Lab+of+Cancer+Genomics
-
 'Jumping Genes' Found to Alter Human Colon Genomes, Offering Insights into Aging and Tumorigenesis
The Korea Advanced Institute of Science and Technology (KAIST) and their collaborators have conducted a groundbreaking study targeting 'jumping genes' in the entire genomes of the human large intestine. Published in Nature on May 18 2023, the research unveils the surprising activity of 'Long interspersed nuclear element-1 (L1),' a type of jumping gene previously thought to be mostly dormant in human genomes. The study shows that L1 genes can become activated and disrupt genomic functions throughout an individual's lifetime, particularly in the colorectal epithelium.
(Paper Title: Widespread somatic L1 retrotransposition in normal colorectal epithelium, https://www.nature.com/articles/s41586-023-06046-z)
With approximately 500,000 L1 jumping genes, accounting for 17% of the human genome, they have long been recognized for their contribution to the evolution of the human species by introducing 'disruptive innovation' to genome sequences. Until now, it was believed that most L1 elements had lost their ability to jump in normal tissues of modern humans. However, this study reveals that some L1 jumping genes can be widely activated in normal cells, leading to the accumulation of genomic mutations over an individual's lifetime. The rate of L1 jumping and resulting genomic changes vary among different cell types, with a notable concentration observed in aged colon epithelial cells. The study illustrates that every colonic epithelial cell experiences an L1 jumping event by the age of 40 on average.
The research, led by co-first authors Chang Hyun Nam (a graduate student at KAIST) and Dr. Jeonghwan Youk (former graduate student at KAIST and assistant clinical professor at Seoul National University Hospital), involved the analysis of whole-genome sequences from 899 single cells obtained from skin (fibroblasts), blood, and colon epithelial tissues collected from 28 individuals. The study uncovers the activation of L1 jumping genes in normal cells, resulting in the gradual accumulation of genomic mutations over time. Additionally, the team explored epigenomic (DNA methylation) sequences to understand the mechanism behind L1 jumping gene activation. They found that cells with activated L1 jumping genes exhibit epigenetic instability, suggesting the critical role of epigenetic changes in regulating L1 jumping gene activity. Most of these epigenomic instabilities were found to arise during the early stages of embryogenesis. The study provides valuable insights into the aging process and the development of diseases in human colorectal tissues.
"This study illustrates that genomic damage in normal cells is acquired not only through exposure to carcinogens but also through the activity of endogenous components whose impact was previously unclear. Genomes of apparently healthy aged cells, particularly in the colorectal epithelium, become mosaic due to the activity of L1 jumping genes," said Prof. Young Seok Ju at KAIST.
"We emphasize the essential and ongoing collaboration among researchers in clinical medicine and basic medical sciences," said Prof. Min Jung Kim of the Department of Surgery at Seoul National University Hospital. "This case highlights the critical role of systematically collected human tissues from clinical settings in unraveling the complex process of disease development in humans."
"I am delighted that the research team's advancements in single-cell genome technology have come to fruition. We will persistently strive to lead in single-cell genome technology," said Prof. Hyun Woo Kwon of the Department of Nuclear Medicine at Korea University School of Medicine.
The research team received support from the Research Leader Program and the Young Researcher Program of the National Research Foundation of Korea, a grant from the MD-PhD/Medical Scientist Training Program through the Korea Health Industry Development Institute, and the Suh Kyungbae Foundation.
< Figure 1. Experimental design of the study >
< Figure 2. Schematic diagram illustrating factors influencing the soL1R landscape. >
Genetic composition of rc-L1s is inherited from the parents. The methylation landscape of rc-L1 promoters is predominantly determined by global DNA demethylation, followed by remethylation processes in the developmental stages. Then, when an rc-L1 is promoter demethylated in a specific cell lineage, the source expresses L1 transcripts thus making possible the induction of soL1Rs.
