system
-
 Why Do Plants Attack Themselves? The Secret of Genetic Conflict Revealed
<Professor Ji-Joon Song of the KAIST Department of Biological Sciences>
Plants, with their unique immune systems, sometimes launch 'autoimmune responses' by mistakenly identifying their own protein structures as pathogens. In particular, 'hybrid necrosis,' a phenomenon where descendant plants fail to grow healthily and perish after cross-breeding different varieties, has long been a difficult challenge for botanists and agricultural researchers. In response, an international research team has successfully elucidated the mechanism inducing plant autoimmune responses and proposed a novel strategy for cultivar improvement that can predict and avoid these reactions.
Professor Ji-Joon Song's research team at KAIST, in collaboration with teams from the National University of Singapore (NUS) and the University of Oxford, announced on the 21st of July that they have elucidated the structure and function of the 'DM3' protein complex, which triggers plant autoimmune responses, using cryo-electron microscopy (Cryo-EM) technology.
This research is drawing attention because it identifies defects in protein structure as the cause of hybrid necrosis, which occurs due to an abnormal reaction of immune receptors during cross-breeding between plant hybrids.
This protein (DM3) is originally an enzyme involved in the plant's immune response, but problems arise when the structure of the DM3 protein is damaged in a specific protein combination called 'DANGEROUS MIX (DM)'.
Notably, one variant of DM3, the 'DM3Col-0' variant, forms a stable complex with six proteins and is recognized as normal, thus not triggering an immune response. In contrast, another 'DM3Hh-0' variant has improper binding between its six proteins, causing the plant to recognize it as an 'abnormal state' and trigger an immune alarm, leading to autoimmunity.
The research team visualized this structure using atomic-resolution cryo-electron microscopy (Cryo-EM) and revealed that the immune-inducing ability is not due to the enzymatic function of the DM3 protein, but rather to 'differences in protein binding affinity.'
<Figure 1. Mechanism of Plant Autoimmunity Triggered by the Collapse of the DM3 Protein Complex>
This demonstrates that plants can initiate an immune response by recognizing not only 'external pathogens' but also 'internal protein structures' when they undergo abnormal changes, treating them as if they were pathogens.
The study shows how sensitively the plant immune system changes and triggers autoimmune responses when genes are mixed and protein structures change during the cross-breeding of different plant varieties. It significantly advanced the understanding of genetic incompatibility that can occur during natural cross-breeding and cultivar improvement processes.
Dr. Gijeong Kim, the co-first author, stated, "Through international research collaboration, we presented a new perspective on understanding the plant immune system by leveraging the autoimmune phenomenon, completing a high-quality study that encompasses structural biochemistry, genetics, and cell biological experiments."
Professor Ji-Joon Song of the KAIST Department of Biological Sciences, who led the research, said, "The fact that the immune system can detect not only external pathogens but also structural abnormalities in its own proteins will set a new standard for plant biotechnology and crop breeding strategies. Cryo-electron microscopy-based structural analysis will be an important tool for understanding the essence of gene interactions."
This research, with Professor Ji-Joon Song and Professor Eunyoung Chae of the University of Oxford as co-corresponding authors, Dr. Gijeong Kim (currently a postdoctoral researcher at the University of Zurich) and Dr. Wei-Lin Wan of the National University of Singapore as co-first authors, and Ph.D candidate Nayun Kim, as the second author, was published on July 17th in Molecular Cell, a sister journal of the international academic journal Cell.
This research was supported by the KAIST Grand Challenge 30 project.
Article Title: Structural determinants of DANGEROUS MIX 3, an alpha/beta hydrolase that triggers NLR-mediated genetic incompatibility in plants DOI: https://doi.org/10.1016/j.molcel.2025.06.021
2025.07.21 View 245
Why Do Plants Attack Themselves? The Secret of Genetic Conflict Revealed
<Professor Ji-Joon Song of the KAIST Department of Biological Sciences>
Plants, with their unique immune systems, sometimes launch 'autoimmune responses' by mistakenly identifying their own protein structures as pathogens. In particular, 'hybrid necrosis,' a phenomenon where descendant plants fail to grow healthily and perish after cross-breeding different varieties, has long been a difficult challenge for botanists and agricultural researchers. In response, an international research team has successfully elucidated the mechanism inducing plant autoimmune responses and proposed a novel strategy for cultivar improvement that can predict and avoid these reactions.
Professor Ji-Joon Song's research team at KAIST, in collaboration with teams from the National University of Singapore (NUS) and the University of Oxford, announced on the 21st of July that they have elucidated the structure and function of the 'DM3' protein complex, which triggers plant autoimmune responses, using cryo-electron microscopy (Cryo-EM) technology.
This research is drawing attention because it identifies defects in protein structure as the cause of hybrid necrosis, which occurs due to an abnormal reaction of immune receptors during cross-breeding between plant hybrids.
This protein (DM3) is originally an enzyme involved in the plant's immune response, but problems arise when the structure of the DM3 protein is damaged in a specific protein combination called 'DANGEROUS MIX (DM)'.
Notably, one variant of DM3, the 'DM3Col-0' variant, forms a stable complex with six proteins and is recognized as normal, thus not triggering an immune response. In contrast, another 'DM3Hh-0' variant has improper binding between its six proteins, causing the plant to recognize it as an 'abnormal state' and trigger an immune alarm, leading to autoimmunity.
The research team visualized this structure using atomic-resolution cryo-electron microscopy (Cryo-EM) and revealed that the immune-inducing ability is not due to the enzymatic function of the DM3 protein, but rather to 'differences in protein binding affinity.'
<Figure 1. Mechanism of Plant Autoimmunity Triggered by the Collapse of the DM3 Protein Complex>
This demonstrates that plants can initiate an immune response by recognizing not only 'external pathogens' but also 'internal protein structures' when they undergo abnormal changes, treating them as if they were pathogens.
The study shows how sensitively the plant immune system changes and triggers autoimmune responses when genes are mixed and protein structures change during the cross-breeding of different plant varieties. It significantly advanced the understanding of genetic incompatibility that can occur during natural cross-breeding and cultivar improvement processes.
Dr. Gijeong Kim, the co-first author, stated, "Through international research collaboration, we presented a new perspective on understanding the plant immune system by leveraging the autoimmune phenomenon, completing a high-quality study that encompasses structural biochemistry, genetics, and cell biological experiments."
Professor Ji-Joon Song of the KAIST Department of Biological Sciences, who led the research, said, "The fact that the immune system can detect not only external pathogens but also structural abnormalities in its own proteins will set a new standard for plant biotechnology and crop breeding strategies. Cryo-electron microscopy-based structural analysis will be an important tool for understanding the essence of gene interactions."
This research, with Professor Ji-Joon Song and Professor Eunyoung Chae of the University of Oxford as co-corresponding authors, Dr. Gijeong Kim (currently a postdoctoral researcher at the University of Zurich) and Dr. Wei-Lin Wan of the National University of Singapore as co-first authors, and Ph.D candidate Nayun Kim, as the second author, was published on July 17th in Molecular Cell, a sister journal of the international academic journal Cell.
This research was supported by the KAIST Grand Challenge 30 project.
Article Title: Structural determinants of DANGEROUS MIX 3, an alpha/beta hydrolase that triggers NLR-mediated genetic incompatibility in plants DOI: https://doi.org/10.1016/j.molcel.2025.06.021
2025.07.21 View 245 -
 KAIST Develops Robots That React to Danger Like Humans
<(From left) Ph.D candidate See-On Park, Professor Jongwon Lee, and Professor Shinhyun Choi>
In the midst of the co-development of artificial intelligence and robotic advancements, developing technologies that enable robots to efficiently perceive and respond to their surroundings like humans has become a crucial task. In this context, Korean researchers are gaining attention for newly implementing an artificial sensory nervous system that mimics the sensory nervous system of living organisms without the need for separate complex software or circuitry. This breakthrough technology is expected to be applied in fields such as in ultra-small robots and robotic prosthetics, where intelligent and energy-efficient responses to external stimuli are essential.
