Synthetic+biology
-
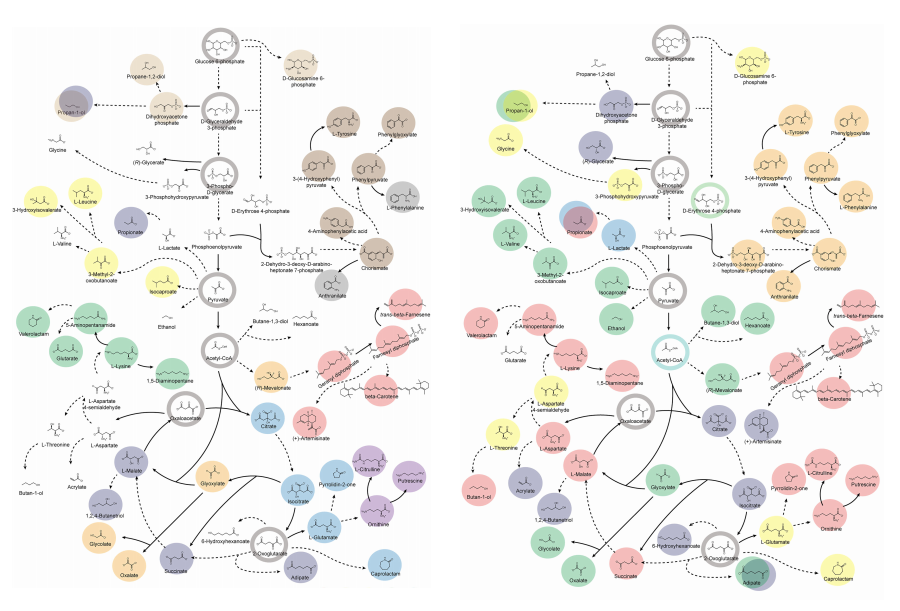 KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3839
KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3839 -
 VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 11965
VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 11965 -
 E. coli Engineered to Grow on CO₂ and Formic Acid as Sole Carbon Sources
- An E. coli strain that can grow to a relatively high cell density solely on CO₂ and formic acid was developed by employing metabolic engineering. -
Most biorefinery processes have relied on the use of biomass as a raw material for the production of chemicals and materials. Even though the use of CO₂ as a carbon source in biorefineries is desirable, it has not been possible to make common microbial strains such as E. coli grow on CO₂.
Now, a metabolic engineering research group at KAIST has developed a strategy to grow an E. coli strain to higher cell density solely on CO₂ and formic acid. Formic acid is a one carbon carboxylic acid, and can be easily produced from CO₂ using a variety of methods. Since it is easier to store and transport than CO₂, formic acid can be considered a good liquid-form alternative of CO₂.
With support from the C1 Gas Refinery R&D Center and the Ministry of Science and ICT, a research team led by Distinguished Professor Sang Yup Lee stepped up their work to develop an engineered E. coli strain capable of growing up to 11-fold higher cell density than those previously reported, using CO₂ and formic acid as sole carbon sources. This work was published in Nature Microbiology on September 28.
Despite the recent reports by several research groups on the development of E. coli strains capable of growing on CO₂ and formic acid, the maximum cell growth remained too low (optical density of around 1) and thus the production of chemicals from CO₂ and formic acid has been far from realized.
The team previously reported the reconstruction of the tetrahydrofolate cycle and reverse glycine cleavage pathway to construct an engineered E. coli strain that can sustain growth on CO₂ and formic acid. To further enhance the growth, the research team introduced the previously designed synthetic CO₂ and formic acid assimilation pathway, and two formate dehydrogenases.
Metabolic fluxes were also fine-tuned, the gluconeogenic flux enhanced, and the levels of cytochrome bo3 and bd-I ubiquinol oxidase for ATP generation were optimized. This engineered E. coli strain was able to grow to a relatively high OD600 of 7~11, showing promise as a platform strain growing solely on CO₂ and formic acid.
Professor Lee said, “We engineered E. coli that can grow to a higher cell density only using CO₂ and formic acid. We think that this is an important step forward, but this is not the end. The engineered strain we developed still needs further engineering so that it can grow faster to a much higher density.”
Professor Lee’s team is continuing to develop such a strain. “In the future, we would be delighted to see the production of chemicals from an engineered E. coli strain using CO₂ and formic acid as sole carbon sources,” he added.
-Profile:Distinguished Professor Sang Yup Leehttp://mbel.kaist.ac.krDepartment of Chemical and Biomolecular EngineeringKAIST
2020.09.29 View 12651
E. coli Engineered to Grow on CO₂ and Formic Acid as Sole Carbon Sources
- An E. coli strain that can grow to a relatively high cell density solely on CO₂ and formic acid was developed by employing metabolic engineering. -
Most biorefinery processes have relied on the use of biomass as a raw material for the production of chemicals and materials. Even though the use of CO₂ as a carbon source in biorefineries is desirable, it has not been possible to make common microbial strains such as E. coli grow on CO₂.
