Interactions
-
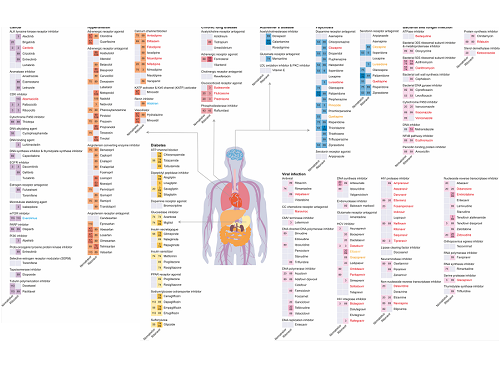 KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 9508
KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 9508 -
 Professor Juho Kim’s Team Wins Best Paper Award at ACM CHI 2022
The research team led by Professor Juho Kim from the KAIST School of Computing won a Best Paper Award and an Honorable Mention Award at the Association for Computing Machinery Conference on Human Factors in Computing Systems (ACM CHI) held between April 30 and May 6.
ACM CHI is the world’s most recognized conference in the field of human computer interactions (HCI), and is ranked number one out of all HCI-related journals and conferences based on Google Scholar’s h-5 index. Best paper awards are given to works that rank in the top one percent, and honorable mention awards are given to the top five percent of the papers accepted by the conference.
Professor Juho Kim presented a total of seven papers at ACM CHI 2022, and tied for the largest number of papers. A total of 19 papers were affiliated with KAIST, putting it fifth out of all participating institutes and thereby proving KAIST’s competence in research.
One of Professor Kim’s research teams composed of Jeongyeon Kim (first author, MS graduate) from the School of Computing, MS candidate Yubin Choi from the School of Electrical Engineering, and Dr. Meng Xia (post-doctoral associate in the School of Computing, currently a post-doctoral associate at Carnegie Mellon University) received a best paper award for their paper, “Mobile-Friendly Content Design for MOOCs: Challenges, Requirements, and Design Opportunities”.
The study analyzed the difficulties experienced by learners watching video-based educational content in a mobile environment and suggests guidelines for solutions. The research team analyzed 134 survey responses and 21 interviews, and revealed that texts that are too small or overcrowded are what mainly brings down the legibility of video contents. Additionally, lighting, noise, and surrounding environments that change frequently are also important factors that may disturb a learning experience.
Based on these findings, the team analyzed the aptness of 41,722 frames from 101 video lectures for mobile environments, and confirmed that they generally show low levels of adequacy. For instance, in the case of text sizes, only 24.5% of the frames were shown to be adequate for learning in mobile environments. To overcome this issue, the research team suggested a guideline that may improve the legibility of video contents and help overcome the difficulties arising from mobile learning environments.
The importance of and dependency on video-based learning continue to rise, especially in the wake of the pandemic, and it is meaningful that this research suggested a means to analyze and tackle the difficulties of users that learn from the small screens of mobile devices. Furthermore, the paper also suggested technology that can solve problems related to video-based learning through human-AI collaborations, enhancing existing video lectures and improving learning experiences. This technology can be applied to various video-based platforms and content creation.
Meanwhile, a research team composed of Ph.D. candidate Tae Soo Kim (first author), MS candidate DaEun Choi, and Ph.D. candidate Yoonseo Choi from the School of Computing received an honorable mention award for their paper, “Stylette: styling the Web with Natural Language”.
The research team developed a novel interface technology that allows nonexperts who are unfamiliar with technical jargon to edit website features through speech. People often find it difficult to use or find the information they need from various websites due to accessibility issues, device-related constraints, inconvenient design, style preferences, etc. However, it is not easy for laymen to edit website features without expertise in programming or design, and most end up just putting up with the inconveniences. But what if the system could read the intentions of its users from their everyday language like “emphasize this part a little more”, or “I want a more modern design”, and edit the features automatically?
