National+Academy+of+Sciences+of+the+United+States
-
 KAIST Researchers Suggest an Extraordinary Alternative to Petroleum-based PET - Bacteria!
< (From left) Dr. Cindy Pricilia, Ph.D. Candidate Cheon Woo Moon, Distinguished Professor Sang Yup Lee >
Currently, the world is suffering from environmental problems caused by plastic waste. The KAIST research team has succeeded in producing a microbial-based plastic that is biodegradable and can replace existing PET bottles, making it a hot topic.
Our university announced on the 7th of November that the research team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering has succeeded in developing a microbial strain that efficiently produces pseudoaromatic polyester monomer to replace polyethylene terephthalate (PET) using systems metabolic engineering.
Pseudoaromatic dicarboxylic acids have better physical properties and higher biodegradability than aromatic polyester (PET) when synthesized as polymers, and are attracting attention as an eco-friendly monomer* that can be synthesized into polymers. The production of pseudoaromatic dicarboxylic acids through chemical methods has the problems of low yield and selectivity, complex reaction conditions, and the generation of hazardous waste.
*Monomer: A material for making polymers, which is used to synthesize polymers by polymerizing monomers together
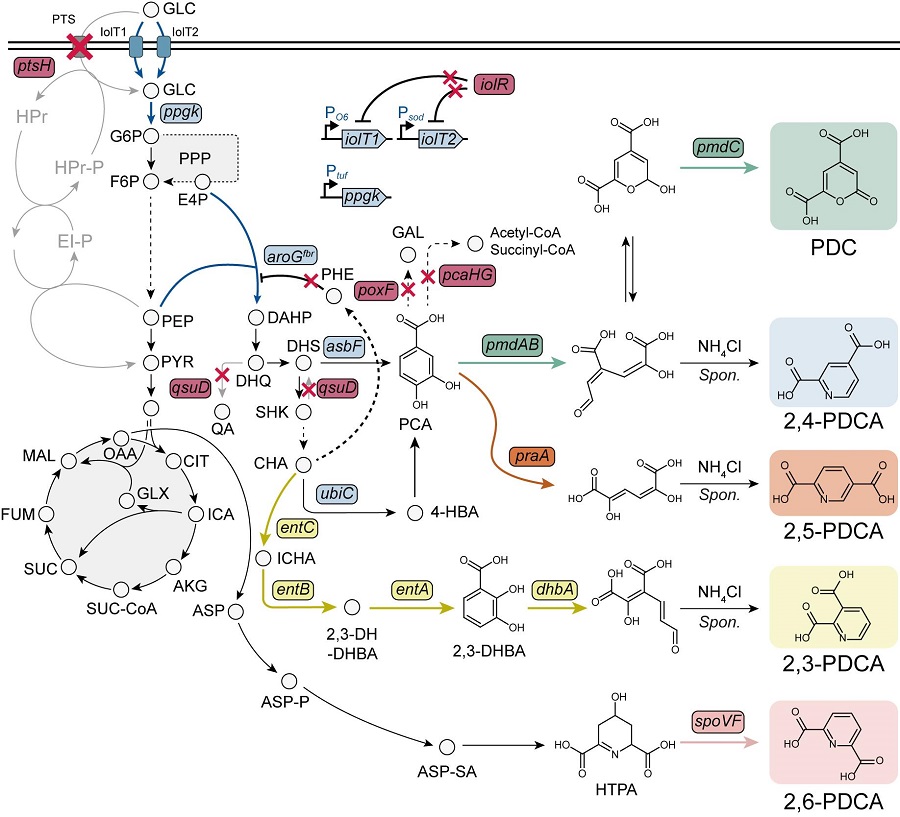
< Figure. Overview of pseudoaromatic dicarboxylic acid production using metabolically engineered C. glutamicum. >
To solve this problem, Professor Sang Yup Lee's research team used metabolic engineering to develop a microbial strain that efficiently produces five types of pseudoaromatic dicarboxylic acids, including 2-pyrone-4,6-dicarboxylic acid and four types of pyridine dicarboxylic acids (2,3-, 2,4-, 2,5-, 2,6-pyridine dicarboxylic acids), in Corynebacterium, a bacterium mainly used for amino acid production.
The research team used metabolic engineering techniques to build a platform microbial strain that enhances the metabolic flow of protocatechuic acid, which is used as a precursor for several pseudoaromatic dicarboxylic acids, and prevents the loss of precursors.
Based on this, the genetic manipulation target was discovered through transcriptome analysis, producing 76.17 g/L of 2-pyrone-4,6-dicarboxylic acid, and by newly discovering and constructing three types of pyridine dicarboxylic acid production metabolic pathways, successfully producing 2.79 g/L of 2,3-pyridine dicarboxylic acid, 0.49 g/L of 2,4-pyridine dicarboxylic acid, and 1.42 g/L of 2,5-pyridine dicarboxylic acid.
