Nature+Biotechnology
-
 Doctoral Student Receives the Best Paper Award from the International Metabolic Engineering Conference 2016
So Young Choi, a Ph.D. candidate at the Department of Chemical and Biomolecular Engineering at KAIST, received the Student and Young Investigator Poster Award at the 11th International Metabolic Engineering Conference held in Awaji, Japan on June 26-30.
Choi received the award for her research on one-step fermentative production of Poly(lactate-co-glycolate) (PLGA) from carbohydrates in Escherichia coli, which was published in the April 2016 issue of Nature Biotechnology.
In her paper, she presented a novel technology to synthesize PLGA, a non-natural copolymer, through a biological production process. Because of its biodegradability, non-toxicity, and biocompatibility, PLGA is widely used in biomedical and therapeutic applications, including surgical sutures, prosthetic devices, drug delivery, and tissue engineering.
Employing a metabolic engineering approach, Choi manipulated the metabolic pathway of an Escherichia coli bacterium to convert glucose and xylose into the biosynthesis of PLGA within the cell. Previously, PLGA could be obtained only through chemical synthesis.
Choi said, “I’m thrilled to receive an award from a flagship conference of my research field. Mindful of this recognition, I will continue my research to produce meaningful results, thereby contributing to the development of science and technology in Korea.”
The International Metabolic Engineering Conference is a leading professional gathering where state-of-the-art developments and achievements made in the field of metabolic engineering are shared. With the participation of about 400 professionals from all around the world, the conference participants discussed this year’s theme of “Design, Synthesis and System Integration for Metabolic Engineering.”
2016.07.07 View 12443
Doctoral Student Receives the Best Paper Award from the International Metabolic Engineering Conference 2016
So Young Choi, a Ph.D. candidate at the Department of Chemical and Biomolecular Engineering at KAIST, received the Student and Young Investigator Poster Award at the 11th International Metabolic Engineering Conference held in Awaji, Japan on June 26-30.
Choi received the award for her research on one-step fermentative production of Poly(lactate-co-glycolate) (PLGA) from carbohydrates in Escherichia coli, which was published in the April 2016 issue of Nature Biotechnology.
In her paper, she presented a novel technology to synthesize PLGA, a non-natural copolymer, through a biological production process. Because of its biodegradability, non-toxicity, and biocompatibility, PLGA is widely used in biomedical and therapeutic applications, including surgical sutures, prosthetic devices, drug delivery, and tissue engineering.
Employing a metabolic engineering approach, Choi manipulated the metabolic pathway of an Escherichia coli bacterium to convert glucose and xylose into the biosynthesis of PLGA within the cell. Previously, PLGA could be obtained only through chemical synthesis.
Choi said, “I’m thrilled to receive an award from a flagship conference of my research field. Mindful of this recognition, I will continue my research to produce meaningful results, thereby contributing to the development of science and technology in Korea.”
The International Metabolic Engineering Conference is a leading professional gathering where state-of-the-art developments and achievements made in the field of metabolic engineering are shared. With the participation of about 400 professionals from all around the world, the conference participants discussed this year’s theme of “Design, Synthesis and System Integration for Metabolic Engineering.”
2016.07.07 View 12443 -
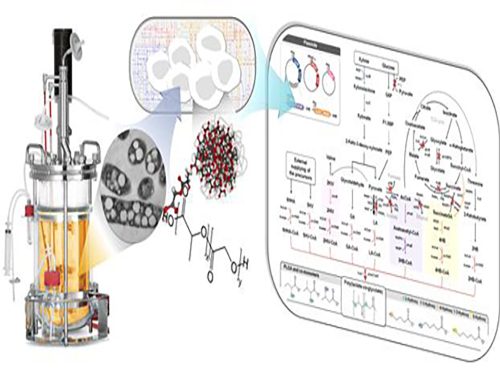 Non-Natural Biomedical Polymers Produced from Microorganisms
KAIST researchers have developed metabolically engineered Escherichia coli strains to synthesize non-natural, biomedically important polymers including poly(lactate-co-glycolate) (PLGA), previously considered impossible to obtain from biobased materials.
Renewable non-food biomass could potentially replace petrochemical raw materials to produce energy sources, useful chemicals, or a vast array of petroleum-based end products such as plastics, lubricants, paints, fertilizers, and vitamin capsules. In recent years, biorefineries which transform non-edible biomass into fuel, heat, power, chemicals, and materials have received a great deal of attention as a sustainable alternative to decreasing the reliance on fossil fuels.
A research team headed by Distinguished Professor Sang Yup Lee of the Chemical and Biomolecular Engineering Department at KAIST has established a biorefinery system to create non-natural polymers from natural sources, allowing various plastics to be made in an environmentally-friendly and sustainable manner. The research results were published online in Nature Biotechnology on March 7, 2016. The print version will be issued in April 2016.
The research team adopted a systems metabolic engineering approach to develop a microorganism that can produce diverse non-natural, biomedically important polymers and succeeded in synthesizing poly(lactate-co-glycolate) (PLGA), a copolymer of two different polymer monomers, lactic and glycolic acid. PLGA is biodegradable, biocompatible, and non-toxic, and has been widely used in biomedical and therapeutic applications such as surgical sutures, prosthetic devices, drug delivery, and tissue engineering.
Inspired by the biosynthesis process for polyhydroxyalkanoates (PHAs), biologically-derived polyesters produced in nature by the bacterial fermentation of sugar or lipids, the research team designed a metabolic pathway for the biosynthesis of PLGA through microbial fermentation directly from carbohydrates in Escherichia coli (E. coli) strains.