2023.05.22 View 8250
'Jumping Genes' Found to Alter Human Colon Genomes, Offering Insights into Aging and Tumorigenesis
The Korea Advanced Institute of Science and Technology (KAIST) and their collaborators have conducted a groundbreaking study targeting 'jumping genes' in the entire genomes of the human large intestine. Published in Nature on May 18 2023, the research unveils the surprising activity of 'Long interspersed nuclear element-1 (L1),' a type of jumping gene previously thought to be mostly dormant in human genomes. The study shows that L1 genes can become activated and disrupt genomic functions throughout an individual's lifetime, particularly in the colorectal epithelium.
(Paper Title: Widespread somatic L1 retrotransposition in normal colorectal epithelium, https://www.nature.com/articles/s41586-023-06046-z)
With approximately 500,000 L1 jumping genes, accounting for 17% of the human genome, they have long been recognized for their contribution to the evolution of the human species by introducing 'disruptive innovation' to genome sequences. Until now, it was believed that most L1 elements had lost their ability to jump in normal tissues of modern humans. However, this study reveals that some L1 jumping genes can be widely activated in normal cells, leading to the accumulation of genomic mutations over an individual's lifetime. The rate of L1 jumping and resulting genomic changes vary among different cell types, with a notable concentration observed in aged colon epithelial cells. The study illustrates that every colonic epithelial cell experiences an L1 jumping event by the age of 40 on average.
The research, led by co-first authors Chang Hyun Nam (a graduate student at KAIST) and Dr. Jeonghwan Youk (former graduate student at KAIST and assistant clinical professor at Seoul National University Hospital), involved the analysis of whole-genome sequences from 899 single cells obtained from skin (fibroblasts), blood, and colon epithelial tissues collected from 28 individuals. The study uncovers the activation of L1 jumping genes in normal cells, resulting in the gradual accumulation of genomic mutations over time. Additionally, the team explored epigenomic (DNA methylation) sequences to understand the mechanism behind L1 jumping gene activation. They found that cells with activated L1 jumping genes exhibit epigenetic instability, suggesting the critical role of epigenetic changes in regulating L1 jumping gene activity. Most of these epigenomic instabilities were found to arise during the early stages of embryogenesis. The study provides valuable insights into the aging process and the development of diseases in human colorectal tissues.
"This study illustrates that genomic damage in normal cells is acquired not only through exposure to carcinogens but also through the activity of endogenous components whose impact was previously unclear. Genomes of apparently healthy aged cells, particularly in the colorectal epithelium, become mosaic due to the activity of L1 jumping genes," said Prof. Young Seok Ju at KAIST.
"We emphasize the essential and ongoing collaboration among researchers in clinical medicine and basic medical sciences," said Prof. Min Jung Kim of the Department of Surgery at Seoul National University Hospital. "This case highlights the critical role of systematically collected human tissues from clinical settings in unraveling the complex process of disease development in humans."
"I am delighted that the research team's advancements in single-cell genome technology have come to fruition. We will persistently strive to lead in single-cell genome technology," said Prof. Hyun Woo Kwon of the Department of Nuclear Medicine at Korea University School of Medicine.
The research team received support from the Research Leader Program and the Young Researcher Program of the National Research Foundation of Korea, a grant from the MD-PhD/Medical Scientist Training Program through the Korea Health Industry Development Institute, and the Suh Kyungbae Foundation.
< Figure 1. Experimental design of the study >
< Figure 2. Schematic diagram illustrating factors influencing the soL1R landscape. >
Genetic composition of rc-L1s is inherited from the parents. The methylation landscape of rc-L1 promoters is predominantly determined by global DNA demethylation, followed by remethylation processes in the developmental stages. Then, when an rc-L1 is promoter demethylated in a specific cell lineage, the source expresses L1 transcripts thus making possible the induction of soL1Rs.
2023.05.22 View 8250 -
 Genomic Data Reveals New Insights into Human Embryonic Development
KAIST researchers have used whole-genome sequencing to track the development from a single fertilized-egg to a human body
Genomic scientists at KAIST have revealed new insights into the process of human embryonic development using large-scale, whole-genome sequencing of cells and tissues from adult humans. The study, published in Nature on Aug.25, is the first to analyse somatic mutations in normal tissue across multiple organs within and between humans.