KAIST (President Kwang Hyung Lee) announced on July15th that a joint research team led by Endowed Chair Professor Shinhyun Choi of the School of Electrical Engineering at KAIST and Professor Jongwon Lee of the Department of Semiconductor Convergence at Chungnam National University (President Jung Kyum Kim) developed a next-generation neuromorphic semiconductor-based artificial sensory nervous system. This system mimics the functions of a living organism's sensory nervous system, and enables a new type of robotic system that can efficiently responds to external stimuli.
In nature, animals — including humans — ignore safe or familiar stimuli and selectively react sensitively to important or dangerous ones. This selective response helps prevent unnecessary energy consumption while maintaining rapid awareness of critical signals. For instance, the sound of an air conditioner or the feel of clothing against the skin soon become familiar and are disregarded. However, if someone calls your name or a sharp object touches your skin, a rapid focus and response occur. These behaviors are regulated by the 'habituation' and 'sensitization' functions in the sensory nervous system. Attempts have been consistently made to apply these sensory nervous system functions of living organisms in order to create robots that efficiently respond to external environments like humans.
However, implementing complex neural characteristics such as habituation and sensitization in robots has faced difficulties in miniaturization and energy efficiency due to the need for separate software or complex circuitry. In particular, there have been attempts to utilize memristors, a neuromorphic semiconductor. A memristor is a next-generation electrical device, which has been widely utilized as an artificial synapse due to its ability to store analog value in the form of device resistance. However, existing memristors had limitations in mimicking the complex characteristics of the nervous system because they only allowed simple monotonic changes in conductivity.
To overcome these limitations, the research team developed a new memristor capable of reproducing complex neural response patterns such as habituation and sensitization within a single device. By introducing additional layer inside the memristor that alter conductivity in opposite directions, the device can more realistically emulate the dynamic synaptic behaviors of a real nervous system — for example, decreasing its response to repeated safe stimuli but quickly regaining sensitivity when a danger signal is detected.
<New memristor mimicking functions of sensory nervous system such as habituation/sensitization>
Using this new memristor, the research team built an artificial sensory nervous system capable of recognizing touch and pain, an applied it to a robotic hand to test its performance. When safe tactile stimuli were repeatedly applied, the robot hand, which initially reacted sensitively to unfamiliar tactile stimuli, gradually showed habituation characteristics by ignoring the stimuli. Later, when stimuli were applied along with an electric shock, it recognized this as a danger signal and showed sensitization characteristics by reacting sensitively again. Through this, it was experimentally proven that robots can efficiently respond to stimuli like humans without separate complex software or processors, verifying the possibility of developing energy-efficient neuro-inspired robots.
<Robot arm with memristor-based artificial sensory nervous system>
See-On Park, researcher at KAIST, stated, "By mimicking the human sensory nervous system with next-generation semiconductors, we have opened up the possibility of implementing a new concept of robots that are smarter and more energy-efficient in responding to external environments." He added, "This technology is expected to be utilized in various fusion fields of next-generation semiconductors and robotics, such as ultra-small robots, military robots, and medical robots like robotic prosthetics".
This research was published online on July 1st in the international journal 'Nature Communications,' with Ph.D candidate See-On Park as the first author.
Paper Title: Experimental demonstration of third-order memristor-based artificial sensory nervous system for neuro-inspired robotics
DOI: https://doi.org/10.1038/s41467-025-60818-x
This research was supported by the Korea National Research Foundation's Next-Generation Intelligent Semiconductor Technology Development Project, the Mid-Career Researcher Program, the PIM Artificial Intelligence Semiconductor Core Technology Development Project, the Excellent New Researcher Program, and the Nano Convergence Technology Division, National Nanofab Center's (NNFC) Nano-Medical Device Project.
2025.07.16 View 489
KAIST Develops Robots That React to Danger Like Humans
<(From left) Ph.D candidate See-On Park, Professor Jongwon Lee, and Professor Shinhyun Choi>
In the midst of the co-development of artificial intelligence and robotic advancements, developing technologies that enable robots to efficiently perceive and respond to their surroundings like humans has become a crucial task. In this context, Korean researchers are gaining attention for newly implementing an artificial sensory nervous system that mimics the sensory nervous system of living organisms without the need for separate complex software or circuitry. This breakthrough technology is expected to be applied in fields such as in ultra-small robots and robotic prosthetics, where intelligent and energy-efficient responses to external stimuli are essential.
KAIST (President Kwang Hyung Lee) announced on July15th that a joint research team led by Endowed Chair Professor Shinhyun Choi of the School of Electrical Engineering at KAIST and Professor Jongwon Lee of the Department of Semiconductor Convergence at Chungnam National University (President Jung Kyum Kim) developed a next-generation neuromorphic semiconductor-based artificial sensory nervous system. This system mimics the functions of a living organism's sensory nervous system, and enables a new type of robotic system that can efficiently responds to external stimuli.
In nature, animals — including humans — ignore safe or familiar stimuli and selectively react sensitively to important or dangerous ones. This selective response helps prevent unnecessary energy consumption while maintaining rapid awareness of critical signals. For instance, the sound of an air conditioner or the feel of clothing against the skin soon become familiar and are disregarded. However, if someone calls your name or a sharp object touches your skin, a rapid focus and response occur. These behaviors are regulated by the 'habituation' and 'sensitization' functions in the sensory nervous system. Attempts have been consistently made to apply these sensory nervous system functions of living organisms in order to create robots that efficiently respond to external environments like humans.
However, implementing complex neural characteristics such as habituation and sensitization in robots has faced difficulties in miniaturization and energy efficiency due to the need for separate software or complex circuitry. In particular, there have been attempts to utilize memristors, a neuromorphic semiconductor. A memristor is a next-generation electrical device, which has been widely utilized as an artificial synapse due to its ability to store analog value in the form of device resistance. However, existing memristors had limitations in mimicking the complex characteristics of the nervous system because they only allowed simple monotonic changes in conductivity.
To overcome these limitations, the research team developed a new memristor capable of reproducing complex neural response patterns such as habituation and sensitization within a single device. By introducing additional layer inside the memristor that alter conductivity in opposite directions, the device can more realistically emulate the dynamic synaptic behaviors of a real nervous system — for example, decreasing its response to repeated safe stimuli but quickly regaining sensitivity when a danger signal is detected.
<New memristor mimicking functions of sensory nervous system such as habituation/sensitization>
Using this new memristor, the research team built an artificial sensory nervous system capable of recognizing touch and pain, an applied it to a robotic hand to test its performance. When safe tactile stimuli were repeatedly applied, the robot hand, which initially reacted sensitively to unfamiliar tactile stimuli, gradually showed habituation characteristics by ignoring the stimuli. Later, when stimuli were applied along with an electric shock, it recognized this as a danger signal and showed sensitization characteristics by reacting sensitively again. Through this, it was experimentally proven that robots can efficiently respond to stimuli like humans without separate complex software or processors, verifying the possibility of developing energy-efficient neuro-inspired robots.
<Robot arm with memristor-based artificial sensory nervous system>
See-On Park, researcher at KAIST, stated, "By mimicking the human sensory nervous system with next-generation semiconductors, we have opened up the possibility of implementing a new concept of robots that are smarter and more energy-efficient in responding to external environments." He added, "This technology is expected to be utilized in various fusion fields of next-generation semiconductors and robotics, such as ultra-small robots, military robots, and medical robots like robotic prosthetics".
This research was published online on July 1st in the international journal 'Nature Communications,' with Ph.D candidate See-On Park as the first author.
Paper Title: Experimental demonstration of third-order memristor-based artificial sensory nervous system for neuro-inspired robotics
DOI: https://doi.org/10.1038/s41467-025-60818-x
This research was supported by the Korea National Research Foundation's Next-Generation Intelligent Semiconductor Technology Development Project, the Mid-Career Researcher Program, the PIM Artificial Intelligence Semiconductor Core Technology Development Project, the Excellent New Researcher Program, and the Nano Convergence Technology Division, National Nanofab Center's (NNFC) Nano-Medical Device Project.