The team had previously reported a recombinant E. coli producing PLGA by using the glyoxylate shunt pathway for the generation of glycolate from glucose, which was disclosed in their patents KR10-1575585-0000 (filing date of March 11, 2011), US08883463 and JP5820363. However, they discovered that the polymer content and glycolate fraction of PLGA could not be significantly enhanced via further engineering techniques. Thus, in this research, the team introduced a heterologous pathway to produce glycolate from xylose and succeeded in developing the recombinant E. coli producing PLGA and various novel copolymers much more efficiently.
In order to produce PLGA by microbial fermentation directly from carbohydrates, the team incorporated external and engineered enzymes as catalysts to co-polymerize PLGA while establishing a few additional metabolic pathways for the biosynthesis to produce a range of different non-natural polymers, some for the first time. This bio-based synthetic process for PLGA and other polymers could substitute for the existing complicated chemical production that involves the preparation and purification of precursors, chemical polymerization processes, and the elimination of metal catalysts.
Professor Lee and his team performed in silico genome-scale metabolic simulations of the E. coli cell to predict and analyze changes in the metabolic fluxes of cells which were caused by the introduction of external metabolic pathways. Based on these results, genes are manipulated to optimize metabolic fluxes by eliminating the genes responsible for byproducts formation and enhancing the expression levels of certain genes, thereby achieving the effective production of target polymers as well as stimulating cell growth.
The team utilized the structural basis of broad substrate specificity of the key synthesizing enzyme, PHA synthase, to incorporate various co-monomers with main and side chains of different lengths. These monomers were produced inside the cell by metabolic engineering, and then copolymerized to improve the material properties of PLGA. As a result, a variety of PLGA copolymers with different monomer compositions such as the US Food and Drug Administration (FDA)-approved monomers, 3-hydroxyburate, 4-hydroxyburate, and 6-hydroxyhexanoate, were produced. Newly applied bioplastics such as 5-hydroxyvalerate and 2-hydroxyisovalerate were also made.
The team employed a systems metabolic engineering application which, according to the researchers, is the first successful example of biological production of PGLA and several novel copolymers from renewable biomass by one-step direct fermentation of metabolically engineered E.coli.
Professor Lee said, “We presented important findings that non-natural polymers, such as PLGA which is commonly used for drug delivery or biomedical devices, were produced by a metabolically engineered gut bacterium. Our research is meaningful in that it proposes a platform strategy in metabolic engineering, which can be further utilized in the development of numerous non-natural, useful polymers.”
Director Ilsub Baek at the Platform Technology Division of the Ministry of Science, ICT and Future Planning of Korea, who oversees the Technology Development Program to Solve Climate Change, said, “Professor Lee has led one of our research projects, the Systems Metabolic Engineering for Biorefineries, which began as part of the Ministry’s Technology Development Program to Solve Climate Change. He and his team have continuously achieved promising results and been attracting greater interest from the global scientific community. As climate change technology grows more important, this research on the biological production of non-natural, high value polymers will have a great impact on science and industry.”
The title of the research paper is “One-step Fermentative Production of Poly(lactate-co-glycolate) from Carbohydrates in Escherichia coli (DOI: 10.1038/nbt.3485).” The lead authors are So Young Choi, a Ph.D. candidate in the Department of Chemical and Biomolecular Engineering at KAIST, and Si Jae Park, Assistant Professor of the Environmental Engineering and Energy Department at Myongji University. Won Jun Kim and Jung Eun Yang, both doctoral students in the Department of Chemical and Biomolecular Engineering at KAIST, also participated in the research.
This research was supported by the Technology Development Program to Solve Climate Change’s research project titled “Systems Metabolic Engineering for Biorefineries” from the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea (NRF-2012M1A2A2026556).
Figure: Production of PLGA and Other Non-Natural Copolymers
This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced a variety of PLGAs with different monomer compositions, proposing the chemosynthetic process of non-natural polymers from biomass. The non-natural polymer PLGA and its other copolymers, which were produced by engineered bacteria developed by taking a systems metabolic engineering approach, accumulate in granule forms within a cell.
2016.03.08 View 13185
Non-Natural Biomedical Polymers Produced from Microorganisms
KAIST researchers have developed metabolically engineered Escherichia coli strains to synthesize non-natural, biomedically important polymers including poly(lactate-co-glycolate) (PLGA), previously considered impossible to obtain from biobased materials.
Renewable non-food biomass could potentially replace petrochemical raw materials to produce energy sources, useful chemicals, or a vast array of petroleum-based end products such as plastics, lubricants, paints, fertilizers, and vitamin capsules. In recent years, biorefineries which transform non-edible biomass into fuel, heat, power, chemicals, and materials have received a great deal of attention as a sustainable alternative to decreasing the reliance on fossil fuels.
A research team headed by Distinguished Professor Sang Yup Lee of the Chemical and Biomolecular Engineering Department at KAIST has established a biorefinery system to create non-natural polymers from natural sources, allowing various plastics to be made in an environmentally-friendly and sustainable manner. The research results were published online in Nature Biotechnology on March 7, 2016. The print version will be issued in April 2016.