An adult human body comprises trillions of cells of more than 200 types. How a human develops from a single fertilized egg to a fully grown adult is a fundamental question in biomedical science. Due to the ethical challenges of performing studies on human embryos, however, the details of this process remain largely unknown.
To overcome these issues, the research team took a different approach. They analysed genetic mutations in cells taken from adult human post-mortem tissue. Specifically, they identified mutations that occur spontaneously in early developmental cell divisions. These mutations, also called genomic scars, act like unique genetic fingerprints that can be used to trace the embryonic development process.
The study, which looked at 334 single-cell colonies and 379 tissue samples from seven recently deceased human body donors, is the largest single-cell, whole-genome analysis carried out to date. The researchers examined the genomic scars of each individual in order to reconstruct their early embryonic cellular dynamics.
The result revealed several key characteristics of the human embryonic development process. Firstly, mutation rates are higher in the first cell division, but then decrease to approximately one mutation per cell during later cell division. Secondly, early cells contributed unequally to the development of the embryo in all informative donors, for example, at the two-cell stage, one of the cells always left more progeny cells than the other. The ratio of this was different from person to person, implying that the process varies between individuals and is not fully deterministic.
The researchers were also able to deduce the timing of when cells begin to differentiate into individual organ-specific cells. They found that within three days of fertilization, embryonic cells began to be distributed asymmetrically into tissues for the left and right sides of the body, followed by differentiation into three germ layers, and then differentiation into specific tissues and organs.
“It is an impressive scientific achievement that, within 20 years of the completion of human genome project, genomic technology has advanced to the extent that we are now able to accurately identify mutations in a single-cell genome,” said Professor Young Seok Ju from the Graduate School of Medical Science and Engineering at KAIST. “This technology will enable us to track human embryogenesis at even higher resolutions in the future.”
The techniques used in this study could be used to improve our understanding of rare diseases caused by abnormalities in embryonic development, and to design new precision diagnostics and treatments for patients.
The research was completed in collaboration with Kyungpook National University Hospital, the Korea Institute of Science and Technology Information, Catholic University of Korea School of Medicine, Genome Insights Inc, and Immune Square Inc. This work was supported by the Suh Kyungbae Foundation, the Ministry of Health and Welfare of Korea, the National Research Foundastion of Korea.
-PublicationSeongyeol Park, Nanda Mali, Ryul Kim et al. ‘Clonal dynamics in early human embryogenesis inferred from somatic mutation’ Nature Online ahead of print, Aug. 25, 2021 (https://doi.org/10.1038/s41586-021-03786-8)
-ProfileProfessor Young Seok JuLab of Cancer Genomics (https://www.julab.kaist.ac.kr/)Graduate School of Medical Science and EngineeringKAIST
2021.08.31 View 9445
Genomic Data Reveals New Insights into Human Embryonic Development
KAIST researchers have used whole-genome sequencing to track the development from a single fertilized-egg to a human body
Genomic scientists at KAIST have revealed new insights into the process of human embryonic development using large-scale, whole-genome sequencing of cells and tissues from adult humans. The study, published in Nature on Aug.25, is the first to analyse somatic mutations in normal tissue across multiple organs within and between humans.
An adult human body comprises trillions of cells of more than 200 types. How a human develops from a single fertilized egg to a fully grown adult is a fundamental question in biomedical science. Due to the ethical challenges of performing studies on human embryos, however, the details of this process remain largely unknown.
To overcome these issues, the research team took a different approach. They analysed genetic mutations in cells taken from adult human post-mortem tissue. Specifically, they identified mutations that occur spontaneously in early developmental cell divisions. These mutations, also called genomic scars, act like unique genetic fingerprints that can be used to trace the embryonic development process.
The study, which looked at 334 single-cell colonies and 379 tissue samples from seven recently deceased human body donors, is the largest single-cell, whole-genome analysis carried out to date. The researchers examined the genomic scars of each individual in order to reconstruct their early embryonic cellular dynamics.