Based on this question, Professor Kim’s research team developed ‘Stylette’, a system in which AI analyses its users’ speech expressed in their natural language and automatically recommends a new style that best fits their intentions. The research team created a new system by putting together language AI, visual AI, and user interface technologies. On the linguistic side, a large-scale language model AI converts the intentions of the users expressed through their everyday language into adequate style elements. On the visual side, computer vision AI compares 1.7 million existing web design features and recommends a style adequate for the current website. In an experiment where 40 nonexperts were asked to edit a website design, the subjects that used this system showed double the success rate in a time span that was 35% shorter compared to the control group.
It is meaningful that this research proposed a practical case in which AI technology constructs intuitive interactions with users. The developed technology can be applied to existing design applications and web browsers in a plug-in format, and can be utilized to improve websites or for advertisements by collecting the natural intention data of users on a large scale.
2022.06.13 View 9686
Professor Juho Kim’s Team Wins Best Paper Award at ACM CHI 2022
The research team led by Professor Juho Kim from the KAIST School of Computing won a Best Paper Award and an Honorable Mention Award at the Association for Computing Machinery Conference on Human Factors in Computing Systems (ACM CHI) held between April 30 and May 6.
ACM CHI is the world’s most recognized conference in the field of human computer interactions (HCI), and is ranked number one out of all HCI-related journals and conferences based on Google Scholar’s h-5 index. Best paper awards are given to works that rank in the top one percent, and honorable mention awards are given to the top five percent of the papers accepted by the conference.
Professor Juho Kim presented a total of seven papers at ACM CHI 2022, and tied for the largest number of papers. A total of 19 papers were affiliated with KAIST, putting it fifth out of all participating institutes and thereby proving KAIST’s competence in research.
One of Professor Kim’s research teams composed of Jeongyeon Kim (first author, MS graduate) from the School of Computing, MS candidate Yubin Choi from the School of Electrical Engineering, and Dr. Meng Xia (post-doctoral associate in the School of Computing, currently a post-doctoral associate at Carnegie Mellon University) received a best paper award for their paper, “Mobile-Friendly Content Design for MOOCs: Challenges, Requirements, and Design Opportunities”.
The study analyzed the difficulties experienced by learners watching video-based educational content in a mobile environment and suggests guidelines for solutions. The research team analyzed 134 survey responses and 21 interviews, and revealed that texts that are too small or overcrowded are what mainly brings down the legibility of video contents. Additionally, lighting, noise, and surrounding environments that change frequently are also important factors that may disturb a learning experience.
Based on these findings, the team analyzed the aptness of 41,722 frames from 101 video lectures for mobile environments, and confirmed that they generally show low levels of adequacy. For instance, in the case of text sizes, only 24.5% of the frames were shown to be adequate for learning in mobile environments. To overcome this issue, the research team suggested a guideline that may improve the legibility of video contents and help overcome the difficulties arising from mobile learning environments.
The importance of and dependency on video-based learning continue to rise, especially in the wake of the pandemic, and it is meaningful that this research suggested a means to analyze and tackle the difficulties of users that learn from the small screens of mobile devices. Furthermore, the paper also suggested technology that can solve problems related to video-based learning through human-AI collaborations, enhancing existing video lectures and improving learning experiences. This technology can be applied to various video-based platforms and content creation.
Meanwhile, a research team composed of Ph.D. candidate Tae Soo Kim (first author), MS candidate DaEun Choi, and Ph.D. candidate Yoonseo Choi from the School of Computing received an honorable mention award for their paper, “Stylette: styling the Web with Natural Language”.
The research team developed a novel interface technology that allows nonexperts who are unfamiliar with technical jargon to edit website features through speech. People often find it difficult to use or find the information they need from various websites due to accessibility issues, device-related constraints, inconvenient design, style preferences, etc. However, it is not easy for laymen to edit website features without expertise in programming or design, and most end up just putting up with the inconveniences. But what if the system could read the intentions of its users from their everyday language like “emphasize this part a little more”, or “I want a more modern design”, and edit the features automatically?