The research team adopted a systems metabolic engineering approach to develop a microorganism that can produce diverse non-natural, biomedically important polymers and succeeded in synthesizing poly(lactate-co-glycolate) (PLGA), a copolymer of two different polymer monomers, lactic and glycolic acid. PLGA is biodegradable, biocompatible, and non-toxic, and has been widely used in biomedical and therapeutic applications such as surgical sutures, prosthetic devices, drug delivery, and tissue engineering.
Inspired by the biosynthesis process for polyhydroxyalkanoates (PHAs), biologically-derived polyesters produced in nature by the bacterial fermentation of sugar or lipids, the research team designed a metabolic pathway for the biosynthesis of PLGA through microbial fermentation directly from carbohydrates in Escherichia coli (E. coli) strains.
The team had previously reported a recombinant E. coli producing PLGA by using the glyoxylate shunt pathway for the generation of glycolate from glucose, which was disclosed in their patents KR10-1575585-0000 (filing date of March 11, 2011), US08883463 and JP5820363. However, they discovered that the polymer content and glycolate fraction of PLGA could not be significantly enhanced via further engineering techniques. Thus, in this research, the team introduced a heterologous pathway to produce glycolate from xylose and succeeded in developing the recombinant E. coli producing PLGA and various novel copolymers much more efficiently.
In order to produce PLGA by microbial fermentation directly from carbohydrates, the team incorporated external and engineered enzymes as catalysts to co-polymerize PLGA while establishing a few additional metabolic pathways for the biosynthesis to produce a range of different non-natural polymers, some for the first time. This bio-based synthetic process for PLGA and other polymers could substitute for the existing complicated chemical production that involves the preparation and purification of precursors, chemical polymerization processes, and the elimination of metal catalysts.
Professor Lee and his team performed in silico genome-scale metabolic simulations of the E. coli cell to predict and analyze changes in the metabolic fluxes of cells which were caused by the introduction of external metabolic pathways. Based on these results, genes are manipulated to optimize metabolic fluxes by eliminating the genes responsible for byproducts formation and enhancing the expression levels of certain genes, thereby achieving the effective production of target polymers as well as stimulating cell growth.
The team utilized the structural basis of broad substrate specificity of the key synthesizing enzyme, PHA synthase, to incorporate various co-monomers with main and side chains of different lengths. These monomers were produced inside the cell by metabolic engineering, and then copolymerized to improve the material properties of PLGA. As a result, a variety of PLGA copolymers with different monomer compositions such as the US Food and Drug Administration (FDA)-approved monomers, 3-hydroxyburate, 4-hydroxyburate, and 6-hydroxyhexanoate, were produced. Newly applied bioplastics such as 5-hydroxyvalerate and 2-hydroxyisovalerate were also made.
The team employed a systems metabolic engineering application which, according to the researchers, is the first successful example of biological production of PGLA and several novel copolymers from renewable biomass by one-step direct fermentation of metabolically engineered E.coli.
Professor Lee said, “We presented important findings that non-natural polymers, such as PLGA which is commonly used for drug delivery or biomedical devices, were produced by a metabolically engineered gut bacterium. Our research is meaningful in that it proposes a platform strategy in metabolic engineering, which can be further utilized in the development of numerous non-natural, useful polymers.”
Director Ilsub Baek at the Platform Technology Division of the Ministry of Science, ICT and Future Planning of Korea, who oversees the Technology Development Program to Solve Climate Change, said, “Professor Lee has led one of our research projects, the Systems Metabolic Engineering for Biorefineries, which began as part of the Ministry’s Technology Development Program to Solve Climate Change. He and his team have continuously achieved promising results and been attracting greater interest from the global scientific community. As climate change technology grows more important, this research on the biological production of non-natural, high value polymers will have a great impact on science and industry.”
The title of the research paper is “One-step Fermentative Production of Poly(lactate-co-glycolate) from Carbohydrates in Escherichia coli (DOI: 10.1038/nbt.3485).” The lead authors are So Young Choi, a Ph.D. candidate in the Department of Chemical and Biomolecular Engineering at KAIST, and Si Jae Park, Assistant Professor of the Environmental Engineering and Energy Department at Myongji University. Won Jun Kim and Jung Eun Yang, both doctoral students in the Department of Chemical and Biomolecular Engineering at KAIST, also participated in the research.
This research was supported by the Technology Development Program to Solve Climate Change’s research project titled “Systems Metabolic Engineering for Biorefineries” from the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea (NRF-2012M1A2A2026556).
Figure: Production of PLGA and Other Non-Natural Copolymers
This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced a variety of PLGAs with different monomer compositions, proposing the chemosynthetic process of non-natural polymers from biomass. The non-natural polymer PLGA and its other copolymers, which were produced by engineered bacteria developed by taking a systems metabolic engineering approach, accumulate in granule forms within a cell.
2016.03.08 View 13185 -
 Nature Biotechnology Nominates Sang Yup Lee of KAIST for Top 20 Translational Researchers of 2014
Nature Biotechnology, recognized as the most prestigious journal in the field of biotechnology, has released today its list of the Top 20 Translational Researchers of 2014. Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST (Korea Advanced Institute of Science and Technology) ranked seventh in the list. He is the only Asian researcher listed.
The journal, in partnership with IP Checkups, a patent analytics firm, presents an annual ranking of researchers based on their paper and patent output. The list includes, among others, each researcher’s most-cited patent in the past five years and their H index, a measurement to evaluate the impact of a researcher’s published work utilizing citation analysis. (More details can be found at http://www.nature.com/bioent/2015/150801/full/bioe.2015.9.html.)