The result revealed several key characteristics of the human embryonic development process. Firstly, mutation rates are higher in the first cell division, but then decrease to approximately one mutation per cell during later cell division. Secondly, early cells contributed unequally to the development of the embryo in all informative donors, for example, at the two-cell stage, one of the cells always left more progeny cells than the other. The ratio of this was different from person to person, implying that the process varies between individuals and is not fully deterministic.
The researchers were also able to deduce the timing of when cells begin to differentiate into individual organ-specific cells. They found that within three days of fertilization, embryonic cells began to be distributed asymmetrically into tissues for the left and right sides of the body, followed by differentiation into three germ layers, and then differentiation into specific tissues and organs.
“It is an impressive scientific achievement that, within 20 years of the completion of human genome project, genomic technology has advanced to the extent that we are now able to accurately identify mutations in a single-cell genome,” said Professor Young Seok Ju from the Graduate School of Medical Science and Engineering at KAIST. “This technology will enable us to track human embryogenesis at even higher resolutions in the future.”
The techniques used in this study could be used to improve our understanding of rare diseases caused by abnormalities in embryonic development, and to design new precision diagnostics and treatments for patients.
The research was completed in collaboration with Kyungpook National University Hospital, the Korea Institute of Science and Technology Information, Catholic University of Korea School of Medicine, Genome Insights Inc, and Immune Square Inc. This work was supported by the Suh Kyungbae Foundation, the Ministry of Health and Welfare of Korea, the National Research Foundastion of Korea.
-PublicationSeongyeol Park, Nanda Mali, Ryul Kim et al. ‘Clonal dynamics in early human embryogenesis inferred from somatic mutation’ Nature Online ahead of print, Aug. 25, 2021 (https://doi.org/10.1038/s41586-021-03786-8)
-ProfileProfessor Young Seok JuLab of Cancer Genomics (https://www.julab.kaist.ac.kr/)Graduate School of Medical Science and EngineeringKAIST
2021.08.31 View 9445 -
 Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke
(Professor Young Seok Ju at the Graduate School of Medical Science and Engineering)
Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancers develop.
Researchers at KAIST, Seoul National University and their collaborators confirmed that gene fusions in non-smokers mostly occur early on, sometimes as early as childhood or adolescence, and on average about three decades before cancer is diagnosed. The study showed that these mutant lung cells, harboring oncogenic seeds, remain dormant for several decades until a number of further mutations accumulate sufficiently for progression into cancer. This is the first study to reveal the landscape of genome structural variations in lung adenocarcinoma.
Lung cancer is the leading cause of cancer-related deaths worldwide, and lung adenocarcinoma is its most common type. Most lung adenocarcinomas are associated with chronic smoking, but about a fourth develop in non-smokers. Precisely what happens in non-smokers for this cancer to develop is not clearly understood.
Researchers analyzed the genomes of 138 lung adenocarcinoma patients, including smokers and non-smokers, with whole-genome sequencing technologies. They explored DNA damage that induced neoplastic transformation.
Lung adenocarcinomas that originated from chronic smoking, referred to as signature 4-high (S4-high) cancers in the study, showed several distinguishing features compared to smoking-unrelated cancers (S4-low).
People in the S4-high group were largely older, men and had more frequent mutations in a cancer-related gene called KRAS. Cancer genomes in the S4-high group were hypermutated with simple mutational classes, such as the substitution, insertion, or deletion of a single base, the building block of DNA.
But the story was very different in the S4-low group. Generally, mutational profiles in this group were much more silent than the S4-high group. However, all cancer-related gene fusions, which are abnormally activated from the merging of two originally separate genes, were exclusively observed in the S4-low group.
The patterns of genomic structural changes underlying gene fusions suggest that about three in four cases of gene fusions emerged from a single cellular crisis causing massive genomic fragmentation and subsequent imprecise repair in normal lung epithelium.
Most strikingly, these major genomic rearrangements, which led to the development of lung adenocarcinoma, are very likely to be acquired decades before cancer diagnosis. The researchers used genomic archaeology techniques to trace the timing of when the catastrophes took place.