2025.07.16 View 489 -
 KAIST-UIUC researchers develop a treatment platform to disable the ‘biofilm’ shield of superbugs
< (From left) Ph.D. Candidate Joo Hun Lee (co-author), Professor Hyunjoon Kong (co-corresponding author) and Postdoctoral Researcher Yujin Ahn (co-first author) from the Department of Chemical and Biomolecular Engineering of the University of Illinois at Urbana-Champaign and Ju Yeon Chung (co-first author) from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung (co-corresponding author) from the Department of Biological Sciences of KAIST >
A major cause of hospital-acquired infections, the super bacteria Methicillin-resistant Staphylococcus aureus (MRSA), not only exhibits strong resistance to existing antibiotics but also forms a dense biofilm that blocks the effects of external treatments. To meet this challenge, KAIST researchers, in collaboration with an international team, successfully developed a platform that utilizes microbubbles to deliver gene-targeted nanoparticles capable of break ing down the biofilms, offering an innovative solution for treating infections resistant to conventional antibiotics.
KAIST (represented by President Kwang Hyung Lee) announced on May 29 that a research team led by Professor Hyun Jung Chung from the Department of Biological Sciences, in collaboration with Professor Hyunjoon Kong's team at the University of Illinois, has developed a microbubble-based nano-gene delivery platform (BTN MB) that precisely delivers gene suppressors into bacteria to effectively remove biofilms formed by MRSA.
The research team first designed short DNA oligonucleotides that simultaneously suppress three major MRSA genes, related to—biofilm formation (icaA), cell division (ftsZ), and antibiotic resistance (mecA)—and engineered nanoparticles (BTN) to effectively deliver them into the bacteria.
< Figure 1. Effective biofilm treatment using biofilm-targeting nanoparticles controlled by microbubbler system. Schematic illustration of BTN delivery with microbubbles (MB), enabling effective permeation of ASOs targeting bacterial genes within biofilms infecting skin wounds. Gene silencing of targets involved in biofilm formation, bacterial proliferation, and antibiotic resistance leads to effective biofilm removal and antibacterial efficacy in vivo. >
In addition, microbubbles (MB) were used to increase the permeability of the microbial membrane, specifically the biofilm formed by MRSA. By combining these two technologies, the team implemented a dual-strike strategy that fundamentally blocks bacterial growth and prevents resistance acquisition.
This treatment system operates in two stages. First, the MBs induce pressure changes within the bacterial biofilm, allowing the BTNs to penetrate. Then, the BTNs slip through the gaps in the biofilm and enter the bacteria, delivering the gene suppressors precisely. This leads to gene regulation within MRSA, simultaneously blocking biofilm regeneration, cell proliferation, and antibiotic resistance expression.
In experiments conducted in a porcine skin model and a mouse wound model infected with MRSA biofilm, the BTN MB treatment group showed a significant reduction in biofilm thickness, as well as remarkable decreases in bacterial count and inflammatory responses.
< Figure 2. (a) Schematic illustration on the evaluation of treatment efficacy of BTN-MB gene therapy. (b) Reduction in MRSA biofilm mass via simultaneous inhibition of multiple genes. (c, d) Antibacterial efficacy of BTN-MB over time in a porcine skin infection biofilm model. (e) Schematic of the experimental setup to verify antibacterial efficacy in a mouse skin wound infection model. (f) Wound healing effects in mice. (g) Antibacterial effects at the wound site. (h) Histological analysis results. >
These results are difficult to achieve with conventional antibiotic monotherapy and demonstrate the potential for treating a wide range of resistant bacterial infections.
Professor Hyun Jung Chung of KAIST, who led the research, stated, “This study presents a new therapeutic solution that combines nanotechnology, gene suppression, and physical delivery strategies to address superbug infections that existing antibiotics cannot resolve. We will continue our research with the aim of expanding its application to systemic infections and various other infectious diseases.”
< (From left) Ju Yeon Chung from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung from the Department of Biological Sciences >
The study was co-first authored by Ju Yeon Chung, a graduate student in the Department of Biological Sciences at KAIST, and Dr. Yujin Ahn from the University of Illinois. The study was published online on May 19 in the journal, Advanced Functional Materials.
※ Paper Title: Microbubble-Controlled Delivery of Biofilm-Targeting Nanoparticles to Treat MRSA Infection ※ DOI: https://doi.org/10.1002/adfm.202508291
This study was supported by the National Research Foundation and the Ministry of Health and Welfare, Republic of Korea; and the National Science Foundation and National Institutes of Health, USA.
2025.05.29 View 3201
KAIST-UIUC researchers develop a treatment platform to disable the ‘biofilm’ shield of superbugs
< (From left) Ph.D. Candidate Joo Hun Lee (co-author), Professor Hyunjoon Kong (co-corresponding author) and Postdoctoral Researcher Yujin Ahn (co-first author) from the Department of Chemical and Biomolecular Engineering of the University of Illinois at Urbana-Champaign and Ju Yeon Chung (co-first author) from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung (co-corresponding author) from the Department of Biological Sciences of KAIST >
A major cause of hospital-acquired infections, the super bacteria Methicillin-resistant Staphylococcus aureus (MRSA), not only exhibits strong resistance to existing antibiotics but also forms a dense biofilm that blocks the effects of external treatments. To meet this challenge, KAIST researchers, in collaboration with an international team, successfully developed a platform that utilizes microbubbles to deliver gene-targeted nanoparticles capable of break ing down the biofilms, offering an innovative solution for treating infections resistant to conventional antibiotics.
KAIST (represented by President Kwang Hyung Lee) announced on May 29 that a research team led by Professor Hyun Jung Chung from the Department of Biological Sciences, in collaboration with Professor Hyunjoon Kong's team at the University of Illinois, has developed a microbubble-based nano-gene delivery platform (BTN MB) that precisely delivers gene suppressors into bacteria to effectively remove biofilms formed by MRSA.
The research team first designed short DNA oligonucleotides that simultaneously suppress three major MRSA genes, related to—biofilm formation (icaA), cell division (ftsZ), and antibiotic resistance (mecA)—and engineered nanoparticles (BTN) to effectively deliver them into the bacteria.
< Figure 1. Effective biofilm treatment using biofilm-targeting nanoparticles controlled by microbubbler system. Schematic illustration of BTN delivery with microbubbles (MB), enabling effective permeation of ASOs targeting bacterial genes within biofilms infecting skin wounds. Gene silencing of targets involved in biofilm formation, bacterial proliferation, and antibiotic resistance leads to effective biofilm removal and antibacterial efficacy in vivo. >
In addition, microbubbles (MB) were used to increase the permeability of the microbial membrane, specifically the biofilm formed by MRSA. By combining these two technologies, the team implemented a dual-strike strategy that fundamentally blocks bacterial growth and prevents resistance acquisition.
This treatment system operates in two stages. First, the MBs induce pressure changes within the bacterial biofilm, allowing the BTNs to penetrate. Then, the BTNs slip through the gaps in the biofilm and enter the bacteria, delivering the gene suppressors precisely. This leads to gene regulation within MRSA, simultaneously blocking biofilm regeneration, cell proliferation, and antibiotic resistance expression.
In experiments conducted in a porcine skin model and a mouse wound model infected with MRSA biofilm, the BTN MB treatment group showed a significant reduction in biofilm thickness, as well as remarkable decreases in bacterial count and inflammatory responses.
< Figure 2. (a) Schematic illustration on the evaluation of treatment efficacy of BTN-MB gene therapy. (b) Reduction in MRSA biofilm mass via simultaneous inhibition of multiple genes. (c, d) Antibacterial efficacy of BTN-MB over time in a porcine skin infection biofilm model. (e) Schematic of the experimental setup to verify antibacterial efficacy in a mouse skin wound infection model. (f) Wound healing effects in mice. (g) Antibacterial effects at the wound site. (h) Histological analysis results. >
These results are difficult to achieve with conventional antibiotic monotherapy and demonstrate the potential for treating a wide range of resistant bacterial infections.
Professor Hyun Jung Chung of KAIST, who led the research, stated, “This study presents a new therapeutic solution that combines nanotechnology, gene suppression, and physical delivery strategies to address superbug infections that existing antibiotics cannot resolve. We will continue our research with the aim of expanding its application to systemic infections and various other infectious diseases.”
< (From left) Ju Yeon Chung from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung from the Department of Biological Sciences >
The study was co-first authored by Ju Yeon Chung, a graduate student in the Department of Biological Sciences at KAIST, and Dr. Yujin Ahn from the University of Illinois. The study was published online on May 19 in the journal, Advanced Functional Materials.
