biomarker
-
 A Way for Smartwatches to Detect Depression Risks Devised by KAIST and U of Michigan Researchers
- A international joint research team of KAIST and the University of Michigan developed a digital biomarker for predicting symptoms of depression based on data collected by smartwatches
- It has the potential to be used as a medical technology to replace the economically burdensome fMRI measurement test
- It is expected to expand the scope of digital health data analysis
The CORONA virus pandemic also brought about a pandemic of mental illness. Approximately one billion people worldwide suffer from various psychiatric conditions. Korea is one of more serious cases, with approximately 1.8 million patients exhibiting depression and anxiety disorders, and the total number of patients with clinical mental diseases has increased by 37% in five years to approximately 4.65 million. A joint research team from Korea and the US has developed a technology that uses biometric data collected through wearable devices to predict tomorrow's mood and, further, to predict the possibility of developing symptoms of depression.
< Figure 1. Schematic diagram of the research results. Based on the biometric data collected by a smartwatch, a mathematical algorithm that solves the inverse problem to estimate the brain's circadian phase and sleep stages has been developed. This algorithm can estimate the degrees of circadian disruption, and these estimates can be used as the digital biomarkers to predict depression risks. >
KAIST (President Kwang Hyung Lee) announced on the 15th of January that the research team under Professor Dae Wook Kim from the Department of Brain and Cognitive Sciences and the team under Professor Daniel B. Forger from the Department of Mathematics at the University of Michigan in the United States have developed a technology to predict symptoms of depression such as sleep disorders, depression, loss of appetite, overeating, and decreased concentration in shift workers from the activity and heart rate data collected from smartwatches.
According to WHO, a promising new treatment direction for mental illness focuses on the sleep and circadian timekeeping system located in the hypothalamus of the brain, which directly affect impulsivity, emotional responses, decision-making, and overall mood.
However, in order to measure endogenous circadian rhythms and sleep states, blood or saliva must be drawn every 30 minutes throughout the night to measure changes in the concentration of the melatonin hormone in our bodies and polysomnography (PSG) must be performed. As such treatments requires hospitalization and most psychiatric patients only visit for outpatient treatment, there has been no significant progress in developing treatment methods that take these two factors into account. In addition, the cost of the PSG test, which is approximately $1000, leaves mental health treatment considering sleep and circadian rhythms out of reach for the socially disadvantaged.
The solution to overcome these problems is to employ wearable devices for the easier collection of biometric data such as heart rate, body temperature, and activity level in real time without spatial constraints. However, current wearable devices have the limitation of providing only indirect information on biomarkers required by medical staff, such as the phase of the circadian clock.
The joint research team developed a filtering technology that accurately estimates the phase of the circadian clock, which changes daily, such as heart rate and activity time series data collected from a smartwatch. This is an implementation of a digital twin that precisely describes the circadian rhythm in the brain, and it can be used to estimate circadian rhythm disruption.
< Figure 2. The suprachiasmatic nucleus located in the hypothalamus of the brain is the central biological clock that regulates the 24-hour physiological rhythm and plays a key role in maintaining the body’s circadian rhythm. If the phase of this biological clock is disrupted, it affects various parts of the brain, which can cause psychiatric conditions such as depression. >
The possibility of using the digital twin of this circadian clock to predict the symptoms of depression was verified through collaboration with the research team of Professor Srijan Sen of the Michigan Neuroscience Institute and Professor Amy Bohnert of the Department of Psychiatry of the University of Michigan.
The collaborative research team conducted a large-scale prospective cohort study involving approximately 800 shift workers and showed that the circadian rhythm disruption digital biomarker estimated through the technology can predict tomorrow's mood as well as six symptoms, including sleep problems, appetite changes, decreased concentration, and suicidal thoughts, which are representative symptoms of depression.
< Figure 3. The circadian rhythm of hormones such as melatonin regulates various physiological functions and behaviors such as heart rate and activity level. These physiological and behavioral signals can be measured in daily life through wearable devices. In order to estimate the body’s circadian rhythm inversely based on the measured biometric signals, a mathematical algorithm is needed. This algorithm plays a key role in accurately identifying the characteristics of circadian rhythms by extracting hidden physiological patterns from biosignals. >
Professor Dae Wook Kim said, "It is very meaningful to be able to conduct research that provides a clue for ways to apply wearable biometric data using mathematics that have not previously been utilized for actual disease management." He added, "We expect that this research will be able to present continuous and non-invasive mental health monitoring technology. This is expected to present a new paradigm for mental health care. By resolving some of the major problems socially disadvantaged people may face in current treatment practices, they may be able to take more active steps when experiencing symptoms of depression, such as seeking counsel before things get out of hand."