Researchers started this study seven years ago when they discovered the expression of the KIF5B-RET gene fusion in lung adenocarcinoma for the first time. Professor Young-Seok Ju, co-lead author from the Graduate School of Medical Science and Engineering at KAIST says, “It is remarkable that oncogenesis can begin by a massive shattering of chromosomes early in life. Our study immediately raises a new question: What induces the mutational catastrophe in our normal lung epithelium.”
Professor Young Tae Kim, co-lead author from Seoul National University says, “We hope this work will help us get one step closer to precision medicine for lung cancer patients.”
The research team plans to further focus on the molecular mechanisms that stimulate complex rearrangements in the body, through screening the genomic structures of fusion genes in other cancer types.
This study was supported by the National Research Foundation of Korea (NRF), Korea Health Industry Development Institute (KHIDI), Suh Kyungbae Foundation, the College of Medicine Research Foundations at Seoul National University and others.
Figure.
(Smoking-unrelated oncogenesis of lung cancers by gene fusions)
Publication.
Jake June-Koo Lee, Seongyeol Park et al., Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma
Cell 177, June 13 2019, online publication ahead of print at May 30, 2019
https://doi.org/10.1016/j.cell.2019.05.013
Profile: Prof Young Seok Ju, MD, PhD
ysju@kaist.ac.kr
http://julab.kaist.ac.kr
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon 34141, Korea
Profile: Prof Young Tae Kim, MD, PhD
ytkim@snu.ac.kr
Professor
Seoul National University Cancer Research Institute
Department of Thoracic and Cardiovascular Surgery
Seoul National University Hospital Seoul 03080, Korea
2019.05.31 View 57444
Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke
(Professor Young Seok Ju at the Graduate School of Medical Science and Engineering)
Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancers develop.
Researchers at KAIST, Seoul National University and their collaborators confirmed that gene fusions in non-smokers mostly occur early on, sometimes as early as childhood or adolescence, and on average about three decades before cancer is diagnosed. The study showed that these mutant lung cells, harboring oncogenic seeds, remain dormant for several decades until a number of further mutations accumulate sufficiently for progression into cancer. This is the first study to reveal the landscape of genome structural variations in lung adenocarcinoma.
Lung cancer is the leading cause of cancer-related deaths worldwide, and lung adenocarcinoma is its most common type. Most lung adenocarcinomas are associated with chronic smoking, but about a fourth develop in non-smokers. Precisely what happens in non-smokers for this cancer to develop is not clearly understood.
Researchers analyzed the genomes of 138 lung adenocarcinoma patients, including smokers and non-smokers, with whole-genome sequencing technologies. They explored DNA damage that induced neoplastic transformation.
Lung adenocarcinomas that originated from chronic smoking, referred to as signature 4-high (S4-high) cancers in the study, showed several distinguishing features compared to smoking-unrelated cancers (S4-low).
People in the S4-high group were largely older, men and had more frequent mutations in a cancer-related gene called KRAS. Cancer genomes in the S4-high group were hypermutated with simple mutational classes, such as the substitution, insertion, or deletion of a single base, the building block of DNA.
But the story was very different in the S4-low group. Generally, mutational profiles in this group were much more silent than the S4-high group. However, all cancer-related gene fusions, which are abnormally activated from the merging of two originally separate genes, were exclusively observed in the S4-low group.
The patterns of genomic structural changes underlying gene fusions suggest that about three in four cases of gene fusions emerged from a single cellular crisis causing massive genomic fragmentation and subsequent imprecise repair in normal lung epithelium.
Most strikingly, these major genomic rearrangements, which led to the development of lung adenocarcinoma, are very likely to be acquired decades before cancer diagnosis. The researchers used genomic archaeology techniques to trace the timing of when the catastrophes took place.
Researchers started this study seven years ago when they discovered the expression of the KIF5B-RET gene fusion in lung adenocarcinoma for the first time. Professor Young-Seok Ju, co-lead author from the Graduate School of Medical Science and Engineering at KAIST says, “It is remarkable that oncogenesis can begin by a massive shattering of chromosomes early in life. Our study immediately raises a new question: What induces the mutational catastrophe in our normal lung epithelium.”