※ Paper Title: Microbubble-Controlled Delivery of Biofilm-Targeting Nanoparticles to Treat MRSA Infection ※ DOI: https://doi.org/10.1002/adfm.202508291
This study was supported by the National Research Foundation and the Ministry of Health and Welfare, Republic of Korea; and the National Science Foundation and National Institutes of Health, USA.
2025.05.29 View 3201 -
 KAIST to Develop a Korean-style ChatGPT Platform Specifically Geared Toward Medical Diagnosis and Drug Discovery
On May 23rd, KAIST (President Kwang-Hyung Lee) announced that its Digital Bio-Health AI Research Center (Director: Professor JongChul Ye of KAIST Kim Jaechul Graduate School of AI) has been selected for the Ministry of Science and ICT's 'AI Top-Tier Young Researcher Support Program (AI Star Fellowship Project).' With a total investment of ₩11.5 billion from May 2025 to December 2030, the center will embark on the full-scale development of AI technology and a platform capable of independently inferring and determining the kinds of diseases, and discovering new drugs.
< Photo. On May 20th, a kick-off meeting for the AI Star Fellowship Project was held at KAIST Kim Jaechul Graduate School of AI’s Yangjae Research Center with the KAIST research team and participating organizations of Samsung Medical Center, NAVER Cloud, and HITS. [From left to right in the front row] Professor Jaegul Joo (KAIST), Professor Yoonjae Choi (KAIST), Professor Woo Youn Kim (KAIST/HITS), Professor JongChul Ye (KAIST), Professor Sungsoo Ahn (KAIST), Dr. Haanju Yoo (NAVER Cloud), Yoonho Lee (KAIST), HyeYoon Moon (Samsung Medical Center), Dr. Su Min Kim (Samsung Medical Center) >
This project aims to foster an innovative AI research ecosystem centered on young researchers and develop an inferential AI agent that can utilize and automatically expand specialized knowledge systems in the bio and medical fields.
Professor JongChul Ye of the Kim Jaechul Graduate School of AI will serve as the lead researcher, with young researchers from KAIST including Professors Yoonjae Choi, Kimin Lee, Sungsoo Ahn, and Chanyoung Park, along with mid-career researchers like Professors Jaegul Joo and Woo Youn Kim, jointly undertaking the project. They will collaborate with various laboratories within KAIST to conduct comprehensive research covering the entire cycle from the theoretical foundations of AI inference to its practical application.
Specifically, the main goals include: - Building high-performance inference models that integrate diverse medical knowledge systems to enhance the precision and reliability of diagnosis and treatment. - Developing a convergence inference platform that efficiently combines symbol-based inference with neural network models. - Securing AI technology for new drug development and biomarker discovery based on 'cell ontology.'
Furthermore, through close collaboration with industry and medical institutions such as Samsung Medical Center, NAVER Cloud, and HITS Co., Ltd., the project aims to achieve: - Clinical diagnostic AI utilizing medical knowledge systems. - AI-based molecular target exploration for new drug development. - Commercialization of an extendible AI inference platform.
Professor JongChul Ye, Director of KAIST's Digital Bio-Health AI Research Center, stated, "At a time when competition in AI inference model development is intensifying, it is a great honor for KAIST to lead the development of AI technology specialized in the bio and medical fields with world-class young researchers." He added, "We will do our best to ensure that the participating young researchers reach a world-leading level in terms of research achievements after the completion of this seven-year project starting in 2025."
The AI Star Fellowship is a newly established program where post-doctoral researchers and faculty members within seven years of appointment participate as project leaders (PLs) to independently lead research. Multiple laboratories within a university and demand-side companies form a consortium to operate the program.
Through this initiative, KAIST plans to nurture bio-medical convergence AI talent and simultaneously promote the commercialization of core technologies in collaboration with Samsung Medical Center, NAVER Cloud, and HITS.
2025.05.26 View 4923
KAIST to Develop a Korean-style ChatGPT Platform Specifically Geared Toward Medical Diagnosis and Drug Discovery
On May 23rd, KAIST (President Kwang-Hyung Lee) announced that its Digital Bio-Health AI Research Center (Director: Professor JongChul Ye of KAIST Kim Jaechul Graduate School of AI) has been selected for the Ministry of Science and ICT's 'AI Top-Tier Young Researcher Support Program (AI Star Fellowship Project).' With a total investment of ₩11.5 billion from May 2025 to December 2030, the center will embark on the full-scale development of AI technology and a platform capable of independently inferring and determining the kinds of diseases, and discovering new drugs.
< Photo. On May 20th, a kick-off meeting for the AI Star Fellowship Project was held at KAIST Kim Jaechul Graduate School of AI’s Yangjae Research Center with the KAIST research team and participating organizations of Samsung Medical Center, NAVER Cloud, and HITS. [From left to right in the front row] Professor Jaegul Joo (KAIST), Professor Yoonjae Choi (KAIST), Professor Woo Youn Kim (KAIST/HITS), Professor JongChul Ye (KAIST), Professor Sungsoo Ahn (KAIST), Dr. Haanju Yoo (NAVER Cloud), Yoonho Lee (KAIST), HyeYoon Moon (Samsung Medical Center), Dr. Su Min Kim (Samsung Medical Center) >
This project aims to foster an innovative AI research ecosystem centered on young researchers and develop an inferential AI agent that can utilize and automatically expand specialized knowledge systems in the bio and medical fields.
Professor JongChul Ye of the Kim Jaechul Graduate School of AI will serve as the lead researcher, with young researchers from KAIST including Professors Yoonjae Choi, Kimin Lee, Sungsoo Ahn, and Chanyoung Park, along with mid-career researchers like Professors Jaegul Joo and Woo Youn Kim, jointly undertaking the project. They will collaborate with various laboratories within KAIST to conduct comprehensive research covering the entire cycle from the theoretical foundations of AI inference to its practical application.
Specifically, the main goals include: - Building high-performance inference models that integrate diverse medical knowledge systems to enhance the precision and reliability of diagnosis and treatment. - Developing a convergence inference platform that efficiently combines symbol-based inference with neural network models. - Securing AI technology for new drug development and biomarker discovery based on 'cell ontology.'
Furthermore, through close collaboration with industry and medical institutions such as Samsung Medical Center, NAVER Cloud, and HITS Co., Ltd., the project aims to achieve: - Clinical diagnostic AI utilizing medical knowledge systems. - AI-based molecular target exploration for new drug development. - Commercialization of an extendible AI inference platform.
Professor JongChul Ye, Director of KAIST's Digital Bio-Health AI Research Center, stated, "At a time when competition in AI inference model development is intensifying, it is a great honor for KAIST to lead the development of AI technology specialized in the bio and medical fields with world-class young researchers." He added, "We will do our best to ensure that the participating young researchers reach a world-leading level in terms of research achievements after the completion of this seven-year project starting in 2025."
The AI Star Fellowship is a newly established program where post-doctoral researchers and faculty members within seven years of appointment participate as project leaders (PLs) to independently lead research. Multiple laboratories within a university and demand-side companies form a consortium to operate the program.
Through this initiative, KAIST plans to nurture bio-medical convergence AI talent and simultaneously promote the commercialization of core technologies in collaboration with Samsung Medical Center, NAVER Cloud, and HITS.
2025.05.26 View 4923 -
 KAIST's Pioneering VR Precision Technology & Choreography Tool Receive Spotlights at CHI 2025
Accurate pointing in virtual spaces is essential for seamless interaction. If pointing is not precise, selecting the desired object becomes challenging, breaking user immersion and reducing overall experience quality. KAIST researchers have developed a technology that offers a vivid, lifelike experience in virtual space, alongside a new tool that assists choreographers throughout the creative process.
KAIST (President Kwang-Hyung Lee) announced on May 13th that a research team led by Professor Sang Ho Yoon of the Graduate School of Culture Technology, in collaboration with Professor Yang Zhang of the University of California, Los Angeles (UCLA), has developed the ‘T2IRay’ technology and the ‘ChoreoCraft’ platform, which enables choreographers to work more freely and creatively in virtual reality. These technologies received two Honorable Mention awards, recognizing the top 5% of papers, at CHI 2025*, the best international conference in the field of human-computer interaction, hosted by the Association for Computing Machinery (ACM) from April 25 to May 1.