Now, a metabolic engineering research group at KAIST has developed a strategy to grow an E. coli strain to higher cell density solely on CO₂ and formic acid. Formic acid is a one carbon carboxylic acid, and can be easily produced from CO₂ using a variety of methods. Since it is easier to store and transport than CO₂, formic acid can be considered a good liquid-form alternative of CO₂.
With support from the C1 Gas Refinery R&D Center and the Ministry of Science and ICT, a research team led by Distinguished Professor Sang Yup Lee stepped up their work to develop an engineered E. coli strain capable of growing up to 11-fold higher cell density than those previously reported, using CO₂ and formic acid as sole carbon sources. This work was published in Nature Microbiology on September 28.
Despite the recent reports by several research groups on the development of E. coli strains capable of growing on CO₂ and formic acid, the maximum cell growth remained too low (optical density of around 1) and thus the production of chemicals from CO₂ and formic acid has been far from realized.
The team previously reported the reconstruction of the tetrahydrofolate cycle and reverse glycine cleavage pathway to construct an engineered E. coli strain that can sustain growth on CO₂ and formic acid. To further enhance the growth, the research team introduced the previously designed synthetic CO₂ and formic acid assimilation pathway, and two formate dehydrogenases.
Metabolic fluxes were also fine-tuned, the gluconeogenic flux enhanced, and the levels of cytochrome bo3 and bd-I ubiquinol oxidase for ATP generation were optimized. This engineered E. coli strain was able to grow to a relatively high OD600 of 7~11, showing promise as a platform strain growing solely on CO₂ and formic acid.
Professor Lee said, “We engineered E. coli that can grow to a higher cell density only using CO₂ and formic acid. We think that this is an important step forward, but this is not the end. The engineered strain we developed still needs further engineering so that it can grow faster to a much higher density.”
Professor Lee’s team is continuing to develop such a strain. “In the future, we would be delighted to see the production of chemicals from an engineered E. coli strain using CO₂ and formic acid as sole carbon sources,” he added.
-Profile:Distinguished Professor Sang Yup Leehttp://mbel.kaist.ac.krDepartment of Chemical and Biomolecular EngineeringKAIST
2020.09.29 View 12651 -
 Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 39762
Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 39762 -
 Efficiently Producing Fatty Acids and Biofuels from Glucose
Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production.
The newly developed strain, created by Distinguished Professor Sang Yup Lee and his team, showed the highest efficiency in producing fatty acids and biodiesels ever reported. It will be expected to serve as a new platform to sustainably produce a wide array of fatty acid-based products from glucose and other carbon substrates.
Fossil fuels, which have long been energy resources for our daily lives, are now facing serious challenges: depletion of their reserves and their role in global warming. The production of sustainable bio-based renewable energy has emerged as an essential alternative and many studies to replace fossil fuels are underway. One of the representative examples is biodiesel. Currently, it is mainly being produced through the transesterification of vegetable oils or animal fats.
The research team engineered oleaginous microorganisms, Rhodococcus opacus, to produce fatty acids and their derivatives that can be used as biodiesel from glucose, one of the most abundant and cheap sugars derived from non-edible biomass.
Professor Lee’s team has already engineered Escherichia coli to produce short-chain hydrocarbons, which can be used as gasoline (published in Nature as the cover paper in 2013). However, the production efficiency of the short-chain hydrocarbons using E. coli (0.58 g/L) fell short of the levels required for commercialization.
To overcome these issues, the team employed oil-accumulating Rhodococcus opacus as a host strain in this study. First, the team optimized the cultivation conditions of Rhodococcus opacus to maximize the accumulation of oil (triacylglycerol), which serves as a precursor for the biosynthesis of fatty acids and their derivatives. Then, they systematically analyzed the metabolism of the strain and redesigned it to enable higher levels of fatty acids and two kinds of fatty acid-derived biodiesels (fatty acid ethyl esters and long-chain hydrocarbons) to be produced.
They found that the resulting strains produced 50.2, 21.3, and 5.2 g/L of fatty acids, fatty acid ethyl esters, and long-chain hydrocarbons, respectively. These are all the highest concentrations ever reported by microbial fermentations. It is expected that these strains can contribute to the future industrialization of microbial-based biodiesel production.
“This technology creates fatty acids and biodiesel with high efficiency by utilizing lignocellulose, one of the most abundant resources on the Earth, without depending on fossil fuels and vegetable or animal oils. This will provide new opportunities for oil and petroleum industries, which have long relied on fossil fuels, to turn to sustainable and eco-friendly biotechnologies,” said Professor Lee.