Professor Young Tae Kim, co-lead author from Seoul National University says, “We hope this work will help us get one step closer to precision medicine for lung cancer patients.”
The research team plans to further focus on the molecular mechanisms that stimulate complex rearrangements in the body, through screening the genomic structures of fusion genes in other cancer types.
This study was supported by the National Research Foundation of Korea (NRF), Korea Health Industry Development Institute (KHIDI), Suh Kyungbae Foundation, the College of Medicine Research Foundations at Seoul National University and others.
Figure.
(Smoking-unrelated oncogenesis of lung cancers by gene fusions)
Publication.
Jake June-Koo Lee, Seongyeol Park et al., Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma
Cell 177, June 13 2019, online publication ahead of print at May 30, 2019
https://doi.org/10.1016/j.cell.2019.05.013
Profile: Prof Young Seok Ju, MD, PhD
ysju@kaist.ac.kr
http://julab.kaist.ac.kr
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon 34141, Korea
Profile: Prof Young Tae Kim, MD, PhD
ytkim@snu.ac.kr
Professor
Seoul National University Cancer Research Institute
Department of Thoracic and Cardiovascular Surgery
Seoul National University Hospital Seoul 03080, Korea
2019.05.31 View 57444 -
 Professor Ju, to Receive Grants from HFSP
(Professor Young Seok Ju)
Professor Young Seok Ju from the Graduate School of Medical Science and Engineering was selected as a young investigator to receive research funds from the Human Frontiers Science Program.
The Human Frontiers Science Program (HFSP) was founded in 1989 with members of the G7 and European Union to stimulate innovative research in the field of life sciences.
Professor Ju placed third out of the eight teams that were selected from 158 applicants representing 60 countries. He is now the fourth Korean to receive a research grant as a young investigator. Professor Jae Kyoung Kim from the Department of Mathematical Sciences also received this prize last year, hence KAIST has produced grant recipients for two consecutive years.
Professor Ju is a medical doctor specializing in cancer genomics and computer biology. He has been studying somatic mutations and their functional consequences in human cancer in a bioinformatics way. He has published papers in international journals including Nature, Science, Genome Research, and Journal of Clinical Oncology.
With a title ‘Tracing AID/APOBEC- and MSI-mediated hyper-mutagenesis in the clonal evolution of gastric cancer,’ Professor Ju will receive 1.05 million dollars for three years along with Professor Bon-Kyoung Koo from the Institute of Molecular Biotechnology at Austrian Academy of Sciences, and Sinppert Hugo from University Medical Center Utrecht.
Professor Ju said, “As a young investigator, it is my great honor to receive this research fund from this organization. Through this internationally collaborative research, I will carry out groundbreaking research to understand the pathophysiology of cancers at a molecular level.”
2018.04.24 View 9229
Professor Ju, to Receive Grants from HFSP
(Professor Young Seok Ju)
Professor Young Seok Ju from the Graduate School of Medical Science and Engineering was selected as a young investigator to receive research funds from the Human Frontiers Science Program.
The Human Frontiers Science Program (HFSP) was founded in 1989 with members of the G7 and European Union to stimulate innovative research in the field of life sciences.
Professor Ju placed third out of the eight teams that were selected from 158 applicants representing 60 countries. He is now the fourth Korean to receive a research grant as a young investigator. Professor Jae Kyoung Kim from the Department of Mathematical Sciences also received this prize last year, hence KAIST has produced grant recipients for two consecutive years.
Professor Ju is a medical doctor specializing in cancer genomics and computer biology. He has been studying somatic mutations and their functional consequences in human cancer in a bioinformatics way. He has published papers in international journals including Nature, Science, Genome Research, and Journal of Clinical Oncology.
With a title ‘Tracing AID/APOBEC- and MSI-mediated hyper-mutagenesis in the clonal evolution of gastric cancer,’ Professor Ju will receive 1.05 million dollars for three years along with Professor Bon-Kyoung Koo from the Institute of Molecular Biotechnology at Austrian Academy of Sciences, and Sinppert Hugo from University Medical Center Utrecht.