American institutions made up the majority of the list, with 18 universities and research institutes, and the remainder was filled by KAIST in Korea and the Commonwealth Scientific and Industrial Research Organization (CSIRO) in Australia.
Globally known as a leading researcher in systems metabolic engineering, Professor Lee has published more than 500 journal papers and 580 patents. He has received many awards, including the Citation Classic Award, Elmer Gaden Award, Merck Metabolic Engineering Award, ACS Marvin Johnson Award, SIMB Charles Thom Award, POSCO TJ Park Prize, Amgen Biochemical Engineering Award, and the Ho Am Prize in Engineering.
2015.08.27 View 12941
Nature Biotechnology Nominates Sang Yup Lee of KAIST for Top 20 Translational Researchers of 2014
Nature Biotechnology, recognized as the most prestigious journal in the field of biotechnology, has released today its list of the Top 20 Translational Researchers of 2014. Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST (Korea Advanced Institute of Science and Technology) ranked seventh in the list. He is the only Asian researcher listed.
The journal, in partnership with IP Checkups, a patent analytics firm, presents an annual ranking of researchers based on their paper and patent output. The list includes, among others, each researcher’s most-cited patent in the past five years and their H index, a measurement to evaluate the impact of a researcher’s published work utilizing citation analysis. (More details can be found at http://www.nature.com/bioent/2015/150801/full/bioe.2015.9.html.)
American institutions made up the majority of the list, with 18 universities and research institutes, and the remainder was filled by KAIST in Korea and the Commonwealth Scientific and Industrial Research Organization (CSIRO) in Australia.
Globally known as a leading researcher in systems metabolic engineering, Professor Lee has published more than 500 journal papers and 580 patents. He has received many awards, including the Citation Classic Award, Elmer Gaden Award, Merck Metabolic Engineering Award, ACS Marvin Johnson Award, SIMB Charles Thom Award, POSCO TJ Park Prize, Amgen Biochemical Engineering Award, and the Ho Am Prize in Engineering.
2015.08.27 View 12941 -
 An efficient strategy for developing microbial cell factories by employing synthetic small regulatory RNAs
A new metabolic engineering tool that allows fine control of gene expression level by employing synthetic small regulatory RNAs was developed to efficiently construct microbial cell factories producing desired chemicals and materials
Biotechnologists have been working hard to address the climate change and limited fossil resource issues through the development of sustainable processes for the production of chemicals, fuels and materials from renewable non-food biomass. One promising sustainable technology is the use of microbial cell factories for the efficient production of desired chemicals and materials. When microorganisms are isolated from nature, the performance in producing our desired product is rather poor. That is why metabolic engineering is performed to improve the metabolic and cellular characteristics to achieve enhanced production of desired product at high yield and productivity. Since the performance of microbial cell factory is very important in lowering the overall production cost of the bioprocess, many different strategies and tools have been developed for the metabolic engineering of microorganisms.
One of the big challenges in metabolic engineering is to find the best platform organism and to find those genes to be engineered so as to maximize the production efficiency of the desired chemical. Even Escherichia coli, the most widely utilized simple microorganism, has thousands of genes, the expression of which is highly regulated and interconnected to finely control cellular and metabolic activities. Thus, the complexity of cellular genetic interactions is beyond our intuition and thus it is very difficult to find effective target genes to engineer. Together with gene amplification strategy, gene knockout strategy has been an essential tool in metabolic engineering to redirect the pathway fluxes toward our desired product formation. However, experiment to engineer many genes can be rather difficult due to the time and effort required; for example, gene deletion experiment can take a few weeks depending on the microorganisms. Furthermore, as certain genes are essential or play important roles for the survival of a microorganism, gene knockout experiments cannot be performed. Even worse, there are many different microbial strains one can employ. There are more than 50 different E. coli strains that metabolic engineer can consider to use. Since gene knockout experiment is hard-coded (that is, one should repeat the gene knockout experiments for each strain), the result cannot be easily transferred from one strain to another.
A paper published in Nature Biotechnology online today addresses this issue and suggests a new strategy for identifying gene targets to be knocked out or knocked down through the use of synthetic small RNA. A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), a prestigeous science and engineering university in Korea reported that synthetic small RNA can be employed for finely controlling the expression levels of multiple genes at the translation level. Already well-known for their systems metabolic engineering strategies, Professor Lee’s team added one more strategy to efficiently develop microbial cell factories for the production of chemicals and materials.
Gene expression works like this: the hard-coded blueprint (DNA) is transcribed into messenger RNA (mRNA), and the coding information in mRNA is read to produce protein by ribosomes. Conventional genetic engineering approaches have often targeted modification of the blueprint itself (DNA) to alter organism’s physiological characteristics. Again, engineering the blueprint itself takes much time and effort, and in addition, the results obtained cannot be transferred to another organism without repeating the whole set of experiments. This is why Professor Lee and his colleagues aimed at controlling the gene expression level at the translation stage through the use of synthetic small RNA. They created novel RNAs that can regulate the translation of multiple messenger RNAs (mRNA), and consequently varying the expression levels of multiple genes at the same time. Briefly, synthetic regulatory RNAs interrupt gene expression process from DNA to protein by destroying the messenger RNAs to different yet controllable extents. The advantages of taking this strategy of employing synthetic small regulatory RNAs include simple, easy and high-throughput identification of gene knockout or knockdown targets, fine control of gene expression levels, transferability to many different host strains, and possibility of identifying those gene targets that are essential.