< Figure 4. A mathematical algorithm was devised to circumvent the problems of estimating the phase of the brain's biological clock and sleep stages inversely from the biodata collected by a smartwatch. This algorithm can estimate the degree of daily circadian rhythm disruption, and this estimate can be used as a digital biomarker to predict depression symptoms. >
The results of this study, in which Professor Dae Wook Kim of the Department of Brain and Cognitive Sciences at KAIST participated as the joint first author and corresponding author, were published in the online version of the international academic journal npj Digital Medicine on December 5, 2024. (Paper title: The real-world association between digital markers of circadian disruption and mental health risks) DOI: 10.1038/s41746-024-01348-6
This study was conducted with the support of the KAIST's Research Support Program for New Faculty Members, the US National Science Foundation, the US National Institutes of Health, and the US Army Research Institute MURI Program.
2025.01.20 View 5333
A Way for Smartwatches to Detect Depression Risks Devised by KAIST and U of Michigan Researchers
- A international joint research team of KAIST and the University of Michigan developed a digital biomarker for predicting symptoms of depression based on data collected by smartwatches
- It has the potential to be used as a medical technology to replace the economically burdensome fMRI measurement test
- It is expected to expand the scope of digital health data analysis
The CORONA virus pandemic also brought about a pandemic of mental illness. Approximately one billion people worldwide suffer from various psychiatric conditions. Korea is one of more serious cases, with approximately 1.8 million patients exhibiting depression and anxiety disorders, and the total number of patients with clinical mental diseases has increased by 37% in five years to approximately 4.65 million. A joint research team from Korea and the US has developed a technology that uses biometric data collected through wearable devices to predict tomorrow's mood and, further, to predict the possibility of developing symptoms of depression.
< Figure 1. Schematic diagram of the research results. Based on the biometric data collected by a smartwatch, a mathematical algorithm that solves the inverse problem to estimate the brain's circadian phase and sleep stages has been developed. This algorithm can estimate the degrees of circadian disruption, and these estimates can be used as the digital biomarkers to predict depression risks. >
KAIST (President Kwang Hyung Lee) announced on the 15th of January that the research team under Professor Dae Wook Kim from the Department of Brain and Cognitive Sciences and the team under Professor Daniel B. Forger from the Department of Mathematics at the University of Michigan in the United States have developed a technology to predict symptoms of depression such as sleep disorders, depression, loss of appetite, overeating, and decreased concentration in shift workers from the activity and heart rate data collected from smartwatches.
According to WHO, a promising new treatment direction for mental illness focuses on the sleep and circadian timekeeping system located in the hypothalamus of the brain, which directly affect impulsivity, emotional responses, decision-making, and overall mood.
However, in order to measure endogenous circadian rhythms and sleep states, blood or saliva must be drawn every 30 minutes throughout the night to measure changes in the concentration of the melatonin hormone in our bodies and polysomnography (PSG) must be performed. As such treatments requires hospitalization and most psychiatric patients only visit for outpatient treatment, there has been no significant progress in developing treatment methods that take these two factors into account. In addition, the cost of the PSG test, which is approximately $1000, leaves mental health treatment considering sleep and circadian rhythms out of reach for the socially disadvantaged.
The solution to overcome these problems is to employ wearable devices for the easier collection of biometric data such as heart rate, body temperature, and activity level in real time without spatial constraints. However, current wearable devices have the limitation of providing only indirect information on biomarkers required by medical staff, such as the phase of the circadian clock.
The joint research team developed a filtering technology that accurately estimates the phase of the circadian clock, which changes daily, such as heart rate and activity time series data collected from a smartwatch. This is an implementation of a digital twin that precisely describes the circadian rhythm in the brain, and it can be used to estimate circadian rhythm disruption.
< Figure 2. The suprachiasmatic nucleus located in the hypothalamus of the brain is the central biological clock that regulates the 24-hour physiological rhythm and plays a key role in maintaining the body’s circadian rhythm. If the phase of this biological clock is disrupted, it affects various parts of the brain, which can cause psychiatric conditions such as depression. >
The possibility of using the digital twin of this circadian clock to predict the symptoms of depression was verified through collaboration with the research team of Professor Srijan Sen of the Michigan Neuroscience Institute and Professor Amy Bohnert of the Department of Psychiatry of the University of Michigan.
The collaborative research team conducted a large-scale prospective cohort study involving approximately 800 shift workers and showed that the circadian rhythm disruption digital biomarker estimated through the technology can predict tomorrow's mood as well as six symptoms, including sleep problems, appetite changes, decreased concentration, and suicidal thoughts, which are representative symptoms of depression.
< Figure 3. The circadian rhythm of hormones such as melatonin regulates various physiological functions and behaviors such as heart rate and activity level. These physiological and behavioral signals can be measured in daily life through wearable devices. In order to estimate the body’s circadian rhythm inversely based on the measured biometric signals, a mathematical algorithm is needed. This algorithm plays a key role in accurately identifying the characteristics of circadian rhythms by extracting hidden physiological patterns from biosignals. >
Professor Dae Wook Kim said, "It is very meaningful to be able to conduct research that provides a clue for ways to apply wearable biometric data using mathematics that have not previously been utilized for actual disease management." He added, "We expect that this research will be able to present continuous and non-invasive mental health monitoring technology. This is expected to present a new paradigm for mental health care. By resolving some of the major problems socially disadvantaged people may face in current treatment practices, they may be able to take more active steps when experiencing symptoms of depression, such as seeking counsel before things get out of hand."