In addition, the research team confirmed the production of 15.01 g/L through the construction and reinforcement of the 2,6-pyridine dicarboxylic acid biosynthesis pathway, successfully producing a total of five similar aromatic dicarboxylic acids with high efficiency.
In conclusion, the team succeeded in producing 2,4-, 2,5-, and 2,6-pyridine dicarboxylic acids at the world's highest concentration. In particular, 2,4-, 2,5-pyridine dicarboxylic acid achieved production on the scale of g/L, which was previously produced in extremely small amounts (mg/L).
Based on this study, it is expected that it will be applied to various polyester production industrial processes, and it is also expected that it will be actively utilized in research on the production of similar aromatic polyesters.
Professor Sang Yup Lee, the corresponding author, said, “The significance lies in the fact that we have developed an eco-friendly technology that efficiently produces similar aromatic polyester monomers based on microorganisms,” and “This study will help the microorganism-based bio-monomer industry replace the petrochemical-based chemical industry in the future.”
The results of this study were published in the international academic journal, the Proceedings of the National Academy of Sciences of United States of America (PNAS) on October 30th.
※ Paper title: Metabolic engineering of Corynebacterium glutamicum for the production of pyrone and pyridine dicarboxylic acids
※ Author information: Jae Sung Cho (co-first author), Zi Wei Luo (co-first author), Cheon Woo Moon (co-first author), Cindy Prabowo (co-author), Sang Yup Lee (corresponding author) - a total of 5 people
This study was conducted with the support of the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project leader: Professor Sang Yup Lee) from the National Research Foundation supported by the Ministry of Science and Technology and ICT of Korea.
2024.11.08 View 9000
KAIST Researchers Suggest an Extraordinary Alternative to Petroleum-based PET - Bacteria!
< (From left) Dr. Cindy Pricilia, Ph.D. Candidate Cheon Woo Moon, Distinguished Professor Sang Yup Lee >
Currently, the world is suffering from environmental problems caused by plastic waste. The KAIST research team has succeeded in producing a microbial-based plastic that is biodegradable and can replace existing PET bottles, making it a hot topic.
Our university announced on the 7th of November that the research team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering has succeeded in developing a microbial strain that efficiently produces pseudoaromatic polyester monomer to replace polyethylene terephthalate (PET) using systems metabolic engineering.
Pseudoaromatic dicarboxylic acids have better physical properties and higher biodegradability than aromatic polyester (PET) when synthesized as polymers, and are attracting attention as an eco-friendly monomer* that can be synthesized into polymers. The production of pseudoaromatic dicarboxylic acids through chemical methods has the problems of low yield and selectivity, complex reaction conditions, and the generation of hazardous waste.
*Monomer: A material for making polymers, which is used to synthesize polymers by polymerizing monomers together
< Figure. Overview of pseudoaromatic dicarboxylic acid production using metabolically engineered C. glutamicum. >
To solve this problem, Professor Sang Yup Lee's research team used metabolic engineering to develop a microbial strain that efficiently produces five types of pseudoaromatic dicarboxylic acids, including 2-pyrone-4,6-dicarboxylic acid and four types of pyridine dicarboxylic acids (2,3-, 2,4-, 2,5-, 2,6-pyridine dicarboxylic acids), in Corynebacterium, a bacterium mainly used for amino acid production.
The research team used metabolic engineering techniques to build a platform microbial strain that enhances the metabolic flow of protocatechuic acid, which is used as a precursor for several pseudoaromatic dicarboxylic acids, and prevents the loss of precursors.
Based on this, the genetic manipulation target was discovered through transcriptome analysis, producing 76.17 g/L of 2-pyrone-4,6-dicarboxylic acid, and by newly discovering and constructing three types of pyridine dicarboxylic acid production metabolic pathways, successfully producing 2.79 g/L of 2,3-pyridine dicarboxylic acid, 0.49 g/L of 2,4-pyridine dicarboxylic acid, and 1.42 g/L of 2,5-pyridine dicarboxylic acid.
In addition, the research team confirmed the production of 15.01 g/L through the construction and reinforcement of the 2,6-pyridine dicarboxylic acid biosynthesis pathway, successfully producing a total of five similar aromatic dicarboxylic acids with high efficiency.
In conclusion, the team succeeded in producing 2,4-, 2,5-, and 2,6-pyridine dicarboxylic acids at the world's highest concentration. In particular, 2,4-, 2,5-pyridine dicarboxylic acid achieved production on the scale of g/L, which was previously produced in extremely small amounts (mg/L).
Based on this study, it is expected that it will be applied to various polyester production industrial processes, and it is also expected that it will be actively utilized in research on the production of similar aromatic polyesters.
Professor Sang Yup Lee, the corresponding author, said, “The significance lies in the fact that we have developed an eco-friendly technology that efficiently produces similar aromatic polyester monomers based on microorganisms,” and “This study will help the microorganism-based bio-monomer industry replace the petrochemical-based chemical industry in the future.”
The results of this study were published in the international academic journal, the Proceedings of the National Academy of Sciences of United States of America (PNAS) on October 30th.