< (From left) PhD candidates Jina Kim and Kyungeun Jung along with Master's candidate, Hyunyoung Han and Professor Sang Ho Yoon of KAIST Graduate School of Culture Technology and Professor Yang Zhang (top) of UCLA >
T2IRay: Enabling Virtual Input with Precision
T2IRay introduces a novel input method that allows for precise object pointing in virtual environments by expanding traditional thumb-to-index gestures. This approach overcomes previous limitations, such as interruptions or reduced accuracy due to changes in hand position or orientation.
The technology uses a local coordinate system based on finger relationships, ensuring continuous input even as hand positions shift. It accurately captures subtle thumb movements within this coordinate system, integrating natural head movements to allow fluid, intuitive control across a wide range.
< Figure 1. T2IRay framework utilizing the delicate movements of the thumb and index fingers for AR/VR pointing >
Professor Sang Ho Yoon explained, “T2IRay can significantly enhance the user experience in AR/VR by enabling smooth, stable control even when the user’s hands are in motion.”
This study, led by first author Jina Kim, was supported by the Excellent New Researcher Support Project of the National Research Foundation of Korea under the Ministry of Science and ICT, as well as the University ICT Research Center (ITRC) Support Project of the Institute of Information and Communications Technology Planning and Evaluation (IITP).
▴ Paper title: T2IRay: Design of Thumb-to-Index Based Indirect Pointing for Continuous and Robust AR/VR Input▴ Paper link: https://doi.org/10.1145/3706598.3713442
▴ T2IRay demo video: https://youtu.be/ElJlcJbkJPY
ChoreoCraft: Creativity Support through VR for Choreographers
In addition, Professor Yoon’s team developed ‘ChoreoCraft,’ a virtual reality tool designed to support choreographers by addressing the unique challenges they face, such as memorizing complex movements, overcoming creative blocks, and managing subjective feedback.
ChoreoCraft reduces reliance on memory by allowing choreographers to save and refine movements directly within a VR space, using a motion-capture avatar for real-time interaction. It also enhances creativity by suggesting movements that naturally fit with prior choreography and musical elements. Furthermore, the system provides quantitative feedback by analyzing kinematic factors like motion stability and engagement, helping choreographers make data-driven creative decisions.
< Figure 2. ChoreoCraft's approaches to encourage creative process >
Professor Yoon noted, “ChoreoCraft is a tool designed to address the core challenges faced by choreographers, enhancing both creativity and efficiency. In user tests with professional choreographers, it received high marks for its ability to spark creative ideas and provide valuable quantitative feedback.”
This research was conducted in collaboration with doctoral candidate Kyungeun Jung and master’s candidate Hyunyoung Han, alongside the Electronics and Telecommunications Research Institute (ETRI) and One Million Co., Ltd. (CEO Hye-rang Kim), with support from the Cultural and Arts Immersive Service Development Project by the Ministry of Culture, Sports and Tourism.
▴ Paper title: ChoreoCraft: In-situ Crafting of Choreography in Virtual Reality through Creativity Support Tools▴ Paper link: https://doi.org/10.1145/3706598.3714220
▴ ChoreoCraft demo video: https://youtu.be/Ms1fwiSBjjw
*CHI (Conference on Human Factors in Computing Systems): The premier international conference on human-computer interaction, organized by the ACM, was held this year from April 25 to May 1, 2025.
2025.05.13 View 4842
KAIST's Pioneering VR Precision Technology & Choreography Tool Receive Spotlights at CHI 2025
Accurate pointing in virtual spaces is essential for seamless interaction. If pointing is not precise, selecting the desired object becomes challenging, breaking user immersion and reducing overall experience quality. KAIST researchers have developed a technology that offers a vivid, lifelike experience in virtual space, alongside a new tool that assists choreographers throughout the creative process.
KAIST (President Kwang-Hyung Lee) announced on May 13th that a research team led by Professor Sang Ho Yoon of the Graduate School of Culture Technology, in collaboration with Professor Yang Zhang of the University of California, Los Angeles (UCLA), has developed the ‘T2IRay’ technology and the ‘ChoreoCraft’ platform, which enables choreographers to work more freely and creatively in virtual reality. These technologies received two Honorable Mention awards, recognizing the top 5% of papers, at CHI 2025*, the best international conference in the field of human-computer interaction, hosted by the Association for Computing Machinery (ACM) from April 25 to May 1.
< (From left) PhD candidates Jina Kim and Kyungeun Jung along with Master's candidate, Hyunyoung Han and Professor Sang Ho Yoon of KAIST Graduate School of Culture Technology and Professor Yang Zhang (top) of UCLA >
T2IRay: Enabling Virtual Input with Precision
T2IRay introduces a novel input method that allows for precise object pointing in virtual environments by expanding traditional thumb-to-index gestures. This approach overcomes previous limitations, such as interruptions or reduced accuracy due to changes in hand position or orientation.
The technology uses a local coordinate system based on finger relationships, ensuring continuous input even as hand positions shift. It accurately captures subtle thumb movements within this coordinate system, integrating natural head movements to allow fluid, intuitive control across a wide range.
< Figure 1. T2IRay framework utilizing the delicate movements of the thumb and index fingers for AR/VR pointing >
Professor Sang Ho Yoon explained, “T2IRay can significantly enhance the user experience in AR/VR by enabling smooth, stable control even when the user’s hands are in motion.”
This study, led by first author Jina Kim, was supported by the Excellent New Researcher Support Project of the National Research Foundation of Korea under the Ministry of Science and ICT, as well as the University ICT Research Center (ITRC) Support Project of the Institute of Information and Communications Technology Planning and Evaluation (IITP).
▴ Paper title: T2IRay: Design of Thumb-to-Index Based Indirect Pointing for Continuous and Robust AR/VR Input▴ Paper link: https://doi.org/10.1145/3706598.3713442
▴ T2IRay demo video: https://youtu.be/ElJlcJbkJPY
ChoreoCraft: Creativity Support through VR for Choreographers
In addition, Professor Yoon’s team developed ‘ChoreoCraft,’ a virtual reality tool designed to support choreographers by addressing the unique challenges they face, such as memorizing complex movements, overcoming creative blocks, and managing subjective feedback.
ChoreoCraft reduces reliance on memory by allowing choreographers to save and refine movements directly within a VR space, using a motion-capture avatar for real-time interaction. It also enhances creativity by suggesting movements that naturally fit with prior choreography and musical elements. Furthermore, the system provides quantitative feedback by analyzing kinematic factors like motion stability and engagement, helping choreographers make data-driven creative decisions.
< Figure 2. ChoreoCraft's approaches to encourage creative process >
Professor Yoon noted, “ChoreoCraft is a tool designed to address the core challenges faced by choreographers, enhancing both creativity and efficiency. In user tests with professional choreographers, it received high marks for its ability to spark creative ideas and provide valuable quantitative feedback.”
This research was conducted in collaboration with doctoral candidate Kyungeun Jung and master’s candidate Hyunyoung Han, alongside the Electronics and Telecommunications Research Institute (ETRI) and One Million Co., Ltd. (CEO Hye-rang Kim), with support from the Cultural and Arts Immersive Service Development Project by the Ministry of Culture, Sports and Tourism.
▴ Paper title: ChoreoCraft: In-situ Crafting of Choreography in Virtual Reality through Creativity Support Tools▴ Paper link: https://doi.org/10.1145/3706598.3714220
▴ ChoreoCraft demo video: https://youtu.be/Ms1fwiSBjjw
*CHI (Conference on Human Factors in Computing Systems): The premier international conference on human-computer interaction, organized by the ACM, was held this year from April 25 to May 1, 2025.
2025.05.13 View 4842 -
 KAIST & CMU Unveils Amuse, a Songwriting AI-Collaborator to Help Create Music
Wouldn't it be great if music creators had someone to brainstorm with, help them when they're stuck, and explore different musical directions together? Researchers of KAIST and Carnegie Mellon University (CMU) have developed AI technology similar to a fellow songwriter who helps create music.
KAIST (President Kwang-Hyung Lee) has developed an AI-based music creation support system, Amuse, by a research team led by Professor Sung-Ju Lee of the School of Electrical Engineering in collaboration with CMU. The research was presented at the ACM Conference on Human Factors in Computing Systems (CHI), one of the world’s top conferences in human-computer interaction, held in Yokohama, Japan from April 26 to May 1. It received the Best Paper Award, given to only the top 1% of all submissions.