< Figure 4. A mathematical algorithm was devised to circumvent the problems of estimating the phase of the brain's biological clock and sleep stages inversely from the biodata collected by a smartwatch. This algorithm can estimate the degree of daily circadian rhythm disruption, and this estimate can be used as a digital biomarker to predict depression symptoms. >
The results of this study, in which Professor Dae Wook Kim of the Department of Brain and Cognitive Sciences at KAIST participated as the joint first author and corresponding author, were published in the online version of the international academic journal npj Digital Medicine on December 5, 2024. (Paper title: The real-world association between digital markers of circadian disruption and mental health risks) DOI: 10.1038/s41746-024-01348-6
This study was conducted with the support of the KAIST's Research Support Program for New Faculty Members, the US National Science Foundation, the US National Institutes of Health, and the US Army Research Institute MURI Program.
2025.01.20 View 5333 -
 Biomarker Predicts Who Will Have Severe COVID-19
- Airway cell analyses showing an activated immune axis could pinpoint the COVID-19 patients who will most benefit from targeted therapies.-
KAIST researchers have identified key markers that could help pinpoint patients who are bound to get a severe reaction to COVID-19 infection. This would help doctors provide the right treatments at the right time, potentially saving lives. The findings were published in the journal Frontiers in Immunology on August 28.
People’s immune systems react differently to infection with SARS-CoV-2, the virus that causes COVID-19, ranging from mild to severe, life-threatening responses.
To understand the differences in responses, Professor Heung Kyu Lee and PhD candidate Jang Hyun Park from the Graduate School of Medical Science and Engineering at KAIST analysed ribonucleic acid (RNA) sequencing data extracted from individual airway cells of healthy controls and of mildly and severely ill patients with COVID-19. The data was available in a public database previously published by a group of Chinese researchers.
“Our analyses identified an association between immune cells called neutrophils and special cell receptors that bind to the steroid hormone glucocorticoid,” Professor Lee explained. “This finding could be used as a biomarker for predicting disease severity in patients and thus selecting a targeted therapy that can help treat them at an appropriate time,” he added.
Severe illness in COVID-19 is associated with an exaggerated immune response that leads to excessive airway-damaging inflammation. This condition, known as acute respiratory distress syndrome (ARDS), accounts for 70% of deaths in fatal COVID-19 infections.
Scientists already know that this excessive inflammation involves heightened neutrophil recruitment to the airways, but the detailed mechanisms of this reaction are still unclear.
Lee and Park’s analyses found that a group of immune cells called myeloid cells produced excess amounts of neutrophil-recruiting chemicals in severely ill patients, including a cytokine called tumour necrosis factor (TNF) and a chemokine called CXCL8.
Further RNA analyses of neutrophils in severely ill patients showed they were less able to recruit very important T cells needed for attacking the virus. At the same time, the neutrophils produced too many extracellular molecules that normally trap pathogens, but damage airway cells when produced in excess.
The researchers additionally found that the airway cells in severely ill patients were not expressing enough glucocorticoid receptors. This was correlated with increased CXCL8 expression and neutrophil recruitment.
Glucocorticoids, like the well-known drug dexamethasone, are anti-inflammatory agents that could play a role in treating COVID-19. However, using them in early or mild forms of the infection could suppress the necessary immune reactions to combat the virus. But if airway damage has already happened in more severe cases, glucocorticoid treatment would be ineffective.
Knowing who to give this treatment to and when is really important. COVID-19 patients showing reduced glucocorticoid receptor expression, increased CXCL8 expression, and excess neutrophil recruitment to the airways could benefit from treatment with glucocorticoids to prevent airway damage. Further research is needed, however, to confirm the relationship between glucocorticoids and neutrophil inflammation at the protein level.
“Our study could serve as a springboard towards more accurate and reliable COVID-19 treatments,” Professor Lee said.
This work was supported by the National Research Foundation of Korea, and Mobile Clinic Module Project funded by KAIST.
Figure. Low glucocorticoid receptor (GR) expression led to excessive inflammation and lung damage by neutrophils through enhancing the expression of CXCL8 and other cytokines.
Image credit: Professor Heung Kyu Lee, KAIST. Created with Biorender.com.
Image usage restrictions: News organizations may use or redistribute these figures and image, with proper attribution, as part of news coverage of this paper only.