As proof-of-concept demonstration of the usefulness of this strategy, Professor Lee and his colleagues applied it to develop engineered E. coli strains capable of producing an aromatic amino acid tyrosine, which is used for stress symptom relief, food supplements, and precursor for many drugs. They examined a large number of genes in multiple E. coli strains, and developed a highly efficient tyrosine producer. Also, they were able to show that this strategy can be employed to an already metabolically engineered E. coli strain for further improvement by demonstrating the development of highly efficient producer of cadaverine, an important platform chemical for nylon in the chemical industry.
This new strategy, being simple yet very powerful for systems metabolic engineering, is thus expected to facilitate the efficient development of microbial cell factories capable of producing chemicals, fuels and materials from renewable biomass.
Source: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park, and Sang Yup Lee, “Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs”, Nature Biotechnology, doi:10.1038/nbt.2461 (2013)
2013.03.19 View 11775
An efficient strategy for developing microbial cell factories by employing synthetic small regulatory RNAs
A new metabolic engineering tool that allows fine control of gene expression level by employing synthetic small regulatory RNAs was developed to efficiently construct microbial cell factories producing desired chemicals and materials
Biotechnologists have been working hard to address the climate change and limited fossil resource issues through the development of sustainable processes for the production of chemicals, fuels and materials from renewable non-food biomass. One promising sustainable technology is the use of microbial cell factories for the efficient production of desired chemicals and materials. When microorganisms are isolated from nature, the performance in producing our desired product is rather poor. That is why metabolic engineering is performed to improve the metabolic and cellular characteristics to achieve enhanced production of desired product at high yield and productivity. Since the performance of microbial cell factory is very important in lowering the overall production cost of the bioprocess, many different strategies and tools have been developed for the metabolic engineering of microorganisms.
One of the big challenges in metabolic engineering is to find the best platform organism and to find those genes to be engineered so as to maximize the production efficiency of the desired chemical. Even Escherichia coli, the most widely utilized simple microorganism, has thousands of genes, the expression of which is highly regulated and interconnected to finely control cellular and metabolic activities. Thus, the complexity of cellular genetic interactions is beyond our intuition and thus it is very difficult to find effective target genes to engineer. Together with gene amplification strategy, gene knockout strategy has been an essential tool in metabolic engineering to redirect the pathway fluxes toward our desired product formation. However, experiment to engineer many genes can be rather difficult due to the time and effort required; for example, gene deletion experiment can take a few weeks depending on the microorganisms. Furthermore, as certain genes are essential or play important roles for the survival of a microorganism, gene knockout experiments cannot be performed. Even worse, there are many different microbial strains one can employ. There are more than 50 different E. coli strains that metabolic engineer can consider to use. Since gene knockout experiment is hard-coded (that is, one should repeat the gene knockout experiments for each strain), the result cannot be easily transferred from one strain to another.
A paper published in Nature Biotechnology online today addresses this issue and suggests a new strategy for identifying gene targets to be knocked out or knocked down through the use of synthetic small RNA. A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), a prestigeous science and engineering university in Korea reported that synthetic small RNA can be employed for finely controlling the expression levels of multiple genes at the translation level. Already well-known for their systems metabolic engineering strategies, Professor Lee’s team added one more strategy to efficiently develop microbial cell factories for the production of chemicals and materials.
Gene expression works like this: the hard-coded blueprint (DNA) is transcribed into messenger RNA (mRNA), and the coding information in mRNA is read to produce protein by ribosomes. Conventional genetic engineering approaches have often targeted modification of the blueprint itself (DNA) to alter organism’s physiological characteristics. Again, engineering the blueprint itself takes much time and effort, and in addition, the results obtained cannot be transferred to another organism without repeating the whole set of experiments. This is why Professor Lee and his colleagues aimed at controlling the gene expression level at the translation stage through the use of synthetic small RNA. They created novel RNAs that can regulate the translation of multiple messenger RNAs (mRNA), and consequently varying the expression levels of multiple genes at the same time. Briefly, synthetic regulatory RNAs interrupt gene expression process from DNA to protein by destroying the messenger RNAs to different yet controllable extents. The advantages of taking this strategy of employing synthetic small regulatory RNAs include simple, easy and high-throughput identification of gene knockout or knockdown targets, fine control of gene expression levels, transferability to many different host strains, and possibility of identifying those gene targets that are essential.
As proof-of-concept demonstration of the usefulness of this strategy, Professor Lee and his colleagues applied it to develop engineered E. coli strains capable of producing an aromatic amino acid tyrosine, which is used for stress symptom relief, food supplements, and precursor for many drugs. They examined a large number of genes in multiple E. coli strains, and developed a highly efficient tyrosine producer. Also, they were able to show that this strategy can be employed to an already metabolically engineered E. coli strain for further improvement by demonstrating the development of highly efficient producer of cadaverine, an important platform chemical for nylon in the chemical industry.
This new strategy, being simple yet very powerful for systems metabolic engineering, is thus expected to facilitate the efficient development of microbial cell factories capable of producing chemicals, fuels and materials from renewable biomass.
Source: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park, and Sang Yup Lee, “Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs”, Nature Biotechnology, doi:10.1038/nbt.2461 (2013)
2013.03.19 View 11775 -
 New BioFactory Technique Developed using sRNAs
Professor Sang Yup Lee
- published on the online edition of Nature Biotechnology. “Expected as a new strategy for the bio industry that may replace the chemical industry.”-
KAIST Chemical & Biomolecular engineering department’s Professor Sang Yup Lee and his team has developed a new technology that utilizes the synthetic small regulatory RNAs (sRNAs) to implement the BioFactory in a larger scale with more effectiveness.