This paper titled “Engineering of an oleaginous bacterium for the production of fatty acids and fuels” was published in Nature Chemical Biology on June 17.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557).
(Figure: Metabolic engineering for the production of free fatty acids (FFAs), fatty acid ethyl esters (FAEEs), and long-chain hydrocarbons (LCHCs) in Rhodococcus opacus PD630. Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production.)
# # #
Source:
Hye Mi Kim, Tong Un Chae, So Young Choi, Won Jun Kim and Sang Yup Lee. Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nature Chemical Biology ( https://www.nature.com/nchembio/ ) DOI: 10.1038/s41589-019-0295-5
Profile
Dr. Sang Yup Lee
leesy@kaist.ac.kr
Distinguished Professor at the Department of Chemical and Biomolecular Engineering
KAIST
2019.06.19 View 51502
Efficiently Producing Fatty Acids and Biofuels from Glucose
Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production.
The newly developed strain, created by Distinguished Professor Sang Yup Lee and his team, showed the highest efficiency in producing fatty acids and biodiesels ever reported. It will be expected to serve as a new platform to sustainably produce a wide array of fatty acid-based products from glucose and other carbon substrates.
Fossil fuels, which have long been energy resources for our daily lives, are now facing serious challenges: depletion of their reserves and their role in global warming. The production of sustainable bio-based renewable energy has emerged as an essential alternative and many studies to replace fossil fuels are underway. One of the representative examples is biodiesel. Currently, it is mainly being produced through the transesterification of vegetable oils or animal fats.
The research team engineered oleaginous microorganisms, Rhodococcus opacus, to produce fatty acids and their derivatives that can be used as biodiesel from glucose, one of the most abundant and cheap sugars derived from non-edible biomass.
Professor Lee’s team has already engineered Escherichia coli to produce short-chain hydrocarbons, which can be used as gasoline (published in Nature as the cover paper in 2013). However, the production efficiency of the short-chain hydrocarbons using E. coli (0.58 g/L) fell short of the levels required for commercialization.
To overcome these issues, the team employed oil-accumulating Rhodococcus opacus as a host strain in this study. First, the team optimized the cultivation conditions of Rhodococcus opacus to maximize the accumulation of oil (triacylglycerol), which serves as a precursor for the biosynthesis of fatty acids and their derivatives. Then, they systematically analyzed the metabolism of the strain and redesigned it to enable higher levels of fatty acids and two kinds of fatty acid-derived biodiesels (fatty acid ethyl esters and long-chain hydrocarbons) to be produced.
They found that the resulting strains produced 50.2, 21.3, and 5.2 g/L of fatty acids, fatty acid ethyl esters, and long-chain hydrocarbons, respectively. These are all the highest concentrations ever reported by microbial fermentations. It is expected that these strains can contribute to the future industrialization of microbial-based biodiesel production.
“This technology creates fatty acids and biodiesel with high efficiency by utilizing lignocellulose, one of the most abundant resources on the Earth, without depending on fossil fuels and vegetable or animal oils. This will provide new opportunities for oil and petroleum industries, which have long relied on fossil fuels, to turn to sustainable and eco-friendly biotechnologies,” said Professor Lee.
This paper titled “Engineering of an oleaginous bacterium for the production of fatty acids and fuels” was published in Nature Chemical Biology on June 17.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557).
(Figure: Metabolic engineering for the production of free fatty acids (FFAs), fatty acid ethyl esters (FAEEs), and long-chain hydrocarbons (LCHCs) in Rhodococcus opacus PD630. Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production.)
# # #
Source:
Hye Mi Kim, Tong Un Chae, So Young Choi, Won Jun Kim and Sang Yup Lee. Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nature Chemical Biology ( https://www.nature.com/nchembio/ ) DOI: 10.1038/s41589-019-0295-5
Profile
Dr. Sang Yup Lee
leesy@kaist.ac.kr
Distinguished Professor at the Department of Chemical and Biomolecular Engineering
KAIST
2019.06.19 View 51502 -
 Engineered Microbial Production of Grape Flavoring
(Image 1: Engineered bacteria that produce grape flavoring.)
Researchers report a microbial method for producing an artificial grape flavor. Methyl anthranilate (MANT) is a common grape flavoring and odorant compound currently produced through a petroleum-based process that uses large volumes of toxic acid catalysts.
Professor Sang-Yup Lee’s team at the Department of Chemical and Biomolecular Engineering demonstrated production of MANT, a naturally occurring compound, via engineered bacteria. The authors engineered strains of Escherichia coli and Corynebacetrium glutamicum to produce MANT through a plant-based engineered metabolic pathway.