-Publication:
Jang Hyun Park, and Heung Kyu Lee. (2020). Re-analysis of Single Cell Transcriptome Reveals That the NR3C1-CXCL8-Neutrophil Axis Determines the Severity of COVID-19. Frontiers in Immunology, Available online at https://doi.org/10.3389/fimmu.2020.02145
-Profile: Heung Kyu Lee
Associate Professor
heungkyu.lee@kaist.ac.kr
https://www.heungkyulee.kaist.ac.kr/
Laboratory of Host Defenses
Graduate School of Medical Science and Engineering (GSMSE)
The Center for Epidemic Preparedness at KAIST Institute
http://kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
Profile: Jang Hyun Park
PhD Candidate
janghyun.park@kaist.ac.kr
GSMSE, KAIST
2020.09.17 View 16938
Biomarker Predicts Who Will Have Severe COVID-19
- Airway cell analyses showing an activated immune axis could pinpoint the COVID-19 patients who will most benefit from targeted therapies.-
KAIST researchers have identified key markers that could help pinpoint patients who are bound to get a severe reaction to COVID-19 infection. This would help doctors provide the right treatments at the right time, potentially saving lives. The findings were published in the journal Frontiers in Immunology on August 28.
People’s immune systems react differently to infection with SARS-CoV-2, the virus that causes COVID-19, ranging from mild to severe, life-threatening responses.
To understand the differences in responses, Professor Heung Kyu Lee and PhD candidate Jang Hyun Park from the Graduate School of Medical Science and Engineering at KAIST analysed ribonucleic acid (RNA) sequencing data extracted from individual airway cells of healthy controls and of mildly and severely ill patients with COVID-19. The data was available in a public database previously published by a group of Chinese researchers.
“Our analyses identified an association between immune cells called neutrophils and special cell receptors that bind to the steroid hormone glucocorticoid,” Professor Lee explained. “This finding could be used as a biomarker for predicting disease severity in patients and thus selecting a targeted therapy that can help treat them at an appropriate time,” he added.
Severe illness in COVID-19 is associated with an exaggerated immune response that leads to excessive airway-damaging inflammation. This condition, known as acute respiratory distress syndrome (ARDS), accounts for 70% of deaths in fatal COVID-19 infections.
Scientists already know that this excessive inflammation involves heightened neutrophil recruitment to the airways, but the detailed mechanisms of this reaction are still unclear.
Lee and Park’s analyses found that a group of immune cells called myeloid cells produced excess amounts of neutrophil-recruiting chemicals in severely ill patients, including a cytokine called tumour necrosis factor (TNF) and a chemokine called CXCL8.
Further RNA analyses of neutrophils in severely ill patients showed they were less able to recruit very important T cells needed for attacking the virus. At the same time, the neutrophils produced too many extracellular molecules that normally trap pathogens, but damage airway cells when produced in excess.
The researchers additionally found that the airway cells in severely ill patients were not expressing enough glucocorticoid receptors. This was correlated with increased CXCL8 expression and neutrophil recruitment.
Glucocorticoids, like the well-known drug dexamethasone, are anti-inflammatory agents that could play a role in treating COVID-19. However, using them in early or mild forms of the infection could suppress the necessary immune reactions to combat the virus. But if airway damage has already happened in more severe cases, glucocorticoid treatment would be ineffective.
Knowing who to give this treatment to and when is really important. COVID-19 patients showing reduced glucocorticoid receptor expression, increased CXCL8 expression, and excess neutrophil recruitment to the airways could benefit from treatment with glucocorticoids to prevent airway damage. Further research is needed, however, to confirm the relationship between glucocorticoids and neutrophil inflammation at the protein level.
“Our study could serve as a springboard towards more accurate and reliable COVID-19 treatments,” Professor Lee said.
This work was supported by the National Research Foundation of Korea, and Mobile Clinic Module Project funded by KAIST.
Figure. Low glucocorticoid receptor (GR) expression led to excessive inflammation and lung damage by neutrophils through enhancing the expression of CXCL8 and other cytokines.
Image credit: Professor Heung Kyu Lee, KAIST. Created with Biorender.com.
Image usage restrictions: News organizations may use or redistribute these figures and image, with proper attribution, as part of news coverage of this paper only.
-Publication:
Jang Hyun Park, and Heung Kyu Lee. (2020). Re-analysis of Single Cell Transcriptome Reveals That the NR3C1-CXCL8-Neutrophil Axis Determines the Severity of COVID-19. Frontiers in Immunology, Available online at https://doi.org/10.3389/fimmu.2020.02145
-Profile: Heung Kyu Lee
Associate Professor
heungkyu.lee@kaist.ac.kr
https://www.heungkyulee.kaist.ac.kr/
Laboratory of Host Defenses
Graduate School of Medical Science and Engineering (GSMSE)
The Center for Epidemic Preparedness at KAIST Institute
http://kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
Profile: Jang Hyun Park
PhD Candidate
janghyun.park@kaist.ac.kr
GSMSE, KAIST
2020.09.17 View 16938 -
 Blood-Based Multiplexed Diagnostic Sensor Helps to Accurately Detect Alzheimer’s Disease
A research team at KAIST reported clinically accurate multiplexed electrical biosensor for detecting Alzheimer’s disease by measuring its core biomarkers using densely aligned carbon nanotubes.