Based on this question, Professor Kim’s research team developed ‘Stylette’, a system in which AI analyses its users’ speech expressed in their natural language and automatically recommends a new style that best fits their intentions. The research team created a new system by putting together language AI, visual AI, and user interface technologies. On the linguistic side, a large-scale language model AI converts the intentions of the users expressed through their everyday language into adequate style elements. On the visual side, computer vision AI compares 1.7 million existing web design features and recommends a style adequate for the current website. In an experiment where 40 nonexperts were asked to edit a website design, the subjects that used this system showed double the success rate in a time span that was 35% shorter compared to the control group.
It is meaningful that this research proposed a practical case in which AI technology constructs intuitive interactions with users. The developed technology can be applied to existing design applications and web browsers in a plug-in format, and can be utilized to improve websites or for advertisements by collecting the natural intention data of users on a large scale.
2022.06.13 View 9686 -
 Professor Sung-Ju Lee’s Team Wins the Best Paper and the Methods Recognition Awards at the ACM CSCW
A research team led by Professor Sung-Ju Lee at the School of Electrical Engineering won the Best Paper Award and the Methods Recognition Award from ACM CSCW (International Conference on Computer-Supported Cooperative Work and Social Computing) 2021 for their paper “Reflect, not Regret: Understanding Regretful Smartphone Use with App Feature-Level Analysis”.
Founded in 1986, CSCW has been a premier conference on HCI (Human Computer Interaction) and Social Computing. This year, 340 full papers were presented and the best paper awards are given to the top 1% papers of the submitted. Methods Recognition, which is a new award, is given “for strong examples of work that includes well developed, explained, or implemented methods, and methodological innovation.”
Hyunsung Cho (KAIST alumus and currently a PhD candidate at Carnegie Mellon University), Daeun Choi (KAIST undergraduate researcher), Donghwi Kim (KAIST PhD Candidate), Wan Ju Kang (KAIST PhD Candidate), and Professor Eun Kyoung Choe (University of Maryland and KAIST alumna) collaborated on this research.
The authors developed a tool that tracks and analyzes which features of a mobile app (e.g., Instagram’s following post, following story, recommended post, post upload, direct messaging, etc.) are in use based on a smartphone’s User Interface (UI) layout. Utilizing this novel method, the authors revealed which feature usage patterns result in regretful smartphone use.
Professor Lee said, “Although many people enjoy the benefits of smartphones, issues have emerged from the overuse of smartphones. With this feature level analysis, users can reflect on their smartphone usage based on finer grained analysis and this could contribute to digital wellbeing.”
2021.11.22 View 8852
Professor Sung-Ju Lee’s Team Wins the Best Paper and the Methods Recognition Awards at the ACM CSCW
A research team led by Professor Sung-Ju Lee at the School of Electrical Engineering won the Best Paper Award and the Methods Recognition Award from ACM CSCW (International Conference on Computer-Supported Cooperative Work and Social Computing) 2021 for their paper “Reflect, not Regret: Understanding Regretful Smartphone Use with App Feature-Level Analysis”.
Founded in 1986, CSCW has been a premier conference on HCI (Human Computer Interaction) and Social Computing. This year, 340 full papers were presented and the best paper awards are given to the top 1% papers of the submitted. Methods Recognition, which is a new award, is given “for strong examples of work that includes well developed, explained, or implemented methods, and methodological innovation.”
Hyunsung Cho (KAIST alumus and currently a PhD candidate at Carnegie Mellon University), Daeun Choi (KAIST undergraduate researcher), Donghwi Kim (KAIST PhD Candidate), Wan Ju Kang (KAIST PhD Candidate), and Professor Eun Kyoung Choe (University of Maryland and KAIST alumna) collaborated on this research.