The authors tuned the bacterial metabolic pathway by optimizing the levels of AAMT1, the key enzyme in the process. To maximize production of MANT, the authors tested six strategies, including increasing the supply of a precursor compound and enhancing the availability of a co-substrate. The most productive strategy proved to be a two-phase extractive culture, in which MANT was extracted into a solvent. This strategy produced MANT on the scale of 4.47 to 5.74 grams per liter, a significant amount, considering that engineered microbes produce most natural products at a scale of milligrams or micrograms per liter.
According to the authors, the results suggest that MANT and other related molecules produced through industrial processes can be produced at scale by engineered microbes in a manner that would allow them to be marketed as natural one, instead of artificial one.
This study, featured at the Proceeding of the National Academy of Sciences of the USA on May 13, was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT.
(Image 2. Overview of the strategies applied for the microbial production of grape flavoring.)
2019.05.15 View 57427
Engineered Microbial Production of Grape Flavoring
(Image 1: Engineered bacteria that produce grape flavoring.)
Researchers report a microbial method for producing an artificial grape flavor. Methyl anthranilate (MANT) is a common grape flavoring and odorant compound currently produced through a petroleum-based process that uses large volumes of toxic acid catalysts.
Professor Sang-Yup Lee’s team at the Department of Chemical and Biomolecular Engineering demonstrated production of MANT, a naturally occurring compound, via engineered bacteria. The authors engineered strains of Escherichia coli and Corynebacetrium glutamicum to produce MANT through a plant-based engineered metabolic pathway.
The authors tuned the bacterial metabolic pathway by optimizing the levels of AAMT1, the key enzyme in the process. To maximize production of MANT, the authors tested six strategies, including increasing the supply of a precursor compound and enhancing the availability of a co-substrate. The most productive strategy proved to be a two-phase extractive culture, in which MANT was extracted into a solvent. This strategy produced MANT on the scale of 4.47 to 5.74 grams per liter, a significant amount, considering that engineered microbes produce most natural products at a scale of milligrams or micrograms per liter.
According to the authors, the results suggest that MANT and other related molecules produced through industrial processes can be produced at scale by engineered microbes in a manner that would allow them to be marketed as natural one, instead of artificial one.
This study, featured at the Proceeding of the National Academy of Sciences of the USA on May 13, was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT.
(Image 2. Overview of the strategies applied for the microbial production of grape flavoring.)
2019.05.15 View 57427 -
 Professor Hee-Sung Park Named Scientist of May
(Professor Hee-Sung Park)
Professor Hee-Sung Park from the Department of Chemistry was named ‘Scientist of May’ sponsored by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Park was honored in recognition of his developing a tool to engineer designer proteins via diverse chemical modifications. This approach provides a novel platform for investigating numerous diseases such as cancer and dementia.
His research focuses on the production of synthetic proteins and the generation of diverse protein functions as well as the designing and engineering of new translation machinery for genetic code expansion, and the application of synthetic biology techniques for basic cell biology and applied medical science.
Post-translational modifications (PTMs) are constantly taking place during or after protein biosynthesis. PTMs play a vital role in expanding protein functional diversity and, as a result, critically affect numerous biological processes. Abnormal PTMs have been known to trigger various diseases including cancer and dementia. Therefore, this technology enables proteins to reproduce with specific modifications at selected residues and will significantly help establish experimental strategies to investigate fundamental biological mechanisms including the development of targeted cancer therapies.
Professor Park also received 10 million KRW in prize money.
2018.05.04 View 12038
Professor Hee-Sung Park Named Scientist of May
(Professor Hee-Sung Park)
Professor Hee-Sung Park from the Department of Chemistry was named ‘Scientist of May’ sponsored by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Park was honored in recognition of his developing a tool to engineer designer proteins via diverse chemical modifications. This approach provides a novel platform for investigating numerous diseases such as cancer and dementia.
His research focuses on the production of synthetic proteins and the generation of diverse protein functions as well as the designing and engineering of new translation machinery for genetic code expansion, and the application of synthetic biology techniques for basic cell biology and applied medical science.
Post-translational modifications (PTMs) are constantly taking place during or after protein biosynthesis. PTMs play a vital role in expanding protein functional diversity and, as a result, critically affect numerous biological processes. Abnormal PTMs have been known to trigger various diseases including cancer and dementia. Therefore, this technology enables proteins to reproduce with specific modifications at selected residues and will significantly help establish experimental strategies to investigate fundamental biological mechanisms including the development of targeted cancer therapies.
Professor Park also received 10 million KRW in prize money.
2018.05.04 View 12038 -
 Distinguished Professor Lee Elected to the NAS
Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering was elected as a foreign associate to the US National Academy of Sciences (NAS) on May 2. The National Academy of Sciences elected 84 new members and 21 foreign associates in recognition of their distinguished and continuing achievements in their original research. Election to the Academy is widely regarded as one of the highest honors that a scientist can receive.