Alzheimer’s disease is the most prevalent neurodegenerative disorder, affecting one in ten aged over 65 years. Early diagnosis can reduce the risk of suffering the disease by one-third, according to recent reports. However, its early diagnosis remains challenging due to the low accuracy but high cost of diagnosis.
Research team led by Professors Chan Beum Park and Steve Park described an ultrasensitive detection of multiple Alzheimer's disease core biomarker in human plasma. The team have designed the sensor array by employing a densely aligned single-walled carbon nanotube thin films as a transducer.
The representative biomarkers of Alzheimer's disease are beta-amyloid42, beta-amyloid40, total tau protein, phosphorylated tau protein and the concentrations of these biomarkers in human plasma are directly correlated with the pathology of Alzheimer’s disease.
The research team developed a highly sensitive resistive biosensor based on densely aligned carbon nanotubes fabricated by Langmuir-Blodgett method with a low manufacturing cost.
Aligned carbon nanotubes with high density minimizes the tube-to-tube junction resistance compared with randomly distributed carbon nanotubes, which leads to the improvement of sensor sensitivity. To be more specific, this resistive sensor with densely aligned carbon nanotubes exhibits a sensitivity over 100 times higher than that of conventional carbon nanotube-based biosensors.
By measuring the concentrations of four Alzheimer’s disease biomarkers simultaneously Alzheimer patients can be discriminated from health controls with an average sensitivity of 90.0%, a selectivity of 90.0% and an average accuracy of 88.6%.
This work, titled “Clinically accurate diagnosis of Alzheimer’s disease via multiplexed sensing of core biomarkers in human plasma”, were published in Nature Communications on January 8th 2020. The authors include PhD candidate Kayoung Kim and MS candidate Min-Ji Kim.
Professor Steve Park said, “This study was conducted on patients who are already confirmed with Alzheimer’s Disease. For further use in practical setting, it is necessary to test the patients with mild cognitive impairment.” He also emphasized that, “It is essential to establish a nationwide infrastructure, such as mild cognitive impairment cohort study and a dementia cohort study. This would enable the establishment of world-wide research network, and will help various private and public institutions.”
This research was supported by the Ministry of Science and ICT, Human Resource Bank of Chungnam National University Hospital and Chungbuk National University Hospital.
< A schematic diagram of a high-density aligned carbon nanotube-based resistive sensor that distinguishes patients with Alzheimer’s Disease by measuring the concentration of four biomarkers in the blood. >
Profile:
Professor Steve Park
stevepark@kaist.ac.kr
Department of Materials Science and Engineering
http://steveparklab.kaist.ac.kr/
KAIST
Profile:
Professor Chan Beum Park
parkcb at kaist.ac.kr
Department of Materials Science and Engineering
http://biomaterials.kaist.ac.kr/
KAIST
2020.02.07 View 11439
Blood-Based Multiplexed Diagnostic Sensor Helps to Accurately Detect Alzheimer’s Disease
A research team at KAIST reported clinically accurate multiplexed electrical biosensor for detecting Alzheimer’s disease by measuring its core biomarkers using densely aligned carbon nanotubes.
Alzheimer’s disease is the most prevalent neurodegenerative disorder, affecting one in ten aged over 65 years. Early diagnosis can reduce the risk of suffering the disease by one-third, according to recent reports. However, its early diagnosis remains challenging due to the low accuracy but high cost of diagnosis.
Research team led by Professors Chan Beum Park and Steve Park described an ultrasensitive detection of multiple Alzheimer's disease core biomarker in human plasma. The team have designed the sensor array by employing a densely aligned single-walled carbon nanotube thin films as a transducer.
The representative biomarkers of Alzheimer's disease are beta-amyloid42, beta-amyloid40, total tau protein, phosphorylated tau protein and the concentrations of these biomarkers in human plasma are directly correlated with the pathology of Alzheimer’s disease.
The research team developed a highly sensitive resistive biosensor based on densely aligned carbon nanotubes fabricated by Langmuir-Blodgett method with a low manufacturing cost.
Aligned carbon nanotubes with high density minimizes the tube-to-tube junction resistance compared with randomly distributed carbon nanotubes, which leads to the improvement of sensor sensitivity. To be more specific, this resistive sensor with densely aligned carbon nanotubes exhibits a sensitivity over 100 times higher than that of conventional carbon nanotube-based biosensors.
By measuring the concentrations of four Alzheimer’s disease biomarkers simultaneously Alzheimer patients can be discriminated from health controls with an average sensitivity of 90.0%, a selectivity of 90.0% and an average accuracy of 88.6%.