The authors developed a tool that tracks and analyzes which features of a mobile app (e.g., Instagram’s following post, following story, recommended post, post upload, direct messaging, etc.) are in use based on a smartphone’s User Interface (UI) layout. Utilizing this novel method, the authors revealed which feature usage patterns result in regretful smartphone use.
Professor Lee said, “Although many people enjoy the benefits of smartphones, issues have emerged from the overuse of smartphones. With this feature level analysis, users can reflect on their smartphone usage based on finer grained analysis and this could contribute to digital wellbeing.”
2021.11.22 View 8852 -
 Defining the Hund Physics Landscape of Two-Orbital Systems
Researchers identify exotic metals in unexpected quantum systems
Electrons are ubiquitous among atoms, subatomic tokens of energy that can independently change how a system behaves—but they also can change each other. An international research collaboration found that collectively measuring electrons revealed unique and unanticipated findings. The researchers published their results on May 17 in Physical Review Letters.
“It is not feasible to obtain the solution just by tracing the behavior of each individual electron,” said paper author Myung Joon Han, professor of physics at KAIST. “Instead, one should describe or track all the entangled electrons at once. This requires a clever way of treating this entanglement.”
Professor Han and the researchers used a recently developed “many-particle” theory to account for the entangled nature of electrons in solids, which approximates how electrons locally interact with one another to predict their global activity.
Through this approach, the researchers examined systems with two orbitals — the space in which electrons can inhabit. They found that the electrons locked into parallel arrangements within atom sites in solids. This phenomenon, known as Hund’s coupling, results in a Hund’s metal. This metallic phase, which can give rise to such properties as superconductivity, was thought only to exist in three-orbital systems.
“Our finding overturns a conventional viewpoint that at least three orbitals are needed for Hund’s metallicity to emerge,” Professor Han said, noting that two-orbital systems have not been a focus of attention for many physicists. “In addition to this finding of a Hund’s metal, we identified various metallic regimes that can naturally occur in generic, correlated electron materials.”
The researchers found four different correlated metals. One stems from the proximity to a Mott insulator, a state of a solid material that should be conductive but actually prevents conduction due to how the electrons interact. The other three metals form as electrons align their magnetic moments — or phases of producing a magnetic field — at various distances from the Mott insulator. Beyond identifying the metal phases, the researchers also suggested classification criteria to define each metal phase in other systems.
“This research will help scientists better characterize and understand the deeper nature of so-called ‘strongly correlated materials,’ in which the standard theory of solids breaks down due to the presence of strong Coulomb interactions between electrons,” Professor Han said, referring to the force with which the electrons attract or repel each other. These interactions are not typically present in solid materials but appear in materials with metallic phases.
The revelation of metals in two-orbital systems and the ability to determine whole system electron behavior could lead to even more discoveries, according to Professor Han.
“This will ultimately enable us to manipulate and control a variety of electron correlation phenomena,” Professor Han said.
Co-authors include Siheon Ryee from KAIST and Sangkook Choi from the Condensed Matter Physics and Materials Science Department, Brookhaven National Laboratory in the United States. Korea’s National Research Foundation and the U.S. Department of Energy’s (DOE) Office of Science, Basic Energy Sciences, supported this work.
-PublicationSiheon Ryee, Myung Joon Han, and SangKook Choi, 2021.Hund Physics Landscape of Two-Orbital Systems, Physical Review Letters, DOI: 10.1103/PhysRevLett.126.206401
-ProfileProfessor Myung Joon HanDepartment of PhysicsCollege of Natural ScienceKAIST
2021.06.17 View 10515
Defining the Hund Physics Landscape of Two-Orbital Systems
Researchers identify exotic metals in unexpected quantum systems
Electrons are ubiquitous among atoms, subatomic tokens of energy that can independently change how a system behaves—but they also can change each other. An international research collaboration found that collectively measuring electrons revealed unique and unanticipated findings. The researchers published their results on May 17 in Physical Review Letters.