< (From left) Professor Chris Donahue of Carnegie Mellon University, Ph.D. Student Yewon Kim and Professor Sung-Ju Lee of the School of Electrical Engineering >
The system developed by Professor Sung-Ju Lee’s research team, Amuse, is an AI-based system that converts various forms of inspiration such as text, images, and audio into harmonic structures (chord progressions) to support composition.
For example, if a user inputs a phrase, image, or sound clip such as “memories of a warm summer beach”, Amuse automatically generates and suggests chord progressions that match the inspiration.
Unlike existing generative AI, Amuse is differentiated in that it respects the user's creative flow and naturally induces creative exploration through an interactive method that allows flexible integration and modification of AI suggestions.
The core technology of the Amuse system is a generation method that blends two approaches: a large language model creates music code based on the user's prompt and inspiration, while another AI model, trained on real music data, filters out awkward or unnatural results using rejection sampling.
< Figure 1. Amuse system configuration. After extracting music keywords from user input, a large language model-based code progression is generated and refined through rejection sampling (left). Code extraction from audio input is also possible (right). The bottom is an example visualizing the chord structure of the generated code. >
The research team conducted a user study targeting actual musicians and evaluated that Amuse has high potential as a creative companion, or a Co-Creative AI, a concept in which people and AI collaborate, rather than having a generative AI simply put together a song.
The paper, in which a Ph.D. student Yewon Kim and Professor Sung-Ju Lee of KAIST School of Electrical and Electronic Engineering and Carnegie Mellon University Professor Chris Donahue participated, demonstrated the potential of creative AI system design in both academia and industry. ※ Paper title: Amuse: Human-AI Collaborative Songwriting with Multimodal Inspirations DOI: https://doi.org/10.1145/3706598.3713818
※ Research demo video: https://youtu.be/udilkRSnftI?si=FNXccC9EjxHOCrm1
※ Research homepage: https://nmsl.kaist.ac.kr/projects/amuse/
Professor Sung-Ju Lee said, “Recent generative AI technology has raised concerns in that it directly imitates copyrighted content, thereby violating the copyright of the creator, or generating results one-way regardless of the creator’s intention. Accordingly, the research team was aware of this trend, paid attention to what the creator actually needs, and focused on designing an AI system centered on the creator.”
He continued, “Amuse is an attempt to explore the possibility of collaboration with AI while maintaining the initiative of the creator, and is expected to be a starting point for suggesting a more creator-friendly direction in the development of music creation tools and generative AI systems in the future.”
This research was conducted with the support of the National Research Foundation of Korea with funding from the government (Ministry of Science and ICT). (RS-2024-00337007)
2025.05.07 View 6543
KAIST & CMU Unveils Amuse, a Songwriting AI-Collaborator to Help Create Music
Wouldn't it be great if music creators had someone to brainstorm with, help them when they're stuck, and explore different musical directions together? Researchers of KAIST and Carnegie Mellon University (CMU) have developed AI technology similar to a fellow songwriter who helps create music.
KAIST (President Kwang-Hyung Lee) has developed an AI-based music creation support system, Amuse, by a research team led by Professor Sung-Ju Lee of the School of Electrical Engineering in collaboration with CMU. The research was presented at the ACM Conference on Human Factors in Computing Systems (CHI), one of the world’s top conferences in human-computer interaction, held in Yokohama, Japan from April 26 to May 1. It received the Best Paper Award, given to only the top 1% of all submissions.
< (From left) Professor Chris Donahue of Carnegie Mellon University, Ph.D. Student Yewon Kim and Professor Sung-Ju Lee of the School of Electrical Engineering >
The system developed by Professor Sung-Ju Lee’s research team, Amuse, is an AI-based system that converts various forms of inspiration such as text, images, and audio into harmonic structures (chord progressions) to support composition.
For example, if a user inputs a phrase, image, or sound clip such as “memories of a warm summer beach”, Amuse automatically generates and suggests chord progressions that match the inspiration.
Unlike existing generative AI, Amuse is differentiated in that it respects the user's creative flow and naturally induces creative exploration through an interactive method that allows flexible integration and modification of AI suggestions.
The core technology of the Amuse system is a generation method that blends two approaches: a large language model creates music code based on the user's prompt and inspiration, while another AI model, trained on real music data, filters out awkward or unnatural results using rejection sampling.
< Figure 1. Amuse system configuration. After extracting music keywords from user input, a large language model-based code progression is generated and refined through rejection sampling (left). Code extraction from audio input is also possible (right). The bottom is an example visualizing the chord structure of the generated code. >
The research team conducted a user study targeting actual musicians and evaluated that Amuse has high potential as a creative companion, or a Co-Creative AI, a concept in which people and AI collaborate, rather than having a generative AI simply put together a song.
The paper, in which a Ph.D. student Yewon Kim and Professor Sung-Ju Lee of KAIST School of Electrical and Electronic Engineering and Carnegie Mellon University Professor Chris Donahue participated, demonstrated the potential of creative AI system design in both academia and industry. ※ Paper title: Amuse: Human-AI Collaborative Songwriting with Multimodal Inspirations DOI: https://doi.org/10.1145/3706598.3713818
※ Research demo video: https://youtu.be/udilkRSnftI?si=FNXccC9EjxHOCrm1
※ Research homepage: https://nmsl.kaist.ac.kr/projects/amuse/
Professor Sung-Ju Lee said, “Recent generative AI technology has raised concerns in that it directly imitates copyrighted content, thereby violating the copyright of the creator, or generating results one-way regardless of the creator’s intention. Accordingly, the research team was aware of this trend, paid attention to what the creator actually needs, and focused on designing an AI system centered on the creator.”
He continued, “Amuse is an attempt to explore the possibility of collaboration with AI while maintaining the initiative of the creator, and is expected to be a starting point for suggesting a more creator-friendly direction in the development of music creation tools and generative AI systems in the future.”
This research was conducted with the support of the National Research Foundation of Korea with funding from the government (Ministry of Science and ICT). (RS-2024-00337007)
2025.05.07 View 6543 -
 KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 6637
KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 6637 -
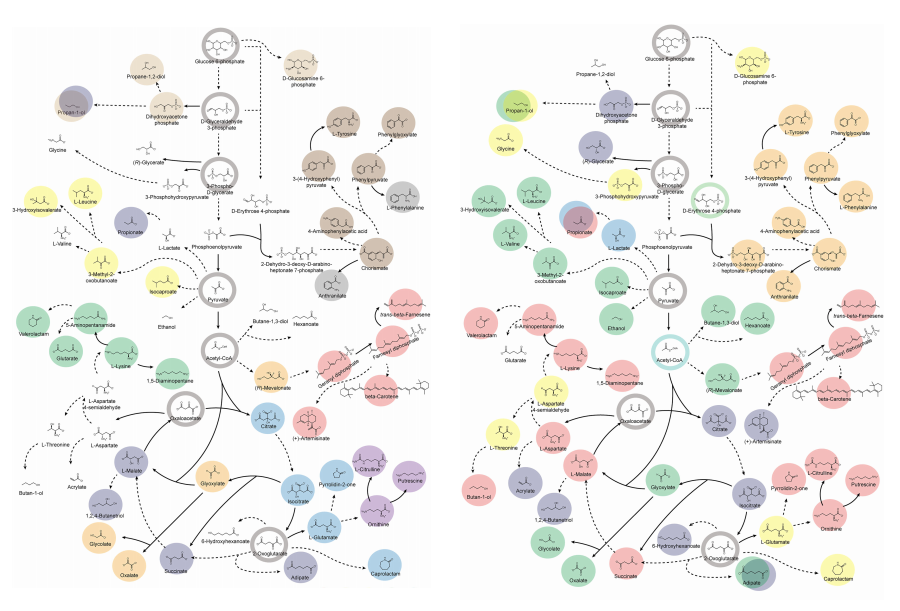 KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 4860
KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 4860 -
 KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 6217
KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 6217 -
 KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 28887
KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 28887 -
 KAIST Develops Neuromorphic Semiconductor Chip that Learns and Corrects Itself
< Photo. The research team of the School of Electrical Engineering posed by the newly deveoped processor. (From center to the right) Professor Young-Gyu Yoon, Integrated Master's and Doctoral Program Students Seungjae Han and Hakcheon Jeong and Professor Shinhyun Choi >
- Professor Shinhyun Choi and Professor Young-Gyu Yoon’s Joint Research Team from the School of Electrical Engineering developed a computing chip that can learn, correct errors, and process AI tasks
- Equipping a computing chip with high-reliability memristor devices with self-error correction functions for real-time learning and image processing
Existing computer systems have separate data processing and storage devices, making them inefficient for processing complex data like AI. A KAIST research team has developed a memristor-based integrated system similar to the way our brain processes information. It is now ready for application in various devices including smart security cameras, allowing them to recognize suspicious activity immediately without having to rely on remote cloud servers, and medical devices with which it can help analyze health data in real time.