This work, titled “Clinically accurate diagnosis of Alzheimer’s disease via multiplexed sensing of core biomarkers in human plasma”, were published in Nature Communications on January 8th 2020. The authors include PhD candidate Kayoung Kim and MS candidate Min-Ji Kim.
Professor Steve Park said, “This study was conducted on patients who are already confirmed with Alzheimer’s Disease. For further use in practical setting, it is necessary to test the patients with mild cognitive impairment.” He also emphasized that, “It is essential to establish a nationwide infrastructure, such as mild cognitive impairment cohort study and a dementia cohort study. This would enable the establishment of world-wide research network, and will help various private and public institutions.”
This research was supported by the Ministry of Science and ICT, Human Resource Bank of Chungnam National University Hospital and Chungbuk National University Hospital.
< A schematic diagram of a high-density aligned carbon nanotube-based resistive sensor that distinguishes patients with Alzheimer’s Disease by measuring the concentration of four biomarkers in the blood. >
Profile:
Professor Steve Park
stevepark@kaist.ac.kr
Department of Materials Science and Engineering
http://steveparklab.kaist.ac.kr/
KAIST
Profile:
Professor Chan Beum Park
parkcb at kaist.ac.kr
Department of Materials Science and Engineering
http://biomaterials.kaist.ac.kr/
KAIST
2020.02.07 View 11439 -
 Professor Il-Doo Kim Named Scientist of the Year by the Journalists
Professor Il-Doo Kim from the Department of Materials Science and Engineering was named the 2019 Scientist of the Year by Korean science journalists. The award was conferred at the 2019 Science Press Night ceremony of the Korea Science Journalists Association (KSJA) on November 29.
Professor Kim focuses on developing nanofiber gas sensors for diagnosing diseases in advance by analyzing exhaled biomarkers with electrospinning technology. His outstanding research was praised and selected as one of the top 10 nanotechnology of 2019 by the Korea Nano Technology Research Society (KoNTRS), the Ministry of Science and ICT (MSIT), and the Ministry of Trade, Industry and Energy (MOTIE).
Professor Kim was honored with the QIAN Baojun Fiber Award, which is awarded every two years by Donghua University in Shanghai, China to recognize outstanding contributions in fiber science and technology. Professor Kim was also elected as an academician of the Asia Pacific Academy of Materials (APAM) on November 21 in Guangzhou, China.
In May, Professor Kim was appointed as an associate editor of ACS Nano, a leading international research journal in the field of nanoscience. In his editorial published in the May issue of ACS Nano, Professor Kim introduced and shared the history of KAIST and its vision for the future with other members of the journal. He hopes this will help with promoting a closer relationship between the members of the journal and KAIST moving forward.
“Above all,” he said in his acceptance speech, “the greatest news for me as an educator is that the first PhD graduate from our lab, Dr. Seonjin Choi, was appointed as the youngest professor in the Division of Materials Science and Engineering at Hanyang University on September 1.”
2019.12.17 View 11950
Professor Il-Doo Kim Named Scientist of the Year by the Journalists
Professor Il-Doo Kim from the Department of Materials Science and Engineering was named the 2019 Scientist of the Year by Korean science journalists. The award was conferred at the 2019 Science Press Night ceremony of the Korea Science Journalists Association (KSJA) on November 29.
Professor Kim focuses on developing nanofiber gas sensors for diagnosing diseases in advance by analyzing exhaled biomarkers with electrospinning technology. His outstanding research was praised and selected as one of the top 10 nanotechnology of 2019 by the Korea Nano Technology Research Society (KoNTRS), the Ministry of Science and ICT (MSIT), and the Ministry of Trade, Industry and Energy (MOTIE).
Professor Kim was honored with the QIAN Baojun Fiber Award, which is awarded every two years by Donghua University in Shanghai, China to recognize outstanding contributions in fiber science and technology. Professor Kim was also elected as an academician of the Asia Pacific Academy of Materials (APAM) on November 21 in Guangzhou, China.
In May, Professor Kim was appointed as an associate editor of ACS Nano, a leading international research journal in the field of nanoscience. In his editorial published in the May issue of ACS Nano, Professor Kim introduced and shared the history of KAIST and its vision for the future with other members of the journal. He hopes this will help with promoting a closer relationship between the members of the journal and KAIST moving forward.
“Above all,” he said in his acceptance speech, “the greatest news for me as an educator is that the first PhD graduate from our lab, Dr. Seonjin Choi, was appointed as the youngest professor in the Division of Materials Science and Engineering at Hanyang University on September 1.”