Professor Lee was also elected in 2010 as a member of the US National Academy of Engineering (NAE) for his leadership in microbial biotechnology and metabolic engineering, including the development of fermentation processes for biodegradable polymers and organic acids. Until 2016, there are only 12 people worldwide who are foreign associates of both NAS and NAE.
He is the first Korean elected to both prestigious academies, the NAS and the NAE in the US. Professor Lee is currently the dean of KAIST Institutes, the world leading institute for multi-and interdisciplinary research. He is also serving as co-chair of the Global Council on Biotechnology and member of the Global Future Council on the Fourth Industrial Revolution, the World Economic Forum.
2017.05.16 View 11294
Distinguished Professor Lee Elected to the NAS
Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering was elected as a foreign associate to the US National Academy of Sciences (NAS) on May 2. The National Academy of Sciences elected 84 new members and 21 foreign associates in recognition of their distinguished and continuing achievements in their original research. Election to the Academy is widely regarded as one of the highest honors that a scientist can receive.
Professor Lee was also elected in 2010 as a member of the US National Academy of Engineering (NAE) for his leadership in microbial biotechnology and metabolic engineering, including the development of fermentation processes for biodegradable polymers and organic acids. Until 2016, there are only 12 people worldwide who are foreign associates of both NAS and NAE.
He is the first Korean elected to both prestigious academies, the NAS and the NAE in the US. Professor Lee is currently the dean of KAIST Institutes, the world leading institute for multi-and interdisciplinary research. He is also serving as co-chair of the Global Council on Biotechnology and member of the Global Future Council on the Fourth Industrial Revolution, the World Economic Forum.
2017.05.16 View 11294 -
 Nature Biotechnology Nominates Sang Yup Lee of KAIST for Top 20 Translational Researchers of 2014
Nature Biotechnology, recognized as the most prestigious journal in the field of biotechnology, has released today its list of the Top 20 Translational Researchers of 2014. Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST (Korea Advanced Institute of Science and Technology) ranked seventh in the list. He is the only Asian researcher listed.
The journal, in partnership with IP Checkups, a patent analytics firm, presents an annual ranking of researchers based on their paper and patent output. The list includes, among others, each researcher’s most-cited patent in the past five years and their H index, a measurement to evaluate the impact of a researcher’s published work utilizing citation analysis. (More details can be found at http://www.nature.com/bioent/2015/150801/full/bioe.2015.9.html.)
American institutions made up the majority of the list, with 18 universities and research institutes, and the remainder was filled by KAIST in Korea and the Commonwealth Scientific and Industrial Research Organization (CSIRO) in Australia.
Globally known as a leading researcher in systems metabolic engineering, Professor Lee has published more than 500 journal papers and 580 patents. He has received many awards, including the Citation Classic Award, Elmer Gaden Award, Merck Metabolic Engineering Award, ACS Marvin Johnson Award, SIMB Charles Thom Award, POSCO TJ Park Prize, Amgen Biochemical Engineering Award, and the Ho Am Prize in Engineering.
2015.08.27 View 12304
Nature Biotechnology Nominates Sang Yup Lee of KAIST for Top 20 Translational Researchers of 2014
Nature Biotechnology, recognized as the most prestigious journal in the field of biotechnology, has released today its list of the Top 20 Translational Researchers of 2014. Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST (Korea Advanced Institute of Science and Technology) ranked seventh in the list. He is the only Asian researcher listed.
The journal, in partnership with IP Checkups, a patent analytics firm, presents an annual ranking of researchers based on their paper and patent output. The list includes, among others, each researcher’s most-cited patent in the past five years and their H index, a measurement to evaluate the impact of a researcher’s published work utilizing citation analysis. (More details can be found at http://www.nature.com/bioent/2015/150801/full/bioe.2015.9.html.)
American institutions made up the majority of the list, with 18 universities and research institutes, and the remainder was filled by KAIST in Korea and the Commonwealth Scientific and Industrial Research Organization (CSIRO) in Australia.
Globally known as a leading researcher in systems metabolic engineering, Professor Lee has published more than 500 journal papers and 580 patents. He has received many awards, including the Citation Classic Award, Elmer Gaden Award, Merck Metabolic Engineering Award, ACS Marvin Johnson Award, SIMB Charles Thom Award, POSCO TJ Park Prize, Amgen Biochemical Engineering Award, and the Ho Am Prize in Engineering.
2015.08.27 View 12304 -
 Professor Sang-Yup Lee Receives the Order of Service Merit Red Stripes from the Korean Government
The government of the Republic of Korea named Professor Sang-Yup Lee of the Department of Chemical and Bio-molecular Engineering at KAIST as the fiftieth recipient of the Order of Service Merit Red Stripes on May 19, 2015.