Professor Ju said, “As a young investigator, it is my great honor to receive this research fund from this organization. Through this internationally collaborative research, I will carry out groundbreaking research to understand the pathophysiology of cancers at a molecular level.”
2018.04.24 View 9229 -
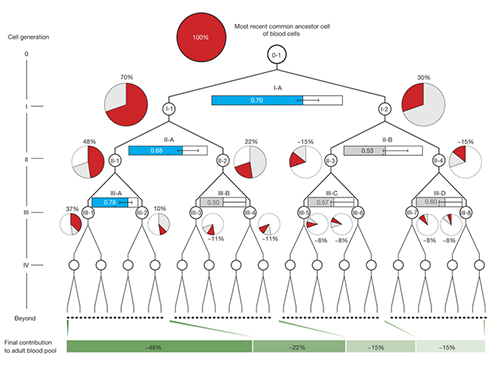 First Mutations in Human Life Discovered
The earliest mutations of human life have been observed by research team led by the Wellcome Trust Sanger Institute and their collaborators. Analyzing genomes from adult cells, the scientists could look back in time to reveal how each embryo developed.
Research team of the Sanger Institute including Professor Young Seok Ju of the Graduate School of Medical Science and Engineering at KAIST published an article of “Somatic Mutations Reveal Asymmetric Cellular Dynamics in the Early Human Embryo” in Nature on March 22.
The study shows that from the two-cell stage of the human embryo, one of these cells becomes more dominant than the other and leads to a higher proportion of the adult body.
A longstanding question for researchers has been what happens in the very early human development as this has proved impossible to study directly. Now, researchers have analyzed the whole genome sequences of blood samples (collected from 279 individuals with breast cancer) and discovered 163 mutations that occurred very early in the embryonic development of those people.
Once identified, the researchers used mutations from the first, second and third divisions of the fertilized egg to calculate which proportion of adult cells resulted from each of the first two cells in the embryo. They found that these first two cells contribute differently to the whole body. One cell gives rise to about 70 percent of the adult body tissues, whereas the other cell has a more minor contribution, leading to about 30percent of the tissues. This skewed contribution continues for some cells in the second and third generation too.
Originally pinpointed in normal blood cells from cancer patients, the researchers then looked for these mutations in cancer samples that had been surgically removed from the patients during treatment. Unlike normal tissues composed of multiple somatic cell clones, a cancer develops from one mutant cell. Therefore, each proposed embryonic mutation should either be present in all of the cancer cells in a tumor, or none of them. This proved to be the case, and by using these cancer samples, the researchers were able to validate that the mutations had originated during early development.
Dr. Young Seok Ju, first author from the Wellcome Trust Sanger Institute and KAIST, said: "This is the first time that anyone has seen where mutations arise in the very early human development. It is like finding a needle in a haystack. There are just a handful of these mutations, compared with millions of inherited genetic variations, and finding them allowed us to track what happened during embryogenesis."
Dr. Inigo Martincorena, from the Sanger Institute, said: "Having identified the mutations, we were able to use statistical analysis to better understand cell dynamics during embryo development. We determined the relative contribution of the first embryonic cells to the adult blood cell pool and found one dominant cell - that led to 70 percent of the blood cells - and one minor cell. We also sequenced normal lymph and breast cells, and the results suggested that the dominant cell also contributes to these other tissues at a similar level. This opens an unprecedented window into the earliest stages of human development."
During this study, the researchers were also able to measure the rate of mutation in early human development for the first time, up to three generations of cell division. Previous researchers had estimated one mutation per cell division, but this study measured three mutations for each cell doubling, in every daughter cell.
Mutations during the development of the embryo occur by two processes - known as mutational signatures 1 and 5. These mutations are fairly randomly distributed through the genome, and the vast majority of them will not affect the developing embryo. However, a mutation that occurs in an important gene can lead to disease such as developmental disorders.