2019.12.17 View 11950 -
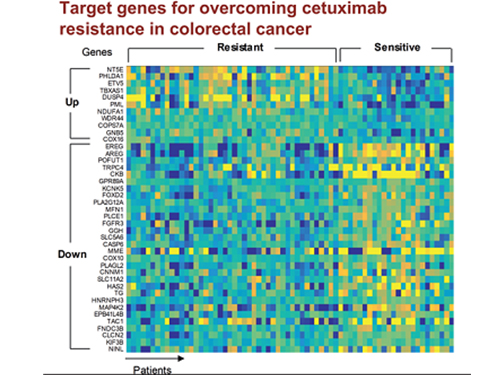 5 Biomarkers for Overcoming Colorectal Cancer Drug Resistance Identified
< Professor Kwang-Hyun Cho's Team >
KAIST researchers have identified five biomarkers that will help them address resistance to cancer-targeting therapeutics. This new treatment strategy will bring us one step closer to precision medicine for patients who showed resistance.
Colorectal cancer is one of the most common types of cancer worldwide. The number of patients has surpassed 1 million, and its five-year survival rate significantly drops to about 20 percent when metastasized. In Korea, the surge of colorectal cancer has been the highest in the last 10 years due to increasing Westernized dietary patterns and obesity. It is expected that the number and mortality rates of colorectal cancer patients will increase sharply as the nation is rapidly facing an increase in its aging population.
Recently, anticancer agents targeting only specific molecules of colon cancer cells have been developed. Unlike conventional anticancer medications, these selectively treat only specific target factors, so they can significantly reduce some of the side-effects of anticancer therapy while enhancing drug efficacy.
Cetuximab is the most well-known FDA approved anticancer medication. It is a biomarker that predicts drug reactivity and utilizes the presence of the ‘KRAS’ gene mutation. Cetuximab is prescribed to patients who don’t carry the KRAS gene mutation.
However, even in patients without the KRAS gene mutation, the response rate of Cetuximab is only about fifty percent, and there is also resistance to drugs after targeted chemotherapy. Compared with conventional chemotherapy alone, the life expectancy only lasts five months on average.
In research featured in the FEBS Journal as the cover paper for the April 7 edition, the KAIST research team led by Professor Kwang-Hyun Cho at the Department of Bio and Brain Engineering presented five additional biomarkers that could increase Cetuximab responsiveness using systems biology approach that combines genomic data analysis, mathematical modeling, and cell experiments. The experimental inhibition of newly discovered biomarkers DUSP4, ETV5, GNB5, NT5E, and PHLDA1 in colorectal cancer cells has been shown to overcome Cetuximab resistance in KRAS-normal genes. The research team confirmed that when suppressing GNB5, one of the new biomarkers, it was shown to overcome resistance to Cetuximab regardless of having a mutation in the KRAS gene.
Professor Cho said, “There has not been an example of colorectal cancer treatment involving regulation of the GNB5 gene.” He continued, “Identifying the principle of drug resistance in cancer cells through systems biology and discovering new biomarkers that could be a new molecular target to overcome drug resistance suggest real potential to actualize precision medicine.”
This study was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT (2017R1A2A1A17069642 and 2015M3A9A7067220).
Image 1. The cover of FEBS Journal for April 2019
2019.05.27 View 60047
5 Biomarkers for Overcoming Colorectal Cancer Drug Resistance Identified
< Professor Kwang-Hyun Cho's Team >
KAIST researchers have identified five biomarkers that will help them address resistance to cancer-targeting therapeutics. This new treatment strategy will bring us one step closer to precision medicine for patients who showed resistance.
Colorectal cancer is one of the most common types of cancer worldwide. The number of patients has surpassed 1 million, and its five-year survival rate significantly drops to about 20 percent when metastasized. In Korea, the surge of colorectal cancer has been the highest in the last 10 years due to increasing Westernized dietary patterns and obesity. It is expected that the number and mortality rates of colorectal cancer patients will increase sharply as the nation is rapidly facing an increase in its aging population.
Recently, anticancer agents targeting only specific molecules of colon cancer cells have been developed. Unlike conventional anticancer medications, these selectively treat only specific target factors, so they can significantly reduce some of the side-effects of anticancer therapy while enhancing drug efficacy.
Cetuximab is the most well-known FDA approved anticancer medication. It is a biomarker that predicts drug reactivity and utilizes the presence of the ‘KRAS’ gene mutation. Cetuximab is prescribed to patients who don’t carry the KRAS gene mutation.
However, even in patients without the KRAS gene mutation, the response rate of Cetuximab is only about fifty percent, and there is also resistance to drugs after targeted chemotherapy. Compared with conventional chemotherapy alone, the life expectancy only lasts five months on average.
In research featured in the FEBS Journal as the cover paper for the April 7 edition, the KAIST research team led by Professor Kwang-Hyun Cho at the Department of Bio and Brain Engineering presented five additional biomarkers that could increase Cetuximab responsiveness using systems biology approach that combines genomic data analysis, mathematical modeling, and cell experiments. The experimental inhibition of newly discovered biomarkers DUSP4, ETV5, GNB5, NT5E, and PHLDA1 in colorectal cancer cells has been shown to overcome Cetuximab resistance in KRAS-normal genes. The research team confirmed that when suppressing GNB5, one of the new biomarkers, it was shown to overcome resistance to Cetuximab regardless of having a mutation in the KRAS gene.
