku. “This serves as an important step toward developing a patient-specific treatment cancer strategy.”
As the researchers used patient samples taken from bone marrow, the next step will be to try to develop a testing method that can identify the problem from just blood samples, which are much easier to acquire from patients.
The team plans to investigate if the analysis can be extended to patients with solid tumors in addition to those with blood cancers.
-Profile
Professor Yoosik Kim
https://qcbio.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering
KAIST
-Publication
Noncanonical immune response to the inhibition of DNA methylation by Staufen1 via stabilization of endogenous retrovirus RNAs, PNAS
2021.05.24 View 12523 -
 What Fuels a “Domino Effect” in Cancer Drug Resistance?
KAIST researchers have identified mechanisms that relay prior acquired resistance to the first-line chemotherapy to the second-line targeted therapy, fueling a “domino effect” in cancer drug resistance. Their study featured in the February 7 edition of Science Advances suggests a new strategy for improving the second-line setting of cancer treatment for patients who showed resistance to anti-cancer drugs.
Resistance to cancer drugs is often managed in the clinic by chemotherapy and targeted therapy. Unlike chemotherapy that works by repressing fast-proliferating cells, targeted therapy blocks a single oncogenic pathway to halt tumor growth. In many cases, targeted therapy is engaged as a maintenance therapy or employed in the second-line after front-line chemotherapy.
A team of researchers led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering and the KAIST Institute for Health Science and Technology (KIHST) has discovered an unexpected resistance signature that occurs between chemotherapy and targeted therapy. The team further identified a set of integrated mechanisms that promotes this kind of sequential therapy resistance.
“There have been multiple clinical accounts reflecting that targeted therapies tend to be least successful in patients who have exhausted all standard treatments,” said the first author of the paper Mark Borris D. Aldonza. He continued, “These accounts ignited our hypothesis that failed responses to some chemotherapies might speed up the evolution of resistance to other drugs, particularly those with specific targets.”
Aldonza and his colleagues extracted large amounts of drug-resistance information from the open-source database the Genomics of Drug Sensitivity in Cancer (GDSC), which contains thousands of drug response data entries from various human cancer cell lines. Their big data analysis revealed that cancer cell lines resistant to chemotherapies classified as anti-mitotic drugs (AMDs), toxins that inhibit overacting cell division, are also resistant to a class of targeted therapies called epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs).
In all of the cancer types analyzed, more than 84 percent of those resistant to AMDs, representatively ‘paclitaxel’, were also resistant to at least nine EGFR-TKIs. In lung, pancreatic, and breast cancers where paclitaxel is often used as a first-line, standard-of-care regimen, greater than 92 percent showed resistance to EGFR-TKIs. Professor Kim said, “It is surprising to see that such collateral resistance can occur specifically between two chemically different classes of drugs.”
To figure out how failed responses to paclitaxel leads to resistance to EGFR-TKIs, the team validated co-resistance signatures that they found in the database by generating and analyzing a subset of slow-doubling, paclitaxel-resistant cancer models called ‘persisters’.
The results demonstrated that paclitaxel-resistant cancers remodel their stress response by first becoming more stem cell-like, evolving the ability to self-renew to adapt to more stressful conditions like drug exposures. More surprisingly, when the researchers characterized the metabolic state of the cells, EGFR-TKI persisters derived from paclitaxel-resistant cancer cells showed high dependencies to energy-producing processes such as glycolysis and glutaminolysis.
“We found that, without an energy stimulus like glucose, these cells transform to becoming more senescent, a characteristic of cells that have arrested cell division. However, this senescence is controlled by stem cell factors, which the paclitaxel-resistant cancers use to escape from this arrested state given a favorable condition to re-grow,” said Aldonza.
Professor Kim explained, “Before this research, there was no reason to expect that acquiring the cancer stem cell phenotype that dramatically leads to a cascade of changes in cellular states affecting metabolism and cell death is linked with drug-specific sequential resistance between two classes of therapies.”
He added, “The expansion of our work to other working models of drug resistance in a much more clinically-relevant setting, perhaps in clinical trials, will take on increasing importance, as sequential treatment strategies will continue to be adapted to various forms of anti-cancer therapy regimens.”
This study was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF-2016R1C1B2009886), and the KAIST Future Systems Healthcare Project (KAISTHEALTHCARE42) funded by the Korean Ministry of Science and ICT (MSIT). Undergraduate student Aldonza participated in this research project and presented the findings as the lead author as part of the Undergraduate Research Participation (URP) Program at KAIST.
< Figure 1. Schematic overview of the study. >
< Figure 2. Big data analysis revealing co-resistance signatures between classes of anti-cancer drugs. >
Publication:
Aldonza et al. (2020) Prior acquired resistance to paclitaxel relays diverse EGFR-targeted therapy persistence mechanisms. Science Advances, Vol. 6, No. 6, eaav7416. Available online at http://dx.doi.org/10.1126/sciadv.aav7416
Profile: Prof. Yoosik Kim, MA, PhD
ysyoosik@kaist.ac.kr
https://qcbio.kaist.ac.kr/
Assistant Professor
Bio Network Analysis Laboratory
Department of Chemical and Biomolecular Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon, Republic of Korea
Profile: Mark Borris D. Aldonza
borris@kaist.ac.kr
Undergraduate Student
Department of Biological Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon, Republic of Korea
(END)
2020.02.10 View 17957
What Fuels a “Domino Effect” in Cancer Drug Resistance?