※ Paper title: Metabolic engineering of Corynebacterium glutamicum for the production of pyrone and pyridine dicarboxylic acids
※ Author information: Jae Sung Cho (co-first author), Zi Wei Luo (co-first author), Cheon Woo Moon (co-first author), Cindy Prabowo (co-author), Sang Yup Lee (corresponding author) - a total of 5 people
This study was conducted with the support of the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project leader: Professor Sang Yup Lee) from the National Research Foundation supported by the Ministry of Science and Technology and ICT of Korea.
2024.11.08 View 9000 -
 X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.07.09 View 16895
X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.07.09 View 16895 -
 A Novel Biosensor to Advance Diverse High-Level Production of Microbial Cell Factories
A research group at KAIST presented a novel biosensor which can produce diverse, high-level microbial cell factories. The biosensor monitors the concentration of products and even intermediates when new strains are being developed. This strategy provides a new platform for manufacturing diverse natural products from renewable resources. The team succeeded in creating four natural products of high-level pharmaceutical importance with this strategy.
Malonyl-CoA is a major building block for many value-added chemicals including diverse natural products with pharmaceutical importance. However, due to the low availability of malonyl-CoA in bacteria, many malonyl-CoA-derived natural products have been produced by chemical synthesis or extraction from natural resources that are harmful to the environment and are unsustainable. For the sustainable biological production of malonyl-CoA-derived natural products, increasing the intracellular malonyl-CoA pool is necessary. To this end, the development of a robust and efficient malonyl-CoA biosensor was required to monitor the concentration of intracellular malonyl-CoA abundance as new strains are developed.
Metabolic engineering researchers at KAIST addressed this issue. This research reports the development of a simple and robust malonyl-CoA biosensor by repurposing a type III polyketide synthase (also known as RppA), which produces flaviolin, a colorimetric indicator of malonyl-CoA. Subsequently, the RppA biosensor was used for the rapid and efficient colorimetric screening of gene manipulation targets enabling enhanced malonyl-CoA abundance. The screened beneficial gene targets were employed for the high-level production of four representative natural products derived from malonyl-CoA. Compared with the previous strategies, which were expensive and time-consuming, the new biosensor could be easily applied to industrially relevant bacteria including Escherichia coli, Pseudomonas putida, and Corynebacterium glutamicum to enable a one-step process.
The study employs synthetic small regulatory RNA (sRNA) technology to rapidly and efficiently reduce endogenous target gene expression for improved malonyl-CoA production. The researchers constructed an E. coli genome-scale synthetic sRNA library targeting 1,858 genes covering all major metabolic genes in E. coli. This library was employed with the RppA biosensor to screen for gene targets which are believed to be beneficial for enhancing malonyl-CoA accumulation upon their expression knockdown.
From this colorimetric screening, 14 gene targets were selected, all of which were successful at significantly increasing the production of four natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin). Although specific examples are demonstrated in E. coli as a host, the researchers showed that the biosensor is also functional in P. putida and C. glutamicum, industrially important representative gram-negative and gram-positive bacteria, respectively. The malonyl-CoA biosensor developed in this research will serve as an efficient platform for the rapid development of strains capable of producing natural products crucial for the pharmaceutical, chemical, cosmetics, and food industries.
An important aspect of this work is that the high-performance strains constructed in this research were developed rapidly and easily by utilizing the simple approach of colorimetric screening, without involving extensive metabolic engineering approaches. 6-Methylsalicylic acid (an antibiotic) could be produced to the highest titer reported for E. coli, and the microbial production of aloesone (a precursor of aloesin, an anti-inflammatory agent/whitening agent) was achieved for the first time.
“A sustainable process for producing diverse natural products using renewable resources is of great interest. This study represents the development of a robust and efficient malonyl-CoA biosensor generally applicable to a wide range of industrially important bacteria. The capability of this biosensor for screening a large library was demonstrated to show that the rapid and efficient construction of high-performance strains is feasible. This research will be useful for further accelerating the development process of strains capable of producing valuable chemicals to industrially relevant levels,” said Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, who led the research.
This study entitled “Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria,” was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on October 02.
PhD students Dongsoo Yang and Won Jun Kim, MS student Shin Hee Ha, research staff Mun Hee Lee, Research Professor Seung Min Yoo, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Dr. Jong Hyun Choi of the Applied Microbiology Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB) participated in this research.
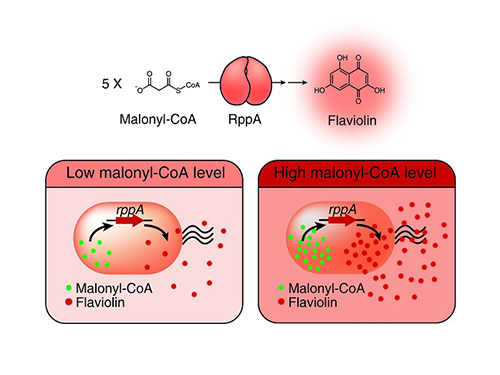
Figure: Type III polyketide synthase (RppA) as a malonyl-CoA biosensor. RppA converts five molecules of malonyl-CoA into one molecule of red-colored flaviolin. This schematic diagram shows the overall conceptualization of the malonyl-CoA biosensor by indicating that higher malonyl-CoA abundance leads to higher production and secretion of flaviolin, resulting in a deeper red color of the culture. This system was employed for the enhanced production of four representative natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin) from engineered E. coli strains.