This medal is awarded to government employees, officials, and teachers in recognition of their contributions to public services including education.
Professor Lee is regarded as a leading scientist in the field of metabolic engineering, genomics, proteomics, metabolomics, and bioinformatics on microorganism producing various primary and secondary metabolites. He contributed significantly to the advancement of bio-based engineering research in Korea.
In addition, his research in microorganism metabolic engineering propelled him to the front of his field, making him the world’s founder of systems metabolic engineering, inventing numerous technologies in strain development.
Professor Lee has received many patent rights in bioprocess engineering. While at KAIST, he applied for 585 patents and registered 227 patents. In particular, he has applied for 135 patents and registered 99 patents in the past five years, successfully turning research results into commercial applications.
Professor Lee said, “I’m glad to contribute to the development of Korean science and technology as a researcher and teacher. I would like to share this honor with my students, master’s and doctoral students in particular, because without their support, it wouldn’t have been possible to pull off the highest level of research results recognized by this medal.”
2015.05.21 View 9821
Professor Sang-Yup Lee Receives the Order of Service Merit Red Stripes from the Korean Government
The government of the Republic of Korea named Professor Sang-Yup Lee of the Department of Chemical and Bio-molecular Engineering at KAIST as the fiftieth recipient of the Order of Service Merit Red Stripes on May 19, 2015.
This medal is awarded to government employees, officials, and teachers in recognition of their contributions to public services including education.
Professor Lee is regarded as a leading scientist in the field of metabolic engineering, genomics, proteomics, metabolomics, and bioinformatics on microorganism producing various primary and secondary metabolites. He contributed significantly to the advancement of bio-based engineering research in Korea.
In addition, his research in microorganism metabolic engineering propelled him to the front of his field, making him the world’s founder of systems metabolic engineering, inventing numerous technologies in strain development.
Professor Lee has received many patent rights in bioprocess engineering. While at KAIST, he applied for 585 patents and registered 227 patents. In particular, he has applied for 135 patents and registered 99 patents in the past five years, successfully turning research results into commercial applications.
Professor Lee said, “I’m glad to contribute to the development of Korean science and technology as a researcher and teacher. I would like to share this honor with my students, master’s and doctoral students in particular, because without their support, it wouldn’t have been possible to pull off the highest level of research results recognized by this medal.”
2015.05.21 View 9821 -
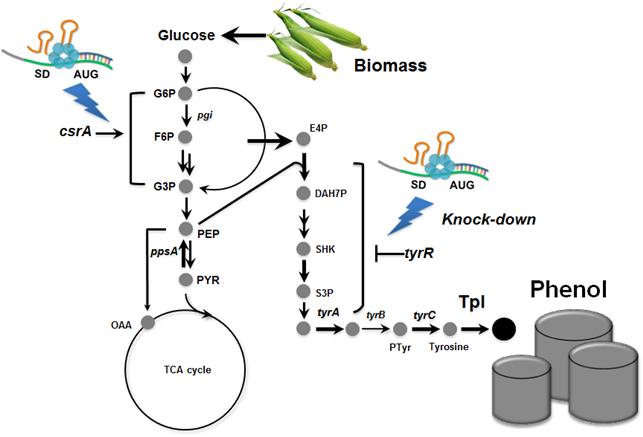 Metabolically engineered E. coli producing phenol
Many chemicals we use in everyday life are derived from fossil resources. Due to the increasing concerns on the use of fossil resources, there has been much interest in producing chemicals from renewable resources through biotechnology.
Phenol is an important commodity chemical, and is a starting material for the production of numerous industrial chemicals and polymers, including bisphenol A and phenolic resins, and others. At present, the production of phenol entirely depends on the chemical synthesis from benzene, and its annual production exceeds 8 million tons worldwide. Microbial production of phenol seems to be a non-viable process considering the high toxicity of phenol to the cell.
In the paper published online in Biotechnology Journal, a Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the successful development of an engineered Escherichia coli (E. coli) strain which can produce phenol from glucose. E. coli has been a workhorse for biological production of various value-added compounds such as succinic acid and 1,4-butanediol in industrial scale. However, due to its low tolerance to phenol, E. coli was not considered a viable host strain for the biological production of phenol.
Professor Lee"s team, a leading research group in metabolic engineering, noted the genetic and physiological differences of various E. coli strains and investigated 18 different E. coli strains with respect to phenol tolerance and engineered all of the 18 strains simultaneously. If the traditional genetic engineering methods were used, this work would have taken years to do. To overcome this challenge, the research team used synthetic small RNA (sRNA) technology they recently developed (Nature Biotechnology, vol 31, pp 170-174, 2013). The sRNA technology allowed the team to screen 18 E. coli strains with respect to the phenol tolerance, and the activities of the metabolic pathway and enzyme involved in the production of phenol. The research team also metabolically engineered the E. coli strains to increase carbon flux toward phenol and finally generated an engineered E. coli strain which can produce phenol from glucose.