“It is not feasible to obtain the solution just by tracing the behavior of each individual electron,” said paper author Myung Joon Han, professor of physics at KAIST. “Instead, one should describe or track all the entangled electrons at once. This requires a clever way of treating this entanglement.”
Professor Han and the researchers used a recently developed “many-particle” theory to account for the entangled nature of electrons in solids, which approximates how electrons locally interact with one another to predict their global activity.
Through this approach, the researchers examined systems with two orbitals — the space in which electrons can inhabit. They found that the electrons locked into parallel arrangements within atom sites in solids. This phenomenon, known as Hund’s coupling, results in a Hund’s metal. This metallic phase, which can give rise to such properties as superconductivity, was thought only to exist in three-orbital systems.
“Our finding overturns a conventional viewpoint that at least three orbitals are needed for Hund’s metallicity to emerge,” Professor Han said, noting that two-orbital systems have not been a focus of attention for many physicists. “In addition to this finding of a Hund’s metal, we identified various metallic regimes that can naturally occur in generic, correlated electron materials.”
The researchers found four different correlated metals. One stems from the proximity to a Mott insulator, a state of a solid material that should be conductive but actually prevents conduction due to how the electrons interact. The other three metals form as electrons align their magnetic moments — or phases of producing a magnetic field — at various distances from the Mott insulator. Beyond identifying the metal phases, the researchers also suggested classification criteria to define each metal phase in other systems.
“This research will help scientists better characterize and understand the deeper nature of so-called ‘strongly correlated materials,’ in which the standard theory of solids breaks down due to the presence of strong Coulomb interactions between electrons,” Professor Han said, referring to the force with which the electrons attract or repel each other. These interactions are not typically present in solid materials but appear in materials with metallic phases.
The revelation of metals in two-orbital systems and the ability to determine whole system electron behavior could lead to even more discoveries, according to Professor Han.
“This will ultimately enable us to manipulate and control a variety of electron correlation phenomena,” Professor Han said.
Co-authors include Siheon Ryee from KAIST and Sangkook Choi from the Condensed Matter Physics and Materials Science Department, Brookhaven National Laboratory in the United States. Korea’s National Research Foundation and the U.S. Department of Energy’s (DOE) Office of Science, Basic Energy Sciences, supported this work.
-PublicationSiheon Ryee, Myung Joon Han, and SangKook Choi, 2021.Hund Physics Landscape of Two-Orbital Systems, Physical Review Letters, DOI: 10.1103/PhysRevLett.126.206401
-ProfileProfessor Myung Joon HanDepartment of PhysicsCollege of Natural ScienceKAIST
2021.06.17 View 10515 -
 Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 13050
Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 13050 -
 Interactions Features KAIST's Human-Computer Interaction Lab
Interactions, a bi-monthly magazine published by the Association for Computing Machinery (ACM), the largest educational and scientific computing society in the world, featured an article introducing Human-Computer Interaction (HCI) Lab at KAIST in the March/April 2015 issue (http://interactions.acm.org/archive/toc/march-april-2015).
Established in 2002, the HCI Lab (http://hcil.kaist.ac.kr/) is run by Professor Geehyuk Lee of the Computer Science Department at KAIST. The lab conducts various research projects to improve the design and operation of physical user interfaces and develops new interaction techniques for new types of computers. For the article, see the link below:
ACM Interactions, March and April 2015
Day in the Lab: Human-Computer Interaction Lab @ KAIST
http://interactions.acm.org/archive/view/march-april-2015/human-computer-interaction-lab-kaist
2015.03.02 View 11501
Interactions Features KAIST's Human-Computer Interaction Lab
Interactions, a bi-monthly magazine published by the Association for Computing Machinery (ACM), the largest educational and scientific computing society in the world, featured an article introducing Human-Computer Interaction (HCI) Lab at KAIST in the March/April 2015 issue (http://interactions.acm.org/archive/toc/march-april-2015).