* BioFactory: Microbial-based production system which creates the desired compound in mass by manipulating the genes of the cell.
In order to solve the problems of modern society, such as environmental pollution caused by the exhaustion of fossil fuels and usage of petrochemical products, an eco-friendly and sustainable bio industry is on the rise. BioFactory development technology has especially attracted the attention world-wide, with its ability to produce bio-energy, pharmaceuticals, eco-friendly materials and more.
For the development of an excellent BioFactory, selection for the gene that produces the desired compounds must be accompanied by finding the microorganism with high production efficiency; however, the previous research method had a complicated and time-consuming problem of having to manipulate the genes of the microorganism one by one.
Professor Sang Yup Lee’s research team, including Dr. Dokyun Na and Dr. Seung Min Yoo, has produced the synthetic sRNAs and utilized it to overcome the technical limitations mentioned above.
In particular, unlike the existing method, this technology using synthetic sRNAs exhibits no strain specificity which can dramatically shorten the experiment that used to take months to just a few days.
The research team applied the synthetic small regulatory RNA technology to the production of the tyrosine*, which is used as the precursor of the medicinal compound, and cadaverine**, widely utilized in a variety of petrochemical products, and has succeeded developing BioFactory with the world’s highest yield rate (21.9g /L, 12.6g / L each).
*tyrosine: amino acid known to control stress and improve concentration
**cadaverine: base material used in many petrochemical products, such as polyurethane
Professor Sang Yup Lee highlighted the significance of this research: “it is expected the synthetic small regulatory RNA technology will stimulate the BioFactory development and also serve as a catalyst which can make the chemical industry, currently represented by its petroleum energy, transform into bio industry.”
The study was carried out with the support of Global Frontier Project (Intelligent Bio-Systems Design and Synthesis Research Unit (Chief Seon Chang Kim)) and the findings have been published on January 20th in the online edition of the worldwide journal Nature Biotechnology.
2013.02.21 View 13762
New BioFactory Technique Developed using sRNAs
Professor Sang Yup Lee
- published on the online edition of Nature Biotechnology. “Expected as a new strategy for the bio industry that may replace the chemical industry.”-
KAIST Chemical & Biomolecular engineering department’s Professor Sang Yup Lee and his team has developed a new technology that utilizes the synthetic small regulatory RNAs (sRNAs) to implement the BioFactory in a larger scale with more effectiveness.
* BioFactory: Microbial-based production system which creates the desired compound in mass by manipulating the genes of the cell.
In order to solve the problems of modern society, such as environmental pollution caused by the exhaustion of fossil fuels and usage of petrochemical products, an eco-friendly and sustainable bio industry is on the rise. BioFactory development technology has especially attracted the attention world-wide, with its ability to produce bio-energy, pharmaceuticals, eco-friendly materials and more.
For the development of an excellent BioFactory, selection for the gene that produces the desired compounds must be accompanied by finding the microorganism with high production efficiency; however, the previous research method had a complicated and time-consuming problem of having to manipulate the genes of the microorganism one by one.
Professor Sang Yup Lee’s research team, including Dr. Dokyun Na and Dr. Seung Min Yoo, has produced the synthetic sRNAs and utilized it to overcome the technical limitations mentioned above.
In particular, unlike the existing method, this technology using synthetic sRNAs exhibits no strain specificity which can dramatically shorten the experiment that used to take months to just a few days.
The research team applied the synthetic small regulatory RNA technology to the production of the tyrosine*, which is used as the precursor of the medicinal compound, and cadaverine**, widely utilized in a variety of petrochemical products, and has succeeded developing BioFactory with the world’s highest yield rate (21.9g /L, 12.6g / L each).
*tyrosine: amino acid known to control stress and improve concentration
**cadaverine: base material used in many petrochemical products, such as polyurethane
Professor Sang Yup Lee highlighted the significance of this research: “it is expected the synthetic small regulatory RNA technology will stimulate the BioFactory development and also serve as a catalyst which can make the chemical industry, currently represented by its petroleum energy, transform into bio industry.”
The study was carried out with the support of Global Frontier Project (Intelligent Bio-Systems Design and Synthesis Research Unit (Chief Seon Chang Kim)) and the findings have been published on January 20th in the online edition of the worldwide journal Nature Biotechnology.
2013.02.21 View 13762 -
 Professor Sang-Yup Lee publishes a requested paper in Nature Biotechnology
Professor Sang-Yup Lee publishes a requested paper in Nature Biotechnology
“The era of commercialized bioplastic is coming”
Disclose an opinion as specialist at a requested paper in Nature Biotechnology, October 2006
A team led by Barbel Friedrich, Professor of Humboldt-Universitat zu Berlin, and Alexander Steinbuchel, Professor of West falische Wilhelms-Universitat Munster, found out the entire genome sequence of the typical bioplastic-producing microorganism ‘Ralstonia eutropha’ and published a paper on it in Nature Biotechnology, October 2006. As the entire genome sequence of the typical bioplactic-producing microorganism has been discovered, it is expected that the efficient production of bioplastic will be available through strain improvement at a more systematic level.