Furthermore, the team developed a biphasic extractive fermentation process to minimize the toxicity of phenol to E. coli cells. Glycerol tributyrate was found to have low toxicity to E. coli and allowed efficient extraction of phenol from the culture broth. Through the biphasic fed-batch fermentation using glycerol tributyrate as an in situ extractant, the final engineered E. coli strain produced phenol to the highest titer and productivity reported (3.8 g/L and 0.18 g/L/h, respectively). The strategy used for the strain development and the fermentation process will serve as a framework for metabolic engineering of microorganisms for the production of toxic chemicals from renewable resources.
This work was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT & Future Planning through the National Research Foundation of Korea.
Process of Phenol Production
2013.11.05 View 11535
Metabolically engineered E. coli producing phenol
Many chemicals we use in everyday life are derived from fossil resources. Due to the increasing concerns on the use of fossil resources, there has been much interest in producing chemicals from renewable resources through biotechnology.
Phenol is an important commodity chemical, and is a starting material for the production of numerous industrial chemicals and polymers, including bisphenol A and phenolic resins, and others. At present, the production of phenol entirely depends on the chemical synthesis from benzene, and its annual production exceeds 8 million tons worldwide. Microbial production of phenol seems to be a non-viable process considering the high toxicity of phenol to the cell.
In the paper published online in Biotechnology Journal, a Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the successful development of an engineered Escherichia coli (E. coli) strain which can produce phenol from glucose. E. coli has been a workhorse for biological production of various value-added compounds such as succinic acid and 1,4-butanediol in industrial scale. However, due to its low tolerance to phenol, E. coli was not considered a viable host strain for the biological production of phenol.
Professor Lee"s team, a leading research group in metabolic engineering, noted the genetic and physiological differences of various E. coli strains and investigated 18 different E. coli strains with respect to phenol tolerance and engineered all of the 18 strains simultaneously. If the traditional genetic engineering methods were used, this work would have taken years to do. To overcome this challenge, the research team used synthetic small RNA (sRNA) technology they recently developed (Nature Biotechnology, vol 31, pp 170-174, 2013). The sRNA technology allowed the team to screen 18 E. coli strains with respect to the phenol tolerance, and the activities of the metabolic pathway and enzyme involved in the production of phenol. The research team also metabolically engineered the E. coli strains to increase carbon flux toward phenol and finally generated an engineered E. coli strain which can produce phenol from glucose.
Furthermore, the team developed a biphasic extractive fermentation process to minimize the toxicity of phenol to E. coli cells. Glycerol tributyrate was found to have low toxicity to E. coli and allowed efficient extraction of phenol from the culture broth. Through the biphasic fed-batch fermentation using glycerol tributyrate as an in situ extractant, the final engineered E. coli strain produced phenol to the highest titer and productivity reported (3.8 g/L and 0.18 g/L/h, respectively). The strategy used for the strain development and the fermentation process will serve as a framework for metabolic engineering of microorganisms for the production of toxic chemicals from renewable resources.
This work was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT & Future Planning through the National Research Foundation of Korea.
Process of Phenol Production
2013.11.05 View 11535 -
 Production of chemicals without petroleum
Systems metabolic engineering of microorganisms allows efficient production of natural and non-natural chemicals from renewable non-food biomass
In our everyday life, we use gasoline, diesel, plastics, rubbers, and numerous chemicals that are derived from fossil oil through petrochemical refinery processes. However, this is not sustainable due to the limited nature of fossil resources. Furthermore, our world is facing problems associated with climate change and other environmental problems due to the increasing use of fossil resources. One solution to address above problems is the use of renewable non-food biomass for the production of chemicals, fuels and materials through biorefineries. Microorganisms are used as biocatalysts for converting biomass to the products of interest. However, when microorganisms are isolated from nature, their efficiencies of producing our desired chemicals and materials are rather low. Metabolic engineering is thus performed to improve cellular characteristics to desired levels. Over the last decade, much advances have been made in systems biology that allows system-wide characterization of cellular networks, both qualitatively and quantitatively, followed by whole-cell level engineering based on these findings. Furthermore, rapid advances in synthetic biology allow design and synthesis of fine controlled metabolic and gene regulatory circuits. The strategies and methods of systems biology and synthetic biology are rapidly integrated with metabolic engineering, thus resulting in "systems metabolic engineering".