2018.10.11 View 10713
A Novel Biosensor to Advance Diverse High-Level Production of Microbial Cell Factories
A research group at KAIST presented a novel biosensor which can produce diverse, high-level microbial cell factories. The biosensor monitors the concentration of products and even intermediates when new strains are being developed. This strategy provides a new platform for manufacturing diverse natural products from renewable resources. The team succeeded in creating four natural products of high-level pharmaceutical importance with this strategy.
Malonyl-CoA is a major building block for many value-added chemicals including diverse natural products with pharmaceutical importance. However, due to the low availability of malonyl-CoA in bacteria, many malonyl-CoA-derived natural products have been produced by chemical synthesis or extraction from natural resources that are harmful to the environment and are unsustainable. For the sustainable biological production of malonyl-CoA-derived natural products, increasing the intracellular malonyl-CoA pool is necessary. To this end, the development of a robust and efficient malonyl-CoA biosensor was required to monitor the concentration of intracellular malonyl-CoA abundance as new strains are developed.
Metabolic engineering researchers at KAIST addressed this issue. This research reports the development of a simple and robust malonyl-CoA biosensor by repurposing a type III polyketide synthase (also known as RppA), which produces flaviolin, a colorimetric indicator of malonyl-CoA. Subsequently, the RppA biosensor was used for the rapid and efficient colorimetric screening of gene manipulation targets enabling enhanced malonyl-CoA abundance. The screened beneficial gene targets were employed for the high-level production of four representative natural products derived from malonyl-CoA. Compared with the previous strategies, which were expensive and time-consuming, the new biosensor could be easily applied to industrially relevant bacteria including Escherichia coli, Pseudomonas putida, and Corynebacterium glutamicum to enable a one-step process.
The study employs synthetic small regulatory RNA (sRNA) technology to rapidly and efficiently reduce endogenous target gene expression for improved malonyl-CoA production. The researchers constructed an E. coli genome-scale synthetic sRNA library targeting 1,858 genes covering all major metabolic genes in E. coli. This library was employed with the RppA biosensor to screen for gene targets which are believed to be beneficial for enhancing malonyl-CoA accumulation upon their expression knockdown.
From this colorimetric screening, 14 gene targets were selected, all of which were successful at significantly increasing the production of four natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin). Although specific examples are demonstrated in E. coli as a host, the researchers showed that the biosensor is also functional in P. putida and C. glutamicum, industrially important representative gram-negative and gram-positive bacteria, respectively. The malonyl-CoA biosensor developed in this research will serve as an efficient platform for the rapid development of strains capable of producing natural products crucial for the pharmaceutical, chemical, cosmetics, and food industries.
An important aspect of this work is that the high-performance strains constructed in this research were developed rapidly and easily by utilizing the simple approach of colorimetric screening, without involving extensive metabolic engineering approaches. 6-Methylsalicylic acid (an antibiotic) could be produced to the highest titer reported for E. coli, and the microbial production of aloesone (a precursor of aloesin, an anti-inflammatory agent/whitening agent) was achieved for the first time.
“A sustainable process for producing diverse natural products using renewable resources is of great interest. This study represents the development of a robust and efficient malonyl-CoA biosensor generally applicable to a wide range of industrially important bacteria. The capability of this biosensor for screening a large library was demonstrated to show that the rapid and efficient construction of high-performance strains is feasible. This research will be useful for further accelerating the development process of strains capable of producing valuable chemicals to industrially relevant levels,” said Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, who led the research.
This study entitled “Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria,” was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on October 02.
PhD students Dongsoo Yang and Won Jun Kim, MS student Shin Hee Ha, research staff Mun Hee Lee, Research Professor Seung Min Yoo, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Dr. Jong Hyun Choi of the Applied Microbiology Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB) participated in this research.
Figure: Type III polyketide synthase (RppA) as a malonyl-CoA biosensor. RppA converts five molecules of malonyl-CoA into one molecule of red-colored flaviolin. This schematic diagram shows the overall conceptualization of the malonyl-CoA biosensor by indicating that higher malonyl-CoA abundance leads to higher production and secretion of flaviolin, resulting in a deeper red color of the culture. This system was employed for the enhanced production of four representative natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin) from engineered E. coli strains.
2018.10.11 View 10713 -
 Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi)
A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements.
The study analyzed the nanomaterial biosynthesis conditions using a Pourbaix diagram to predict the producibility and crystallinity. Researchers studied a Pourbaix diagram to predict the stable chemical species of each element for nanomaterial biosynthesis at varying levels of reduction potential (Eh) and pH. Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, resulting in the biosynthesis of various crystalline nanomaterials that were previously amorphous or not synthesized.
