protein+folding
-
 X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.07.09 View 15832
X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.07.09 View 15832 -
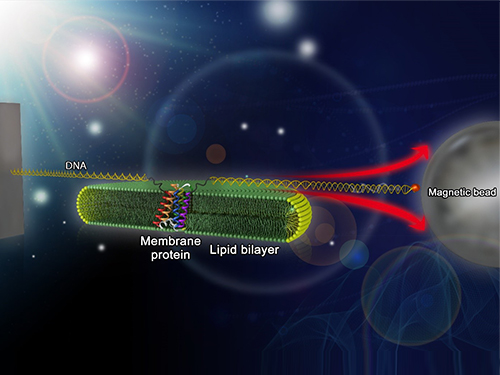 Mapping the Folding Process of a Single Membrane Protein
KAIST and UCLA scientists were able to observe an individual membrane protein fold and unfold by pulling and releasing magnetically trapped protein molecules.
Proteins are huge molecules containing hundreds to thousands of atoms that adopt a unique three dimensional structure, placing chemical groups in just the right place to catalyze reactions or build cellular structures. How all those atoms manage to find the right location - the so-called folding problem - has fascinated molecular biologists since the first structures were seen in the 1950s. Moreover, folding has important medical implications because most genetic defects cause protein misfolding.
About a third of all proteins float around in the cell membrane where they ensure the right chemicals get in the cell in the right amounts. Membrane proteins also provide key information links between the cell and its environment. Indeed, most drugs target membrane proteins. Nevertheless, the folding of membrane proteins has been particularly difficult to study and has rarely been studied in natural environments, leaving the folding process for a large fraction of the protein universe still largely cloaked in mystery.
In a recent issue of Nature Chemical Biology, published on October 19, 2015, a research team led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and James U. Bowie of the Department of Chemistry and Biochemistry at the University of California, Los Angeles (UCLA), report a new method for manipulating the folding of membrane proteins in a membrane environment using a tool called a magnetic tweezer.
Researchers first attach long DNA handles to the ends of the protein. One handle is attached to a glass surface and the other to a magnetic bead. Using a magnet, they can essentially grab the protein and pull on it, inducing it to unfold. By playing with the bead attached to the protein, they can force the protein to unfold or allow it to refold, and watch all this happening by 3D-tracking of the magnetic bead. With this novel strategy, they were able to quantitatively map the folding energy landscape, the folding kinetic rate, and folding intermediates of a membrane protein in a membrane environment for the first time.
“I have been dreaming about this experiment for a decade. To see it work so well is really gratifying,” said Dr. Bowie.
One of the major surprises in the study was that essentially all the atoms of the protein jump into the correct structure together. The researchers expected that the protein structure would come together in a more piecemeal fashion, with different parts of the structure forming separately, but that was not the case. It is possible that nature evolved such a smooth, highly cooperative folding process to prevent partially folded forms that could get into trouble in the crowded cell membrane. On the other hand, the cooperative folding seen here might not apply to other membrane proteins.
“We need to look at more proteins. The technique developed here may allow us to do just that,” said Dr. Yoon.
The single molecule mechanical manipulation technique could enable detailed folding studies of many other membrane proteins. A major barrier to the study of membrane proteins previously is that the proteins tend to stick together and get tangled up, as computer cords lying at your feet tend to do. With the tweezer technique used in this work, the protein cords are held apart from other cords so they can’t get knotted up. It is hoped that the new approach will open up an important part of the protein universe to scrutiny, including many proteins that become misfolded in disease states.
The title of the research paper is “Mapping the energy landscape for second-stage folding of a single membrane protein” (DOI: 10.1038/nchembio.1939).
Picture: Single-molecule magnetic tweezers that induce mechanical unfolding and refolding of a single membrane protein. Since the force applied is parallel to the biological lipid membrane, the unfolding and refolding processes occur within the membrane.
2015.10.20 View 10272
Mapping the Folding Process of a Single Membrane Protein
KAIST and UCLA scientists were able to observe an individual membrane protein fold and unfold by pulling and releasing magnetically trapped protein molecules.
Proteins are huge molecules containing hundreds to thousands of atoms that adopt a unique three dimensional structure, placing chemical groups in just the right place to catalyze reactions or build cellular structures. How all those atoms manage to find the right location - the so-called folding problem - has fascinated molecular biologists since the first structures were seen in the 1950s. Moreover, folding has important medical implications because most genetic defects cause protein misfolding.
About a third of all proteins float around in the cell membrane where they ensure the right chemicals get in the cell in the right amounts. Membrane proteins also provide key information links between the cell and its environment. Indeed, most drugs target membrane proteins. Nevertheless, the folding of membrane proteins has been particularly difficult to study and has rarely been studied in natural environments, leaving the folding process for a large fraction of the protein universe still largely cloaked in mystery.
In a recent issue of Nature Chemical Biology, published on October 19, 2015, a research team led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and James U. Bowie of the Department of Chemistry and Biochemistry at the University of California, Los Angeles (UCLA), report a new method for manipulating the folding of membrane proteins in a membrane environment using a tool called a magnetic tweezer.
Researchers first attach long DNA handles to the ends of the protein. One handle is attached to a glass surface and the other to a magnetic bead. Using a magnet, they can essentially grab the protein and pull on it, inducing it to unfold. By playing with the bead attached to the protein, they can force the protein to unfold or allow it to refold, and watch all this happening by 3D-tracking of the magnetic bead. With this novel strategy, they were able to quantitatively map the folding energy landscape, the folding kinetic rate, and folding intermediates of a membrane protein in a membrane environment for the first time.
“I have been dreaming about this experiment for a decade. To see it work so well is really gratifying,” said Dr. Bowie.
One of the major surprises in the study was that essentially all the atoms of the protein jump into the correct structure together. The researchers expected that the protein structure would come together in a more piecemeal fashion, with different parts of the structure forming separately, but that was not the case. It is possible that nature evolved such a smooth, highly cooperative folding process to prevent partially folded forms that could get into trouble in the crowded cell membrane. On the other hand, the cooperative folding seen here might not apply to other membrane proteins.
“We need to look at more proteins. The technique developed here may allow us to do just that,” said Dr. Yoon.
The single molecule mechanical manipulation technique could enable detailed folding studies of many other membrane proteins. A major barrier to the study of membrane proteins previously is that the proteins tend to stick together and get tangled up, as computer cords lying at your feet tend to do. With the tweezer technique used in this work, the protein cords are held apart from other cords so they can’t get knotted up. It is hoped that the new approach will open up an important part of the protein universe to scrutiny, including many proteins that become misfolded in disease states.
The title of the research paper is “Mapping the energy landscape for second-stage folding of a single membrane protein” (DOI: 10.1038/nchembio.1939).
Picture: Single-molecule magnetic tweezers that induce mechanical unfolding and refolding of a single membrane protein. Since the force applied is parallel to the biological lipid membrane, the unfolding and refolding processes occur within the membrane.
2015.10.20 View 10272