Established in 2002, the HCI Lab (http://hcil.kaist.ac.kr/) is run by Professor Geehyuk Lee of the Computer Science Department at KAIST. The lab conducts various research projects to improve the design and operation of physical user interfaces and develops new interaction techniques for new types of computers. For the article, see the link below:
ACM Interactions, March and April 2015
Day in the Lab: Human-Computer Interaction Lab @ KAIST
http://interactions.acm.org/archive/view/march-april-2015/human-computer-interaction-lab-kaist
2015.03.02 View 11501 -
 ACM Interactions: Demo Hour, March and April 2014 Issue
The
Association for Computing Machinery (ACM), the largest educational and scientific
computing society in the world, publishes a magazine called
Interactions
bi-monthly.
Interactions
is the flagship magazine
for the ACM’s Special Interest Group on Computer-Human Interaction (SIGCHI) with
a global circulation that includes all SIGCHI members.
In
its March and April 2014 issue, the Smart E-book was introduced. It was developed by Sangtae Kim, Jaejeung
Kim, and Soobin Lee at the Information Technology Convergence in KAIST
Institute, KAIST.
For
the article, please go to the link or download the .pdf files below:
Interactions,
March &
April 2014
Demo Hour: Bezel-Flipper
Bezel-Flipper
Interactions_Mar & Apr 2014.pdf
http://interactions.acm.org/archive/view/march-april-2014/demo-hour29
2014.03.28 View 11855
ACM Interactions: Demo Hour, March and April 2014 Issue
The
Association for Computing Machinery (ACM), the largest educational and scientific
computing society in the world, publishes a magazine called
Interactions
bi-monthly.
Interactions
is the flagship magazine
for the ACM’s Special Interest Group on Computer-Human Interaction (SIGCHI) with
a global circulation that includes all SIGCHI members.
In
its March and April 2014 issue, the Smart E-book was introduced. It was developed by Sangtae Kim, Jaejeung
Kim, and Soobin Lee at the Information Technology Convergence in KAIST
Institute, KAIST.
For
the article, please go to the link or download the .pdf files below:
Interactions,
March &
April 2014
Demo Hour: Bezel-Flipper
Bezel-Flipper
Interactions_Mar & Apr 2014.pdf
http://interactions.acm.org/archive/view/march-april-2014/demo-hour29
2014.03.28 View 11855 -
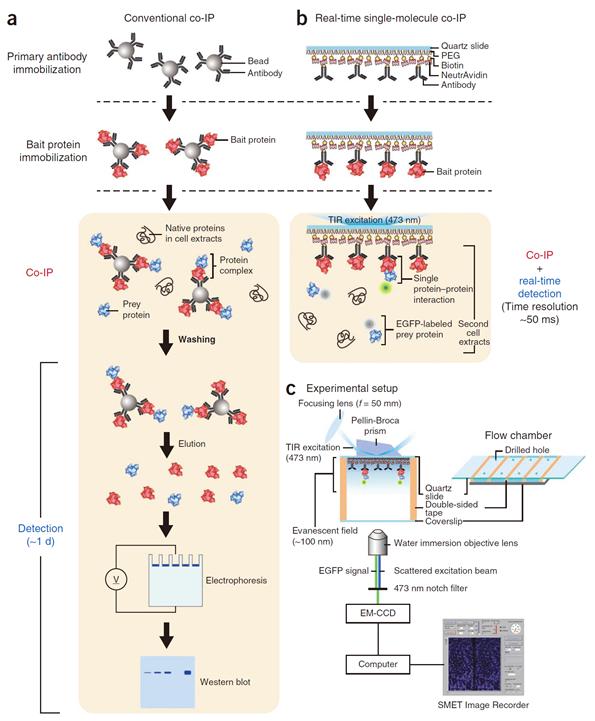 Success in Measuring Protein Interaction at the Molecular Level
Professor Tae Young Yoon
- Live observation of two protein interaction in molecular level successful- The limit in measurement and time resolution of immunoprecipitation technique improved by a hundred thousand fold
KAIST Department of Physics Professor Tae Young Yoon’s research team has successfully observed the interaction of two proteins live on molecular level and the findings were published in the October edition of Nature Protocols.