This strategy was extended to biosynthesize multi-element nanomaterials. Various single and multi-element nanomaterials biosynthesized in this research can potentially serve as new and novel nanomaterials for industrial applications such as catalysts, chemical sensors, biosensors, bioimaging, drug delivery, and cancer therapy.
A research group consisting of PhD candidate Yoojin Choi, Associate Professor Doh Chang Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST and Associate Professor Tae Jung Park of the Department of Chemistry at Chung-Ang University reported the synthesis. This study, entitled “Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials,” was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on May 21.
A recent successful biosynthesis of nanomaterials under mild conditions without requiring physical and chemical treatments has triggered the exploration of the full biosynthesis capacity of a biological system for producing a diverse range of nanomaterials as well as for understanding biosynthesis mechanisms for crystalline versus amorphous nanomaterials.
There has been increased interest in synthesizing various nanomaterials that have not yet been synthesized for various applications including semiconducting materials, enhanced solar cells, biomedical materials, and many others. This research reports the construction of a recombinant E. coli strain that co-expresses metallothionein, a metal binding protein, and phytochelatin synthase that synthesizes the metal-binding peptide phytochelatin for the biosynthesis of various nanomaterials. Subsequently, an E. coli strain was engineered to produce a diverse range of nanomaterials, including those never biosynthesized before, by using 35 individual elements from the periodic table and also by combining multi-elements.
Distinguished Professor Lee said, “An environmentally-friendly and sustainable process is of much interest for producing nanomaterials by not only chemical and physical methods but biological synthesis. Moreover, there has been much attention paid to producing diverse and novel nanomaterials for new industrial applications. This is the first report to predict the biosynthesis of various nanomaterials, by far the largest number of various single- and multi-elements nanomaterials. The strategies used for nanomaterial biosynthesis in this research will be useful for further diversifying the portfolio of nanomaterials that can be manufactured.”
Figure: The biosynthesis of diverse nanomaterials using recombinant E. coli. This schematic diagram shows the overall conceptualization of the biosynthesis of various single and multi-element nanomaterials using recombinant E. coli under incubation with corresponding elemental precursors. The 35 elements that were tested to biosynthesize nanomaterials are shown in black circles on the periodic table.
2018.05.23 View 12452
Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi)
A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements.
The study analyzed the nanomaterial biosynthesis conditions using a Pourbaix diagram to predict the producibility and crystallinity. Researchers studied a Pourbaix diagram to predict the stable chemical species of each element for nanomaterial biosynthesis at varying levels of reduction potential (Eh) and pH. Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, resulting in the biosynthesis of various crystalline nanomaterials that were previously amorphous or not synthesized.
This strategy was extended to biosynthesize multi-element nanomaterials. Various single and multi-element nanomaterials biosynthesized in this research can potentially serve as new and novel nanomaterials for industrial applications such as catalysts, chemical sensors, biosensors, bioimaging, drug delivery, and cancer therapy.
A research group consisting of PhD candidate Yoojin Choi, Associate Professor Doh Chang Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST and Associate Professor Tae Jung Park of the Department of Chemistry at Chung-Ang University reported the synthesis. This study, entitled “Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials,” was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on May 21.
A recent successful biosynthesis of nanomaterials under mild conditions without requiring physical and chemical treatments has triggered the exploration of the full biosynthesis capacity of a biological system for producing a diverse range of nanomaterials as well as for understanding biosynthesis mechanisms for crystalline versus amorphous nanomaterials.
There has been increased interest in synthesizing various nanomaterials that have not yet been synthesized for various applications including semiconducting materials, enhanced solar cells, biomedical materials, and many others. This research reports the construction of a recombinant E. coli strain that co-expresses metallothionein, a metal binding protein, and phytochelatin synthase that synthesizes the metal-binding peptide phytochelatin for the biosynthesis of various nanomaterials. Subsequently, an E. coli strain was engineered to produce a diverse range of nanomaterials, including those never biosynthesized before, by using 35 individual elements from the periodic table and also by combining multi-elements.
Distinguished Professor Lee said, “An environmentally-friendly and sustainable process is of much interest for producing nanomaterials by not only chemical and physical methods but biological synthesis. Moreover, there has been much attention paid to producing diverse and novel nanomaterials for new industrial applications. This is the first report to predict the biosynthesis of various nanomaterials, by far the largest number of various single- and multi-elements nanomaterials. The strategies used for nanomaterial biosynthesis in this research will be useful for further diversifying the portfolio of nanomaterials that can be manufactured.”
Figure: The biosynthesis of diverse nanomaterials using recombinant E. coli. This schematic diagram shows the overall conceptualization of the biosynthesis of various single and multi-element nanomaterials using recombinant E. coli under incubation with corresponding elemental precursors. The 35 elements that were tested to biosynthesize nanomaterials are shown in black circles on the periodic table.
2018.05.23 View 12452 -
 Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 13083
Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 13083 -
 Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin.
Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team.
The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30, 2016.
This interdisciplinary research project was conducted by six research teams from four different countries and covered a wide scope of academic fields, from neurobiology to super resolution optic imaging.
Inositol pyrophosphates such as 5-diphosphoinositol pentakisphos-phate (5-IP7), which naturally occur in corns and beans, are essential metabolites in the body. In particular, inositol hexakisphosphate (IP6) has anti-cancer properties and is thought to have an important role in cell signaling.
Inositol pentakisphosphate (IP7) differs from IP6 by having an additional phosphate group, which was first discovered 20 years ago. IP7 has recently been identified as playing a key role in diabetes and obesity.
Psychopathy and neurodegenerative diseases are known to result from the disrupted balance of inositol pyrophosphates. However, the role and the mechanism of action of IP7 in brain neurons and nerve transmission remained unknown.
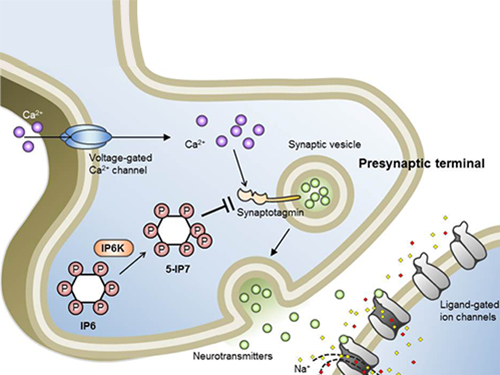
Professor Kim’s team has worked on inositol pyrophosphates for several years and discovered that very small quantities of IP7 control cell-signaling transduction. Professor Yoon of Yonsei University identified IP7 as a much stronger inhibitor of neuron signaling compared to IP6. In particular, IP7 directly suppresses synaptotagmin, one of the key proteins in neuron signaling. Moreover, Professor Kim of Kyung Hee University observed IP7 inhibition in sea horse neurons.
Together, the joint research team identified inositol pyrophosphates as the key switch metabolite of brain-signaling transduction.
The researchers hope that future research on synaptotagmin and IP7 will reveal the mechanism of neuron-signal transduction and thus enable the treatment of neurological disorders.
These research findings were the result of cooperation of various science and technology institutes: KAIST, Yonsei-IBS (Institute for Basic Science), Kyung Hee University, Sungkyunkwan University, KIST, University of Zurich in Switzerland, and Albert-Ludwigs-University Freiburg in Germany.
Schematic Image of Controlling the Synaptic Exocytotic Pathway by 5-IP7 , Helping the Understanding of the Signaling Mechanisms of Inositol Pyrophosphates
2016.07.21 View 12895
Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin.
Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team.
The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30, 2016.
This interdisciplinary research project was conducted by six research teams from four different countries and covered a wide scope of academic fields, from neurobiology to super resolution optic imaging.
Inositol pyrophosphates such as 5-diphosphoinositol pentakisphos-phate (5-IP7), which naturally occur in corns and beans, are essential metabolites in the body. In particular, inositol hexakisphosphate (IP6) has anti-cancer properties and is thought to have an important role in cell signaling.
Inositol pentakisphosphate (IP7) differs from IP6 by having an additional phosphate group, which was first discovered 20 years ago. IP7 has recently been identified as playing a key role in diabetes and obesity.
Psychopathy and neurodegenerative diseases are known to result from the disrupted balance of inositol pyrophosphates. However, the role and the mechanism of action of IP7 in brain neurons and nerve transmission remained unknown.
Professor Kim’s team has worked on inositol pyrophosphates for several years and discovered that very small quantities of IP7 control cell-signaling transduction. Professor Yoon of Yonsei University identified IP7 as a much stronger inhibitor of neuron signaling compared to IP6. In particular, IP7 directly suppresses synaptotagmin, one of the key proteins in neuron signaling. Moreover, Professor Kim of Kyung Hee University observed IP7 inhibition in sea horse neurons.
Together, the joint research team identified inositol pyrophosphates as the key switch metabolite of brain-signaling transduction.
The researchers hope that future research on synaptotagmin and IP7 will reveal the mechanism of neuron-signal transduction and thus enable the treatment of neurological disorders.
These research findings were the result of cooperation of various science and technology institutes: KAIST, Yonsei-IBS (Institute for Basic Science), Kyung Hee University, Sungkyunkwan University, KIST, University of Zurich in Switzerland, and Albert-Ludwigs-University Freiburg in Germany.
Schematic Image of Controlling the Synaptic Exocytotic Pathway by 5-IP7 , Helping the Understanding of the Signaling Mechanisms of Inositol Pyrophosphates
2016.07.21 View 12895 -
 Materials Developed for Sodium Rechargeable Battery by EEWS
The research group of Professor William Goddard III, You-Sung Jung, and Jang-Wook Choi from the Graduate School of Energy, Environment, Water, and Sustainability (EEWS) at KAIST has developed a new sodium-ion rechargeable battery which operates at a high voltage, can be charged, and stably discharges over 10,000 cycles. The research results were published in the online version of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on December 30, 2013.