KAIST (President Kwang Hyung Lee) announced on the 17th of January that the joint research team of Professor Shinhyun Choi and Professor Young-Gyu Yoon of the School of Electrical Engineering has developed a next-generation neuromorphic semiconductor-based ultra-small computing chip that can learn and correct errors on its own.
< Figure 1. Scanning electron microscope (SEM) image of a computing chip equipped with a highly reliable selector-less 32×32 memristor crossbar array (left). Hardware system developed for real-time artificial intelligence implementation (right). >
What is special about this computing chip is that it can learn and correct errors that occur due to non-ideal characteristics that were difficult to solve in existing neuromorphic devices. For example, when processing a video stream, the chip learns to automatically separate a moving object from the background, and it becomes better at this task over time.
This self-learning ability has been proven by achieving accuracy comparable to ideal computer simulations in real-time image processing. The research team's main achievement is that it has completed a system that is both reliable and practical, beyond the development of brain-like components.
The research team has developed the world's first memristor-based integrated system that can adapt to immediate environmental changes, and has presented an innovative solution that overcomes the limitations of existing technology.
< Figure 2. Background and foreground separation results of an image containing non-ideal characteristics of memristor devices (left). Real-time image separation results through on-device learning using the memristor computing chip developed by our research team (right). >
At the heart of this innovation is a next-generation semiconductor device called a memristor*. The variable resistance characteristics of this device can replace the role of synapses in neural networks, and by utilizing it, data storage and computation can be performed simultaneously, just like our brain cells.
*Memristor: A compound word of memory and resistor, next-generation electrical device whose resistance value is determined by the amount and direction of charge that has flowed between the two terminals in the past.
The research team designed a highly reliable memristor that can precisely control resistance changes and developed an efficient system that excludes complex compensation processes through self-learning. This study is significant in that it experimentally verified the commercialization possibility of a next-generation neuromorphic semiconductor-based integrated system that supports real-time learning and inference.
This technology will revolutionize the way artificial intelligence is used in everyday devices, allowing AI tasks to be processed locally without relying on remote cloud servers, making them faster, more privacy-protected, and more energy-efficient.
“This system is like a smart workspace where everything is within arm’s reach instead of having to go back and forth between desks and file cabinets,” explained KAIST researchers Hakcheon Jeong and Seungjae Han, who led the development of this technology. “This is similar to the way our brain processes information, where everything is processed efficiently at once at one spot.”
The research was conducted with Hakcheon Jeong and Seungjae Han, the students of Integrated Master's and Doctoral Program at KAIST School of Electrical Engineering being the co-first authors, the results of which was published online in the international academic journal, Nature Electronics, on January 8, 2025.
*Paper title: Self-supervised video processing with self-calibration on an analogue computing platform based on a selector-less memristor array ( https://doi.org/10.1038/s41928-024-01318-6 )
This research was supported by the Next-Generation Intelligent Semiconductor Technology Development Project, Excellent New Researcher Project and PIM AI Semiconductor Core Technology Development Project of the National Research Foundation of Korea, and the Electronics and Telecommunications Research Institute Research and Development Support Project of the Institute of Information & communications Technology Planning & Evaluation.
2025.01.17 View 9184
KAIST Develops Neuromorphic Semiconductor Chip that Learns and Corrects Itself
< Photo. The research team of the School of Electrical Engineering posed by the newly deveoped processor. (From center to the right) Professor Young-Gyu Yoon, Integrated Master's and Doctoral Program Students Seungjae Han and Hakcheon Jeong and Professor Shinhyun Choi >
- Professor Shinhyun Choi and Professor Young-Gyu Yoon’s Joint Research Team from the School of Electrical Engineering developed a computing chip that can learn, correct errors, and process AI tasks
- Equipping a computing chip with high-reliability memristor devices with self-error correction functions for real-time learning and image processing
Existing computer systems have separate data processing and storage devices, making them inefficient for processing complex data like AI. A KAIST research team has developed a memristor-based integrated system similar to the way our brain processes information. It is now ready for application in various devices including smart security cameras, allowing them to recognize suspicious activity immediately without having to rely on remote cloud servers, and medical devices with which it can help analyze health data in real time.
KAIST (President Kwang Hyung Lee) announced on the 17th of January that the joint research team of Professor Shinhyun Choi and Professor Young-Gyu Yoon of the School of Electrical Engineering has developed a next-generation neuromorphic semiconductor-based ultra-small computing chip that can learn and correct errors on its own.
< Figure 1. Scanning electron microscope (SEM) image of a computing chip equipped with a highly reliable selector-less 32×32 memristor crossbar array (left). Hardware system developed for real-time artificial intelligence implementation (right). >
What is special about this computing chip is that it can learn and correct errors that occur due to non-ideal characteristics that were difficult to solve in existing neuromorphic devices. For example, when processing a video stream, the chip learns to automatically separate a moving object from the background, and it becomes better at this task over time.
This self-learning ability has been proven by achieving accuracy comparable to ideal computer simulations in real-time image processing. The research team's main achievement is that it has completed a system that is both reliable and practical, beyond the development of brain-like components.
The research team has developed the world's first memristor-based integrated system that can adapt to immediate environmental changes, and has presented an innovative solution that overcomes the limitations of existing technology.
< Figure 2. Background and foreground separation results of an image containing non-ideal characteristics of memristor devices (left). Real-time image separation results through on-device learning using the memristor computing chip developed by our research team (right). >
At the heart of this innovation is a next-generation semiconductor device called a memristor*. The variable resistance characteristics of this device can replace the role of synapses in neural networks, and by utilizing it, data storage and computation can be performed simultaneously, just like our brain cells.
*Memristor: A compound word of memory and resistor, next-generation electrical device whose resistance value is determined by the amount and direction of charge that has flowed between the two terminals in the past.
The research team designed a highly reliable memristor that can precisely control resistance changes and developed an efficient system that excludes complex compensation processes through self-learning. This study is significant in that it experimentally verified the commercialization possibility of a next-generation neuromorphic semiconductor-based integrated system that supports real-time learning and inference.
This technology will revolutionize the way artificial intelligence is used in everyday devices, allowing AI tasks to be processed locally without relying on remote cloud servers, making them faster, more privacy-protected, and more energy-efficient.
“This system is like a smart workspace where everything is within arm’s reach instead of having to go back and forth between desks and file cabinets,” explained KAIST researchers Hakcheon Jeong and Seungjae Han, who led the development of this technology. “This is similar to the way our brain processes information, where everything is processed efficiently at once at one spot.”
The research was conducted with Hakcheon Jeong and Seungjae Han, the students of Integrated Master's and Doctoral Program at KAIST School of Electrical Engineering being the co-first authors, the results of which was published online in the international academic journal, Nature Electronics, on January 8, 2025.
*Paper title: Self-supervised video processing with self-calibration on an analogue computing platform based on a selector-less memristor array ( https://doi.org/10.1038/s41928-024-01318-6 )
This research was supported by the Next-Generation Intelligent Semiconductor Technology Development Project, Excellent New Researcher Project and PIM AI Semiconductor Core Technology Development Project of the National Research Foundation of Korea, and the Electronics and Telecommunications Research Institute Research and Development Support Project of the Institute of Information & communications Technology Planning & Evaluation.