Professor Cho said, “There has not been an example of colorectal cancer treatment involving regulation of the GNB5 gene.” He continued, “Identifying the principle of drug resistance in cancer cells through systems biology and discovering new biomarkers that could be a new molecular target to overcome drug resistance suggest real potential to actualize precision medicine.”
This study was supported by the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT (2017R1A2A1A17069642 and 2015M3A9A7067220).
Image 1. The cover of FEBS Journal for April 2019
2019.05.27 View 60047 -
 Successful Development of Excavation System of Biomarkers containing Protein Decomposition Control Enzyme Information
A Korean team of researchers successfully developed a biomarker excavation system named E3Net that excavates biomarkers containing information of the enzymes that control the decomposition of proteins. The development of the system paved the possibility of development of new high quality biomarkers.
*Biomarker: Molecular information of unique patterns derived from genes and proteins that allow the monitoring of physical changes from genetic or environmental causes.
Professor Lee Kwan Soo’s team (Department of Biological Sciences) composed of Doctorate candidate Han Young Woong, Lee Ho Dong Ph.D. and Professor Park Jong Chul published a dissertation in the April edition of Molecular and Cellular Proteomics. (Dissertation Title: A system for exploring E3-mediated regulatory networks of cellular functions).
Professor Lee’s team compiled all available information of the enzyme that controls protein decomposition (E3 enzyme) and successfully compiled the inter-substrate network by extracting information from 20,000 biology related data base dissertations. The result was the development of the E3Net system that analyzes the related cell function and disease.
Cells have a system that produces, destroys, and recycles proteins in response to the ever changing environmental conditions. Error in these processes leads to disease.
Therefore finding the relationship between E3 enzymes that control the decomposition of proteins and the substrates will allow disease curing and prevention to become much easier. E3 enzyme is responsible for 80% of the protein decomposition and is therefore predicted to be related to various diseases.
However the information on E3 enzyme and inter-substrate behavior are spread out among numerous dissertations and data bases which prevented methodological analysis of the role of the related cells and characteristics of the disease itself.
Professor Lee’s team was successful in creating the E3Net that compiled 2,201 pieces of E3 substrate information, 4,896 pieces of substrate information, and 1,671 pieces of inter-substrate relationship information. This compilation allows for the systematic analysis of cells and diseases.
The newly created network is 10 times larger than the existing network and is the first case where it is possible to accurately find the cell function and related diseases.
It is anticipated that the use of the E3Net will allow the excavation of new biomarkers for the development of personalized drug systems.
The research team applied the E3Net to find tens of new candidate biomarkers related to the major modern diseases like diabetes and cancer.
2012.05.30 View 13774
Successful Development of Excavation System of Biomarkers containing Protein Decomposition Control Enzyme Information
A Korean team of researchers successfully developed a biomarker excavation system named E3Net that excavates biomarkers containing information of the enzymes that control the decomposition of proteins. The development of the system paved the possibility of development of new high quality biomarkers.
*Biomarker: Molecular information of unique patterns derived from genes and proteins that allow the monitoring of physical changes from genetic or environmental causes.
Professor Lee Kwan Soo’s team (Department of Biological Sciences) composed of Doctorate candidate Han Young Woong, Lee Ho Dong Ph.D. and Professor Park Jong Chul published a dissertation in the April edition of Molecular and Cellular Proteomics. (Dissertation Title: A system for exploring E3-mediated regulatory networks of cellular functions).
Professor Lee’s team compiled all available information of the enzyme that controls protein decomposition (E3 enzyme) and successfully compiled the inter-substrate network by extracting information from 20,000 biology related data base dissertations. The result was the development of the E3Net system that analyzes the related cell function and disease.
Cells have a system that produces, destroys, and recycles proteins in response to the ever changing environmental conditions. Error in these processes leads to disease.
Therefore finding the relationship between E3 enzymes that control the decomposition of proteins and the substrates will allow disease curing and prevention to become much easier. E3 enzyme is responsible for 80% of the protein decomposition and is therefore predicted to be related to various diseases.
However the information on E3 enzyme and inter-substrate behavior are spread out among numerous dissertations and data bases which prevented methodological analysis of the role of the related cells and characteristics of the disease itself.
Professor Lee’s team was successful in creating the E3Net that compiled 2,201 pieces of E3 substrate information, 4,896 pieces of substrate information, and 1,671 pieces of inter-substrate relationship information. This compilation allows for the systematic analysis of cells and diseases.
The newly created network is 10 times larger than the existing network and is the first case where it is possible to accurately find the cell function and related diseases.
It is anticipated that the use of the E3Net will allow the excavation of new biomarkers for the development of personalized drug systems.
The research team applied the E3Net to find tens of new candidate biomarkers related to the major modern diseases like diabetes and cancer.
2012.05.30 View 13774