Since the material costs of sodium rechargeable batteries is 30 to 40 times lower than lithium batteries, it has received attention as an energy saving tool for smart grids and as the next generation of lithium rechargeable batteries. Until now, sodium-ion rechargeable batteries have had issues with stability when charging and discharging. The research group developed a vanadium-based electrode to solve these problems.
The group said follow-up research will be continued to develop advanced technology on sodium rechargeable batteries as it is still currently in the beginning stages.
The research team: From left to right is Professors William Goddard, You-Sung Jung, and Jang-Wook Choi
2014.01.13 View 12535
Materials Developed for Sodium Rechargeable Battery by EEWS
The research group of Professor William Goddard III, You-Sung Jung, and Jang-Wook Choi from the Graduate School of Energy, Environment, Water, and Sustainability (EEWS) at KAIST has developed a new sodium-ion rechargeable battery which operates at a high voltage, can be charged, and stably discharges over 10,000 cycles. The research results were published in the online version of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on December 30, 2013.
Since the material costs of sodium rechargeable batteries is 30 to 40 times lower than lithium batteries, it has received attention as an energy saving tool for smart grids and as the next generation of lithium rechargeable batteries. Until now, sodium-ion rechargeable batteries have had issues with stability when charging and discharging. The research group developed a vanadium-based electrode to solve these problems.
The group said follow-up research will be continued to develop advanced technology on sodium rechargeable batteries as it is still currently in the beginning stages.
The research team: From left to right is Professors William Goddard, You-Sung Jung, and Jang-Wook Choi
2014.01.13 View 12535 -
 Scaling Laws between Population and Facility Densities Found
A research team led by Prof. Ha-Woong Jeong of the Department of Physics, KAIST, has found a positive correlation between facilities and population densities, university authorities said on Tuesday (Sept. 2). The research was conducted in the cooperation with a research team of Prof. Beom-Jun Kim at Sungkyunkwan University.
The researchers investigated the ideal relation between the population and the facilities within the framework of an economic mechanism governing microdynamics.
In previous studies based on the global optimization of facility positions in minimizing the overall travel distance between people and facilities, the relation between population and facilities should follow a simple law. The new empirical analysis, however, determined that the law is not a fixed value but spreads in a broad range depending on facility types.
To explain this discrepancy, the researchers proposed a model based on economic mechanism that mimics the competitive balance between the profit of the facilities and the social opportunity cost for population.
The results were published in the Proceedings of the National Academy of Sciences of the United States on Aug. 25.
2009.09.04 View 15227
Scaling Laws between Population and Facility Densities Found
A research team led by Prof. Ha-Woong Jeong of the Department of Physics, KAIST, has found a positive correlation between facilities and population densities, university authorities said on Tuesday (Sept. 2). The research was conducted in the cooperation with a research team of Prof. Beom-Jun Kim at Sungkyunkwan University.
The researchers investigated the ideal relation between the population and the facilities within the framework of an economic mechanism governing microdynamics.
In previous studies based on the global optimization of facility positions in minimizing the overall travel distance between people and facilities, the relation between population and facilities should follow a simple law. The new empirical analysis, however, determined that the law is not a fixed value but spreads in a broad range depending on facility types.
To explain this discrepancy, the researchers proposed a model based on economic mechanism that mimics the competitive balance between the profit of the facilities and the social opportunity cost for population.
The results were published in the Proceedings of the National Academy of Sciences of the United States on Aug. 25.
2009.09.04 View 15227 -
 Professor Seong-Ihl Woo Develops New High-Speed Research Method
Professor Seong-Ihl Woo Develops New High-Speed Research Method
Reduce research periods and expenses for thin film materials several ten times
Posted on the online version of Proceedings of National Academy of Sciences of the United States of America (PNAS) on January 9
A team led by Seong-Ihl Woo, a professor of KAIST Department of Chemical & Biomolecular Engineering and the director of the Center for Ultramicrochemical Process Systems, has developed a high-speed research method that can maximize research performances and posted the relevant contents on the online version of Proceedings of National Academy of Sciences of the United States of America (PNAS), a distinguished scientific journal, on January 9, 2007.
Professor Woo’s team has developed a high-speed research method that can fabricate several tens or several thousands of thin films with different compositions (mixing ratio) at the same time and carry out structural analysis and performance evaluation more than ten times faster and accurately, which leads to the shortening of the research processes of thin film materials. This is an epoch-making method that can reduce research periods and expenses several ten times or more, compared to the previous methods.
The qualities of final products of electronic materials, displays, and semi-conductors depend on the features of thin film materials. Averagely, it takes about two weeks or longer to fabricate a functional thin film and analyze and evaluate its performances. In order to fabricate thin film materials in need successfully, more than several thousand times of tests are required.