In the paper published online in Nature Chemical Biology on May 17, Professor Sang Yup Lee and his colleagues at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea present new general strategies of systems metabolic engineering for developing microorganisms for the production of natural and non-natural chemicals from renewable biomass. They first classified the chemicals to be produced into four categories based on whether they have thus far been identified to exist in nature (natural vs. nonnatural) and whether they can be produced by inherent pathways of microorganisms (inherent, noninherent, or created): natural-inherent, natural-noninherent, non-natural-noninherent, and non-natural-created ones. General strategies for systems metabolic engineering of microorganisms for the production of these chemicals using various tools and methods based on omics, genome-scale metabolic modeling and simulation, evolutionary engineering, synthetic biology are suggested with relevant examples. For the production of non-natural chemicals, strategies for the construction of synthetic metabolic pathways are also suggested. Having collected diverse tools and methods for systems metabolic engineering, authors also suggest how to use them and their possible limitations.
Professor Sang Yup Lee said "It is expected that increasing number of chemicals and materials will be produced through biorefineries. We are now equipped with new strategies for developing microbial strains that can produce our desired products at very high efficiencies, thus allowing cost competitiveness to those produced by petrochemical refineries."
Editor of Nature Chemical Biology, Dr. Catherine Goodman, said "It is exciting to see how quickly science is progressing in this field – ideas that used to be science fiction are taking shape in research labs and biorefineries. The article by Professor Lee and his colleagues not only highlights the most advanced techniques and strategies available, but offers critical advice to progress the field as a whole."
The works of Professor Lee have been supported by the Advanced Biomass Center and Intelligent Synthetic Biology Center of Global Frontier Program from the Korean Ministry of Education, Science and Technology through National Research Foundation.
Contact: Dr. Sang Yup Lee, Distinguished Professor and Dean, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
2012.05.23 View 15481
Production of chemicals without petroleum
Systems metabolic engineering of microorganisms allows efficient production of natural and non-natural chemicals from renewable non-food biomass
In our everyday life, we use gasoline, diesel, plastics, rubbers, and numerous chemicals that are derived from fossil oil through petrochemical refinery processes. However, this is not sustainable due to the limited nature of fossil resources. Furthermore, our world is facing problems associated with climate change and other environmental problems due to the increasing use of fossil resources. One solution to address above problems is the use of renewable non-food biomass for the production of chemicals, fuels and materials through biorefineries. Microorganisms are used as biocatalysts for converting biomass to the products of interest. However, when microorganisms are isolated from nature, their efficiencies of producing our desired chemicals and materials are rather low. Metabolic engineering is thus performed to improve cellular characteristics to desired levels. Over the last decade, much advances have been made in systems biology that allows system-wide characterization of cellular networks, both qualitatively and quantitatively, followed by whole-cell level engineering based on these findings. Furthermore, rapid advances in synthetic biology allow design and synthesis of fine controlled metabolic and gene regulatory circuits. The strategies and methods of systems biology and synthetic biology are rapidly integrated with metabolic engineering, thus resulting in "systems metabolic engineering".
In the paper published online in Nature Chemical Biology on May 17, Professor Sang Yup Lee and his colleagues at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea present new general strategies of systems metabolic engineering for developing microorganisms for the production of natural and non-natural chemicals from renewable biomass. They first classified the chemicals to be produced into four categories based on whether they have thus far been identified to exist in nature (natural vs. nonnatural) and whether they can be produced by inherent pathways of microorganisms (inherent, noninherent, or created): natural-inherent, natural-noninherent, non-natural-noninherent, and non-natural-created ones. General strategies for systems metabolic engineering of microorganisms for the production of these chemicals using various tools and methods based on omics, genome-scale metabolic modeling and simulation, evolutionary engineering, synthetic biology are suggested with relevant examples. For the production of non-natural chemicals, strategies for the construction of synthetic metabolic pathways are also suggested. Having collected diverse tools and methods for systems metabolic engineering, authors also suggest how to use them and their possible limitations.
Professor Sang Yup Lee said "It is expected that increasing number of chemicals and materials will be produced through biorefineries. We are now equipped with new strategies for developing microbial strains that can produce our desired products at very high efficiencies, thus allowing cost competitiveness to those produced by petrochemical refineries."
Editor of Nature Chemical Biology, Dr. Catherine Goodman, said "It is exciting to see how quickly science is progressing in this field – ideas that used to be science fiction are taking shape in research labs and biorefineries. The article by Professor Lee and his colleagues not only highlights the most advanced techniques and strategies available, but offers critical advice to progress the field as a whole."
The works of Professor Lee have been supported by the Advanced Biomass Center and Intelligent Synthetic Biology Center of Global Frontier Program from the Korean Ministry of Education, Science and Technology through National Research Foundation.
Contact: Dr. Sang Yup Lee, Distinguished Professor and Dean, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
2012.05.23 View 15481