2025.01.17 View 9184 -
 KAIST Develops Insect-Eye-Inspired Camera Capturing 9,120 Frames Per Second
< (From left) Bio and Brain Engineering PhD Student Jae-Myeong Kwon, Professor Ki-Hun Jeong, PhD Student Hyun-Kyung Kim, PhD Student Young-Gil Cha, and Professor Min H. Kim of the School of Computing >
The compound eyes of insects can detect fast-moving objects in parallel and, in low-light conditions, enhance sensitivity by integrating signals over time to determine motion. Inspired by these biological mechanisms, KAIST researchers have successfully developed a low-cost, high-speed camera that overcomes the limitations of frame rate and sensitivity faced by conventional high-speed cameras.
KAIST (represented by President Kwang Hyung Lee) announced on the 16th of January that a research team led by Professors Ki-Hun Jeong (Department of Bio and Brain Engineering) and Min H. Kim (School of Computing) has developed a novel bio-inspired camera capable of ultra-high-speed imaging with high sensitivity by mimicking the visual structure of insect eyes.
High-quality imaging under high-speed and low-light conditions is a critical challenge in many applications. While conventional high-speed cameras excel in capturing fast motion, their sensitivity decreases as frame rates increase because the time available to collect light is reduced.
To address this issue, the research team adopted an approach similar to insect vision, utilizing multiple optical channels and temporal summation. Unlike traditional monocular camera systems, the bio-inspired camera employs a compound-eye-like structure that allows for the parallel acquisition of frames from different time intervals.
< Figure 1. (A) Vision in a fast-eyed insect. Reflected light from swiftly moving objects sequentially stimulates the photoreceptors along the individual optical channels called ommatidia, of which the visual signals are separately and parallelly processed via the lamina and medulla. Each neural response is temporally summed to enhance the visual signals. The parallel processing and temporal summation allow fast and low-light imaging in dim light. (B) High-speed and high-sensitivity microlens array camera (HS-MAC). A rolling shutter image sensor is utilized to simultaneously acquire multiple frames by channel division, and temporal summation is performed in parallel to realize high speed and sensitivity even in a low-light environment. In addition, the frame components of a single fragmented array image are stitched into a single blurred frame, which is subsequently deblurred by compressive image reconstruction. >
During this process, light is accumulated over overlapping time periods for each frame, increasing the signal-to-noise ratio. The researchers demonstrated that their bio-inspired camera could capture objects up to 40 times dimmer than those detectable by conventional high-speed cameras.
The team also introduced a "channel-splitting" technique to significantly enhance the camera's speed, achieving frame rates thousands of times faster than those supported by the image sensors used in packaging. Additionally, a "compressed image restoration" algorithm was employed to eliminate blur caused by frame integration and reconstruct sharp images.
The resulting bio-inspired camera is less than one millimeter thick and extremely compact, capable of capturing 9,120 frames per second while providing clear images in low-light conditions.
< Figure 2. A high-speed, high-sensitivity biomimetic camera packaged in an image sensor. It is made small enough to fit on a finger, with a thickness of less than 1 mm. >
The research team plans to extend this technology to develop advanced image processing algorithms for 3D imaging and super-resolution imaging, aiming for applications in biomedical imaging, mobile devices, and various other camera technologies.
Hyun-Kyung Kim, a doctoral student in the Department of Bio and Brain Engineering at KAIST and the study's first author, stated, “We have experimentally validated that the insect-eye-inspired camera delivers outstanding performance in high-speed and low-light imaging despite its small size. This camera opens up possibilities for diverse applications in portable camera systems, security surveillance, and medical imaging.”
< Figure 3. Rotating plate and flame captured using the high-speed, high-sensitivity biomimetic camera. The rotating plate at 1,950 rpm was accurately captured at 9,120 fps. In addition, the pinch-off of the flame with a faint intensity of 880 µlux was accurately captured at 1,020 fps. >
This research was published in the international journal Science Advances in January 2025 (Paper Title: “Biologically-inspired microlens array camera for high-speed and high-sensitivity imaging”).
DOI: https://doi.org/10.1126/sciadv.ads3389
This study was supported by the Korea Research Institute for Defense Technology Planning and Advancement (KRIT) of the Defense Acquisition Program Administration (DAPA), the Ministry of Science and ICT, and the Ministry of Trade, Industry and Energy (MOTIE).
2025.01.16 View 8756
KAIST Develops Insect-Eye-Inspired Camera Capturing 9,120 Frames Per Second
< (From left) Bio and Brain Engineering PhD Student Jae-Myeong Kwon, Professor Ki-Hun Jeong, PhD Student Hyun-Kyung Kim, PhD Student Young-Gil Cha, and Professor Min H. Kim of the School of Computing >
The compound eyes of insects can detect fast-moving objects in parallel and, in low-light conditions, enhance sensitivity by integrating signals over time to determine motion. Inspired by these biological mechanisms, KAIST researchers have successfully developed a low-cost, high-speed camera that overcomes the limitations of frame rate and sensitivity faced by conventional high-speed cameras.
KAIST (represented by President Kwang Hyung Lee) announced on the 16th of January that a research team led by Professors Ki-Hun Jeong (Department of Bio and Brain Engineering) and Min H. Kim (School of Computing) has developed a novel bio-inspired camera capable of ultra-high-speed imaging with high sensitivity by mimicking the visual structure of insect eyes.
High-quality imaging under high-speed and low-light conditions is a critical challenge in many applications. While conventional high-speed cameras excel in capturing fast motion, their sensitivity decreases as frame rates increase because the time available to collect light is reduced.
To address this issue, the research team adopted an approach similar to insect vision, utilizing multiple optical channels and temporal summation. Unlike traditional monocular camera systems, the bio-inspired camera employs a compound-eye-like structure that allows for the parallel acquisition of frames from different time intervals.
< Figure 1. (A) Vision in a fast-eyed insect. Reflected light from swiftly moving objects sequentially stimulates the photoreceptors along the individual optical channels called ommatidia, of which the visual signals are separately and parallelly processed via the lamina and medulla. Each neural response is temporally summed to enhance the visual signals. The parallel processing and temporal summation allow fast and low-light imaging in dim light. (B) High-speed and high-sensitivity microlens array camera (HS-MAC). A rolling shutter image sensor is utilized to simultaneously acquire multiple frames by channel division, and temporal summation is performed in parallel to realize high speed and sensitivity even in a low-light environment. In addition, the frame components of a single fragmented array image are stitched into a single blurred frame, which is subsequently deblurred by compressive image reconstruction. >
During this process, light is accumulated over overlapping time periods for each frame, increasing the signal-to-noise ratio. The researchers demonstrated that their bio-inspired camera could capture objects up to 40 times dimmer than those detectable by conventional high-speed cameras.
The team also introduced a "channel-splitting" technique to significantly enhance the camera's speed, achieving frame rates thousands of times faster than those supported by the image sensors used in packaging. Additionally, a "compressed image restoration" algorithm was employed to eliminate blur caused by frame integration and reconstruct sharp images.
The resulting bio-inspired camera is less than one millimeter thick and extremely compact, capable of capturing 9,120 frames per second while providing clear images in low-light conditions.
< Figure 2. A high-speed, high-sensitivity biomimetic camera packaged in an image sensor. It is made small enough to fit on a finger, with a thickness of less than 1 mm. >
The research team plans to extend this technology to develop advanced image processing algorithms for 3D imaging and super-resolution imaging, aiming for applications in biomedical imaging, mobile devices, and various other camera technologies.
Hyun-Kyung Kim, a doctoral student in the Department of Bio and Brain Engineering at KAIST and the study's first author, stated, “We have experimentally validated that the insect-eye-inspired camera delivers outstanding performance in high-speed and low-light imaging despite its small size. This camera opens up possibilities for diverse applications in portable camera systems, security surveillance, and medical imaging.”
< Figure 3. Rotating plate and flame captured using the high-speed, high-sensitivity biomimetic camera. The rotating plate at 1,950 rpm was accurately captured at 9,120 fps. In addition, the pinch-off of the flame with a faint intensity of 880 µlux was accurately captured at 1,020 fps. >
This research was published in the international journal Science Advances in January 2025 (Paper Title: “Biologically-inspired microlens array camera for high-speed and high-sensitivity imaging”).
DOI: https://doi.org/10.1126/sciadv.ads3389
This study was supported by the Korea Research Institute for Defense Technology Planning and Advancement (KRIT) of the Defense Acquisition Program Administration (DAPA), the Ministry of Science and ICT, and the Ministry of Trade, Industry and Energy (MOTIE).
2025.01.16 View 8756