Regarding this paper, Nature Biotechnology requested world-renowned scholar Sang-Yup Lee, LG Chemical Chair-Professor of KAIST Chemical and Biomolecular Engineering Department, an expert analysis on the future of bioplastic production as a result of the deciphering of the genome sequence, and Professor Lee revealed his opinion at ‘News and Views’ in Nature Biotechnology, October 2006, issued on October 10. In the analysis, he insisted, “The deciphering of the genomes of Ralstonia means to pave the way for the improvement of strains at a system level by combining simulation through various omics and imaginary cells and engineering at a genome level. It will be possible to produce plastic with desired properties by altering the components of plastic as desired and produce bioplastic, more efficient and economical than have been reported so far, through the optimization of metabolic flow.”
Professor Lee is a world-renowned scholar in the bioplastic field, who has presented about 70 SCI papers in the field. He created a word ‘Plastic Bacteria’ at Trends in Biotechnology in 1996 and published an expert paper regarding E.Coli Plastic at Nature Biotechnology in 1997. He is now performing a research concerning the improvement of bioplastic-producing strains as an example of a research employing a systematic method for the system biological research and development project of the Ministry of Science and Technology.
The followings are the contents of Professor Lee’s paper concerning microorganism plastic published at ‘News and Views’ in Nature Biotechnology, October 2006.
- Polyhydroxyalkanoate (PHA) is a high molecule that numerous microorganisms accumulate in their own cells as energy storage substance when they are rich in carbonic resources, but poor in the other growth factors. The PHA high molecule is polyester, in which the unit substances (unit chemicals) are ester-bonded, and has been studied worldwide about twenty years before. However, PHA’s worse properties than petrochemical plastic and extremely high production cost have prevented its commercialization. The production cost of PHA was 15 dollars per kg in 1980’s, twenty times higher than the price of polypropylene. Sang-Yup Lee, LG Chemical Chair-Professor of KAIST Chemical & Biomolecular Department’s BK21 Project Group, has performed a research concerning the efficient production of microorganism plastic through the combination of metabolic engineering and fermentation process under the support of the Ministry of Science and Technology, and developed a process that lowers the production cost of PHA to 2-3 dollars per kg. He also has developed PHA-producing bacteria, efficient enough to fill plastic tightly, and named it ‘Plastic Bacteria’.
- The unprecedented rise of oil price for the past two years activated the researches on Bio-based energies and chemical production globally. PHA is also regaining attentions although the researches on it have been withered so far due to its poor economical efficiency and properties. The result of the genome deciphering of the typical plastic-producing microorganism ‘Ralstonia eutropha’ published by a German research team in Nature Biotechnology, October 2006 suggests huge meanings. That is, it will provide a blueprint over the metabolic activities of the bacteria and thus enables more systematic strain improvement.
- Eyeing on these facts, Nature Biotechnology requested Professor Sang-Yup Lee an expert analysis, and Professor Lee analyzed that there would be a dramatic development of microorganism plastic production through the application of the system biological engineering method, which is now being performed actively by Professor Lee at KAIST. In the analysis, Professor Lee revealed, “As the genome sequence has been found out, it becomes possible to establish metabolic network at a genome level, and since simulation becomes available, numberless trial and errors and experiments can be replaced with imaginary experiments rapidly. In addition, It makes the more efficient development of strains possible by fusion-analyzing the omics result such as various transcripts, proteins, metabolites, etc.” He also expected that it would be possible to produce tailor-made PHA having desired properties through metabolic engineering as well as the efficient production of plastic. Besides, he prospected that his research on the production of optically pure hydroxyl carboxyl acid, Professor Lee’s international patent right, would gain driving forces and technical development would be made rapidly at biological hydrogen production, production, dissolution and application of aromatic compounds, etc. by featuring this strain.
- Recently, Metabolic and ADM, U.S. companies, jointly started to produce PHA at a commercialization level, and Brazil having rich natural resources is commercializing PHA, following Bio-ethanol. In addition, Japan and Germany having a bunch of research performance in this field, and Australia having rich biomass are also performing consistent researches on PHA’s commercialization. Professor Lee prospected, “With the finding out of the genome sequence of the typical bioplastic-producing microorganism, competition for commercialization will be fiercer among nations through the development of efficient production systems.”
- Professor Lee prospected that as the efficient production of PHA becomes possible, the production of plastic from various renewable ingredients (cellurose, starch, suger, etc.) through microorganism fermentation would be made practically and the white biotechnologies of existing chemicals would gain more power. He also said, “Korea also will have to try to secure the production technologies and industry of Bio-based chemicals through strategic cooperation with resource powerfuls, etc. on the basis of the technical dominancy in some system metabolic engineering fields.”
- ‘News and Views’ in Nature Biotechnology is a section that publishes analyses of world-renowned specialists in the corresponding fields over the contents of some papers having great influences among papers published in the issue. KAIST Professor Sang-Yup Lee has published his second expert analysis of ‘Deciphering bioplastic production’ in the volume of October 2006, following the first paper ‘Going into the era of E.Coli plastic’.
2006.10.23 View 19562
Professor Sang-Yup Lee publishes a requested paper in Nature Biotechnology
Professor Sang-Yup Lee publishes a requested paper in Nature Biotechnology
“The era of commercialized bioplastic is coming”
Disclose an opinion as specialist at a requested paper in Nature Biotechnology, October 2006
A team led by Barbel Friedrich, Professor of Humboldt-Universitat zu Berlin, and Alexander Steinbuchel, Professor of West falische Wilhelms-Universitat Munster, found out the entire genome sequence of the typical bioplastic-producing microorganism ‘Ralstonia eutropha’ and published a paper on it in Nature Biotechnology, October 2006. As the entire genome sequence of the typical bioplactic-producing microorganism has been discovered, it is expected that the efficient production of bioplastic will be available through strain improvement at a more systematic level.