The existing thin film-fabricating equipment is expensive one demanding high-degree vacuum, such as chemical vapor deposition, sputtering, physical vapor deposition, laser evaporation, and so on. In order to fabricate thin films of various compositions with this equipment, a several million won-worth target (solid-state raw material) and precursors (volatile organic metal compound) pricing several hundreds won per gram are required. Therefore, huge amount of experiment expense is demanded for fabrication of several ten thousands of thin films with various compositions.
Professor Woo’s team has developed ‘combinatorial droplet chemical deposition’ equipment, which does not demand high-degree vacuum and is automated by computers and robots, by using a new high-speed research measure. The equipment is priced at about 1/5 of the existing equipment and easy for maintenance.
This equipment uses cheap reagents, instead of expensive raw materials. Reagents necessary to form required compositions are dissolved in water or proper solvents, and then applied by high frequencies to make several micrometer-scaled droplets (fine liquid droplet). Theses droplets are moved by nitrogen and dropped onto a substrate, which is to be fabricated into a thin film, and then subsequent thermal treatment is applied to the substrate to fabricate a thin film of required composition. At this moment, several tens or several hundreds of thin films with various compositions can be fabricated at the same time by reducing the size of thin film specimens into millimeter scale with the use of shade mask and adjusting vaporization time with masks, the moving speed of which can be adjusted. The expenses for materials necessary for the fabrication of thin films with this equipment amount to several ten thousands won per 100 grams, which is in the range of 1/100 and 1/10 of the previous methods, and the research period can be shortened into one of several tenth.
“If this new method is applied to the development of elements in the fields of core energy, material and health, which have not been discovered by the existing research methods so far, as well as researches in thin film material field, substantial effects will be brought,” said Professor Woo.
‘Combinatorial droplet chemical vaporization’ equipment is pending a domestic patent application and international patent applications at Japan and Germany. This equipment will be produced by order and provided to general researchers.
2007.02.02 View 19331
Professor Seong-Ihl Woo Develops New High-Speed Research Method
Professor Seong-Ihl Woo Develops New High-Speed Research Method
Reduce research periods and expenses for thin film materials several ten times
Posted on the online version of Proceedings of National Academy of Sciences of the United States of America (PNAS) on January 9
A team led by Seong-Ihl Woo, a professor of KAIST Department of Chemical & Biomolecular Engineering and the director of the Center for Ultramicrochemical Process Systems, has developed a high-speed research method that can maximize research performances and posted the relevant contents on the online version of Proceedings of National Academy of Sciences of the United States of America (PNAS), a distinguished scientific journal, on January 9, 2007.
Professor Woo’s team has developed a high-speed research method that can fabricate several tens or several thousands of thin films with different compositions (mixing ratio) at the same time and carry out structural analysis and performance evaluation more than ten times faster and accurately, which leads to the shortening of the research processes of thin film materials. This is an epoch-making method that can reduce research periods and expenses several ten times or more, compared to the previous methods.
The qualities of final products of electronic materials, displays, and semi-conductors depend on the features of thin film materials. Averagely, it takes about two weeks or longer to fabricate a functional thin film and analyze and evaluate its performances. In order to fabricate thin film materials in need successfully, more than several thousand times of tests are required.
The existing thin film-fabricating equipment is expensive one demanding high-degree vacuum, such as chemical vapor deposition, sputtering, physical vapor deposition, laser evaporation, and so on. In order to fabricate thin films of various compositions with this equipment, a several million won-worth target (solid-state raw material) and precursors (volatile organic metal compound) pricing several hundreds won per gram are required. Therefore, huge amount of experiment expense is demanded for fabrication of several ten thousands of thin films with various compositions.
Professor Woo’s team has developed ‘combinatorial droplet chemical deposition’ equipment, which does not demand high-degree vacuum and is automated by computers and robots, by using a new high-speed research measure. The equipment is priced at about 1/5 of the existing equipment and easy for maintenance.
This equipment uses cheap reagents, instead of expensive raw materials. Reagents necessary to form required compositions are dissolved in water or proper solvents, and then applied by high frequencies to make several micrometer-scaled droplets (fine liquid droplet). Theses droplets are moved by nitrogen and dropped onto a substrate, which is to be fabricated into a thin film, and then subsequent thermal treatment is applied to the substrate to fabricate a thin film of required composition. At this moment, several tens or several hundreds of thin films with various compositions can be fabricated at the same time by reducing the size of thin film specimens into millimeter scale with the use of shade mask and adjusting vaporization time with masks, the moving speed of which can be adjusted. The expenses for materials necessary for the fabrication of thin films with this equipment amount to several ten thousands won per 100 grams, which is in the range of 1/100 and 1/10 of the previous methods, and the research period can be shortened into one of several tenth.
“If this new method is applied to the development of elements in the fields of core energy, material and health, which have not been discovered by the existing research methods so far, as well as researches in thin film material field, substantial effects will be brought,” said Professor Woo.
‘Combinatorial droplet chemical vaporization’ equipment is pending a domestic patent application and international patent applications at Japan and Germany. This equipment will be produced by order and provided to general researchers.
2007.02.02 View 19331