KAIST researchers have identified mechanisms that relay prior acquired resistance to the first-line chemotherapy to the second-line targeted therapy, fueling a “domino effect” in cancer drug resistance. Their study featured in the February 7 edition of Science Advances suggests a new strategy for improving the second-line setting of cancer treatment for patients who showed resistance to anti-cancer drugs.
Resistance to cancer drugs is often managed in the clinic by chemotherapy and targeted therapy. Unlike chemotherapy that works by repressing fast-proliferating cells, targeted therapy blocks a single oncogenic pathway to halt tumor growth. In many cases, targeted therapy is engaged as a maintenance therapy or employed in the second-line after front-line chemotherapy.
A team of researchers led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering and the KAIST Institute for Health Science and Technology (KIHST) has discovered an unexpected resistance signature that occurs between chemotherapy and targeted therapy. The team further identified a set of integrated mechanisms that promotes this kind of sequential therapy resistance.
“There have been multiple clinical accounts reflecting that targeted therapies tend to be least successful in patients who have exhausted all standard treatments,” said the first author of the paper Mark Borris D. Aldonza. He continued, “These accounts ignited our hypothesis that failed responses to some chemotherapies might speed up the evolution of resistance to other drugs, particularly those with specific targets.”
Aldonza and his colleagues extracted large amounts of drug-resistance information from the open-source database the Genomics of Drug Sensitivity in Cancer (GDSC), which contains thousands of drug response data entries from various human cancer cell lines. Their big data analysis revealed that cancer cell lines resistant to chemotherapies classified as anti-mitotic drugs (AMDs), toxins that inhibit overacting cell division, are also resistant to a class of targeted therapies called epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs).
In all of the cancer types analyzed, more than 84 percent of those resistant to AMDs, representatively ‘paclitaxel’, were also resistant to at least nine EGFR-TKIs. In lung, pancreatic, and breast cancers where paclitaxel is often used as a first-line, standard-of-care regimen, greater than 92 percent showed resistance to EGFR-TKIs. Professor Kim said, “It is surprising to see that such collateral resistance can occur specifically between two chemically different classes of drugs.”
To figure out how failed responses to paclitaxel leads to resistance to EGFR-TKIs, the team validated co-resistance signatures that they found in the database by generating and analyzing a subset of slow-doubling, paclitaxel-resistant cancer models called ‘persisters’.
The results demonstrated that paclitaxel-resistant cancers remodel their stress response by first becoming more stem cell-like, evolving the ability to self-renew to adapt to more stressful conditions like drug exposures. More surprisingly, when the researchers characterized the metabolic state of the cells, EGFR-TKI persisters derived from paclitaxel-resistant cancer cells showed high dependencies to energy-producing processes such as glycolysis and glutaminolysis.
“We found that, without an energy stimulus like glucose, these cells transform to becoming more senescent, a characteristic of cells that have arrested cell division. However, this senescence is controlled by stem cell factors, which the paclitaxel-resistant cancers use to escape from this arrested state given a favorable condition to re-grow,” said Aldonza.
Professor Kim explained, “Before this research, there was no reason to expect that acquiring the cancer stem cell phenotype that dramatically leads to a cascade of changes in cellular states affecting metabolism and cell death is linked with drug-specific sequential resistance between two classes of therapies.”
He added, “The expansion of our work to other working models of drug resistance in a much more clinically-relevant setting, perhaps in clinical trials, will take on increasing importance, as sequential treatment strategies will continue to be adapted to various forms of anti-cancer therapy regimens.”
This study was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF-2016R1C1B2009886), and the KAIST Future Systems Healthcare Project (KAISTHEALTHCARE42) funded by the Korean Ministry of Science and ICT (MSIT). Undergraduate student Aldonza participated in this research project and presented the findings as the lead author as part of the Undergraduate Research Participation (URP) Program at KAIST.
< Figure 1. Schematic overview of the study. >
< Figure 2. Big data analysis revealing co-resistance signatures between classes of anti-cancer drugs. >
Publication:
Aldonza et al. (2020) Prior acquired resistance to paclitaxel relays diverse EGFR-targeted therapy persistence mechanisms. Science Advances, Vol. 6, No. 6, eaav7416. Available online at http://dx.doi.org/10.1126/sciadv.aav7416
Profile: Prof. Yoosik Kim, MA, PhD
ysyoosik@kaist.ac.kr
https://qcbio.kaist.ac.kr/
Assistant Professor
Bio Network Analysis Laboratory
Department of Chemical and Biomolecular Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon, Republic of Korea
Profile: Mark Borris D. Aldonza
borris@kaist.ac.kr
Undergraduate Student
Department of Biological Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon, Republic of Korea
(END)
2020.02.10 View 17957