Professor Sir Mike Stratton, lead author on the paper and Director of the Sanger Institute, said: "This is a significant step forward in widening the range of biological insights that can be extracted using genome sequences and mutations. Essentially, the mutations are archaeological traces of embryonic development left in our adult tissues, so if we can find and interpret them, we can understand human embryology better. This is just one early insight into human development, with hopefully many more to come in the future."
(Figure 1. Detection of somatic mutations acquired in early human embryogenesis )
(Figure 2. Unequal contributions of early embryonic cells to adult somatic tissues )
2017.03.23 View 8111
First Mutations in Human Life Discovered
The earliest mutations of human life have been observed by research team led by the Wellcome Trust Sanger Institute and their collaborators. Analyzing genomes from adult cells, the scientists could look back in time to reveal how each embryo developed.
Research team of the Sanger Institute including Professor Young Seok Ju of the Graduate School of Medical Science and Engineering at KAIST published an article of “Somatic Mutations Reveal Asymmetric Cellular Dynamics in the Early Human Embryo” in Nature on March 22.
The study shows that from the two-cell stage of the human embryo, one of these cells becomes more dominant than the other and leads to a higher proportion of the adult body.
A longstanding question for researchers has been what happens in the very early human development as this has proved impossible to study directly. Now, researchers have analyzed the whole genome sequences of blood samples (collected from 279 individuals with breast cancer) and discovered 163 mutations that occurred very early in the embryonic development of those people.
Once identified, the researchers used mutations from the first, second and third divisions of the fertilized egg to calculate which proportion of adult cells resulted from each of the first two cells in the embryo. They found that these first two cells contribute differently to the whole body. One cell gives rise to about 70 percent of the adult body tissues, whereas the other cell has a more minor contribution, leading to about 30percent of the tissues. This skewed contribution continues for some cells in the second and third generation too.
Originally pinpointed in normal blood cells from cancer patients, the researchers then looked for these mutations in cancer samples that had been surgically removed from the patients during treatment. Unlike normal tissues composed of multiple somatic cell clones, a cancer develops from one mutant cell. Therefore, each proposed embryonic mutation should either be present in all of the cancer cells in a tumor, or none of them. This proved to be the case, and by using these cancer samples, the researchers were able to validate that the mutations had originated during early development.
Dr. Young Seok Ju, first author from the Wellcome Trust Sanger Institute and KAIST, said: "This is the first time that anyone has seen where mutations arise in the very early human development. It is like finding a needle in a haystack. There are just a handful of these mutations, compared with millions of inherited genetic variations, and finding them allowed us to track what happened during embryogenesis."
Dr. Inigo Martincorena, from the Sanger Institute, said: "Having identified the mutations, we were able to use statistical analysis to better understand cell dynamics during embryo development. We determined the relative contribution of the first embryonic cells to the adult blood cell pool and found one dominant cell - that led to 70 percent of the blood cells - and one minor cell. We also sequenced normal lymph and breast cells, and the results suggested that the dominant cell also contributes to these other tissues at a similar level. This opens an unprecedented window into the earliest stages of human development."
During this study, the researchers were also able to measure the rate of mutation in early human development for the first time, up to three generations of cell division. Previous researchers had estimated one mutation per cell division, but this study measured three mutations for each cell doubling, in every daughter cell.
Mutations during the development of the embryo occur by two processes - known as mutational signatures 1 and 5. These mutations are fairly randomly distributed through the genome, and the vast majority of them will not affect the developing embryo. However, a mutation that occurs in an important gene can lead to disease such as developmental disorders.
Professor Sir Mike Stratton, lead author on the paper and Director of the Sanger Institute, said: "This is a significant step forward in widening the range of biological insights that can be extracted using genome sequences and mutations. Essentially, the mutations are archaeological traces of embryonic development left in our adult tissues, so if we can find and interpret them, we can understand human embryology better. This is just one early insight into human development, with hopefully many more to come in the future."
(Figure 1. Detection of somatic mutations acquired in early human embryogenesis )
(Figure 2. Unequal contributions of early embryonic cells to adult somatic tissues )
2017.03.23 View 8111