Professor Yoon’s research team developed a fluorescent microscope that can observe a single molecule. The team grafted the immunoprecipitation technique, traditionally used in protein interaction analysis, to the microscope to develop a “live molecular level immunoprecipitation technique”. The team successfully and accurately measured the reaction between two proteins by repeated momentary interactions in the unit of tens of milliseconds.
The existing immunoprecipitation technique required at least one day to detect interaction between two proteins. There were limitations in detecting momentary or weak interactions. Also, quantitative analysis of the results was difficult since the image was measured by protein-band strength. The technique could not be used for live observation.
The team aimed to drastically improve the existing technique and to develop accurate method of measurement on molecular level. The newly developed technology can enable observation of protein interaction within one hour. Also, the interaction can be measured live, thus the protein interaction phenomenon can be measured in depth.
Moreover, every programme used in the experiment was developed and distributed by the research team so source energy is secured and created the foundation for global infra.
Professor Tae Young Yoon said, “The newly developed technology does not require additional protein expression or purification. Hence, a very small sample of protein is enough to accurately analyse protein interaction on a kinetic level.” He continued, “Even cancerous protein from the tissue of a cancer patient can be analysed. Thus a platform for customised anti-cancer medicine in the future has been prepared, as well.”
Figure 1. Mimetic diagram comparing the existing immunoprecipitation technique and the newly developed live molecular level immunoprecipitation technique
2013.12.11 View 8892
Success in Measuring Protein Interaction at the Molecular Level
Professor Tae Young Yoon
- Live observation of two protein interaction in molecular level successful- The limit in measurement and time resolution of immunoprecipitation technique improved by a hundred thousand fold
KAIST Department of Physics Professor Tae Young Yoon’s research team has successfully observed the interaction of two proteins live on molecular level and the findings were published in the October edition of Nature Protocols.
Professor Yoon’s research team developed a fluorescent microscope that can observe a single molecule. The team grafted the immunoprecipitation technique, traditionally used in protein interaction analysis, to the microscope to develop a “live molecular level immunoprecipitation technique”. The team successfully and accurately measured the reaction between two proteins by repeated momentary interactions in the unit of tens of milliseconds.
The existing immunoprecipitation technique required at least one day to detect interaction between two proteins. There were limitations in detecting momentary or weak interactions. Also, quantitative analysis of the results was difficult since the image was measured by protein-band strength. The technique could not be used for live observation.
The team aimed to drastically improve the existing technique and to develop accurate method of measurement on molecular level. The newly developed technology can enable observation of protein interaction within one hour. Also, the interaction can be measured live, thus the protein interaction phenomenon can be measured in depth.
Moreover, every programme used in the experiment was developed and distributed by the research team so source energy is secured and created the foundation for global infra.
Professor Tae Young Yoon said, “The newly developed technology does not require additional protein expression or purification. Hence, a very small sample of protein is enough to accurately analyse protein interaction on a kinetic level.” He continued, “Even cancerous protein from the tissue of a cancer patient can be analysed. Thus a platform for customised anti-cancer medicine in the future has been prepared, as well.”
Figure 1. Mimetic diagram comparing the existing immunoprecipitation technique and the newly developed live molecular level immunoprecipitation technique
2013.12.11 View 8892 -
 The thermal fluctuation and elasticity of cell membranes, lipid vesicles, interacting with pore-forming peptides were reported by a research team at KAIST.