Regarding this paper, Nature Biotechnology requested world-renowned scholar Sang-Yup Lee, LG Chemical Chair-Professor of KAIST Chemical and Biomolecular Engineering Department, an expert analysis on the future of bioplastic production as a result of the deciphering of the genome sequence, and Professor Lee revealed his opinion at ‘News and Views’ in Nature Biotechnology, October 2006, issued on October 10. In the analysis, he insisted, “The deciphering of the genomes of Ralstonia means to pave the way for the improvement of strains at a system level by combining simulation through various omics and imaginary cells and engineering at a genome level. It will be possible to produce plastic with desired properties by altering the components of plastic as desired and produce bioplastic, more efficient and economical than have been reported so far, through the optimization of metabolic flow.”
Professor Lee is a world-renowned scholar in the bioplastic field, who has presented about 70 SCI papers in the field. He created a word ‘Plastic Bacteria’ at Trends in Biotechnology in 1996 and published an expert paper regarding E.Coli Plastic at Nature Biotechnology in 1997. He is now performing a research concerning the improvement of bioplastic-producing strains as an example of a research employing a systematic method for the system biological research and development project of the Ministry of Science and Technology.
The followings are the contents of Professor Lee’s paper concerning microorganism plastic published at ‘News and Views’ in Nature Biotechnology, October 2006.
- Polyhydroxyalkanoate (PHA) is a high molecule that numerous microorganisms accumulate in their own cells as energy storage substance when they are rich in carbonic resources, but poor in the other growth factors. The PHA high molecule is polyester, in which the unit substances (unit chemicals) are ester-bonded, and has been studied worldwide about twenty years before. However, PHA’s worse properties than petrochemical plastic and extremely high production cost have prevented its commercialization. The production cost of PHA was 15 dollars per kg in 1980’s, twenty times higher than the price of polypropylene. Sang-Yup Lee, LG Chemical Chair-Professor of KAIST Chemical & Biomolecular Department’s BK21 Project Group, has performed a research concerning the efficient production of microorganism plastic through the combination of metabolic engineering and fermentation process under the support of the Ministry of Science and Technology, and developed a process that lowers the production cost of PHA to 2-3 dollars per kg. He also has developed PHA-producing bacteria, efficient enough to fill plastic tightly, and named it ‘Plastic Bacteria’.
- The unprecedented rise of oil price for the past two years activated the researches on Bio-based energies and chemical production globally. PHA is also regaining attentions although the researches on it have been withered so far due to its poor economical efficiency and properties. The result of the genome deciphering of the typical plastic-producing microorganism ‘Ralstonia eutropha’ published by a German research team in Nature Biotechnology, October 2006 suggests huge meanings. That is, it will provide a blueprint over the metabolic activities of the bacteria and thus enables more systematic strain improvement.
- Eyeing on these facts, Nature Biotechnology requested Professor Sang-Yup Lee an expert analysis, and Professor Lee analyzed that there would be a dramatic development of microorganism plastic production through the application of the system biological engineering method, which is now being performed actively by Professor Lee at KAIST. In the analysis, Professor Lee revealed, “As the genome sequence has been found out, it becomes possible to establish metabolic network at a genome level, and since simulation becomes available, numberless trial and errors and experiments can be replaced with imaginary experiments rapidly. In addition, It makes the more efficient development of strains possible by fusion-analyzing the omics result such as various transcripts, proteins, metabolites, etc.” He also expected that it would be possible to produce tailor-made PHA having desired properties through metabolic engineering as well as the efficient production of plastic. Besides, he prospected that his research on the production of optically pure hydroxyl carboxyl acid, Professor Lee’s international patent right, would gain driving forces and technical development would be made rapidly at biological hydrogen production, production, dissolution and application of aromatic compounds, etc. by featuring this strain.
- Recently, Metabolic and ADM, U.S. companies, jointly started to produce PHA at a commercialization level, and Brazil having rich natural resources is commercializing PHA, following Bio-ethanol. In addition, Japan and Germany having a bunch of research performance in this field, and Australia having rich biomass are also performing consistent researches on PHA’s commercialization. Professor Lee prospected, “With the finding out of the genome sequence of the typical bioplastic-producing microorganism, competition for commercialization will be fiercer among nations through the development of efficient production systems.”
- Professor Lee prospected that as the efficient production of PHA becomes possible, the production of plastic from various renewable ingredients (cellurose, starch, suger, etc.) through microorganism fermentation would be made practically and the white biotechnologies of existing chemicals would gain more power. He also said, “Korea also will have to try to secure the production technologies and industry of Bio-based chemicals through strategic cooperation with resource powerfuls, etc. on the basis of the technical dominancy in some system metabolic engineering fields.”
- ‘News and Views’ in Nature Biotechnology is a section that publishes analyses of world-renowned specialists in the corresponding fields over the contents of some papers having great influences among papers published in the issue. KAIST Professor Sang-Yup Lee has published his second expert analysis of ‘Deciphering bioplastic production’ in the volume of October 2006, following the first paper ‘Going into the era of E.Coli plastic’.
2006.10.23 View 19562