A research team from KAIST, consisted of Sung-Min Choi, Professor of Nuclear and Quantum Engineering Department, and Ji-Hwan Lee, a doctoral student in the Department, published a paper on the “thermal fluctuation and elasticity of lipid vesicles interacting with pore-forming peptides.” The paper was carried by Physical Review Letters, an internationally renowned peer-review journal on physics on July 16, 2010.
Cell membranes, which consist of lipid bilayers, play important roles in cells as barriers to maintain concentrations and matrices to host membrane proteins. During cellular processes such as cell fission and fusion, the cell membranes undergo various morphological changes governed by the interplay between protein and lipid membranes. There have been many theoretical and experimental approaches to understand cellular processes driven by protein-lipid membrane interactions. However, it is not fully established how the membrane elastic properties, which play an important role in membrane deformation, are affected by the protein-membrane interactions.
Antimicrobial peptides are one of the most common examples of proteins that modify membrane morphology. While the pore-forming mechanisms of antimicrobial peptides in lipid bilayers have been widely investigated, there have been only a few attempts to understand the mechanisms in terms of membrane elastic properties. In particular, the effects of pore formation on the membrane fluctuation and elastic properties, which provide key information to understand the mechanism of antimicrobial peptide activity, have not been reported yet. The research team reports the thermal fluctuation and elasticity of lipid vesicles interacting with pore-forming peptides, which were measured by neutron spin-echo spectroscopy.
The results of this study are expected to pay an important role in understanding the elastic behavior and morphological changes of cell membranes induced by protein-membrane interactions, and may provide new insights for developing new theoretical models for membrane fluctuations which include the membrane mediated interaction between protein patches.
(a) (b)
Figure
(a) Schematics for bound melittin and pores in lipid bilayers
(b) P NMR signal ratio (with/without Mn2+) of DOPC LUV-melittin vs P/L at 30˚C. The dashed line is a guide for eyes.
2010.07.23 View 13944
The thermal fluctuation and elasticity of cell membranes, lipid vesicles, interacting with pore-forming peptides were reported by a research team at KAIST.
A research team from KAIST, consisted of Sung-Min Choi, Professor of Nuclear and Quantum Engineering Department, and Ji-Hwan Lee, a doctoral student in the Department, published a paper on the “thermal fluctuation and elasticity of lipid vesicles interacting with pore-forming peptides.” The paper was carried by Physical Review Letters, an internationally renowned peer-review journal on physics on July 16, 2010.
Cell membranes, which consist of lipid bilayers, play important roles in cells as barriers to maintain concentrations and matrices to host membrane proteins. During cellular processes such as cell fission and fusion, the cell membranes undergo various morphological changes governed by the interplay between protein and lipid membranes. There have been many theoretical and experimental approaches to understand cellular processes driven by protein-lipid membrane interactions. However, it is not fully established how the membrane elastic properties, which play an important role in membrane deformation, are affected by the protein-membrane interactions.
Antimicrobial peptides are one of the most common examples of proteins that modify membrane morphology. While the pore-forming mechanisms of antimicrobial peptides in lipid bilayers have been widely investigated, there have been only a few attempts to understand the mechanisms in terms of membrane elastic properties. In particular, the effects of pore formation on the membrane fluctuation and elastic properties, which provide key information to understand the mechanism of antimicrobial peptide activity, have not been reported yet. The research team reports the thermal fluctuation and elasticity of lipid vesicles interacting with pore-forming peptides, which were measured by neutron spin-echo spectroscopy.
The results of this study are expected to pay an important role in understanding the elastic behavior and morphological changes of cell membranes induced by protein-membrane interactions, and may provide new insights for developing new theoretical models for membrane fluctuations which include the membrane mediated interaction between protein patches.
(a) (b)
Figure
(a) Schematics for bound melittin and pores in lipid bilayers
(b) P NMR signal ratio (with/without Mn2+) of DOPC LUV-melittin vs P/L at 30˚C. The dashed line is a guide for eyes.
2010.07.23 View 13944