Escherichia+coli
-
 KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
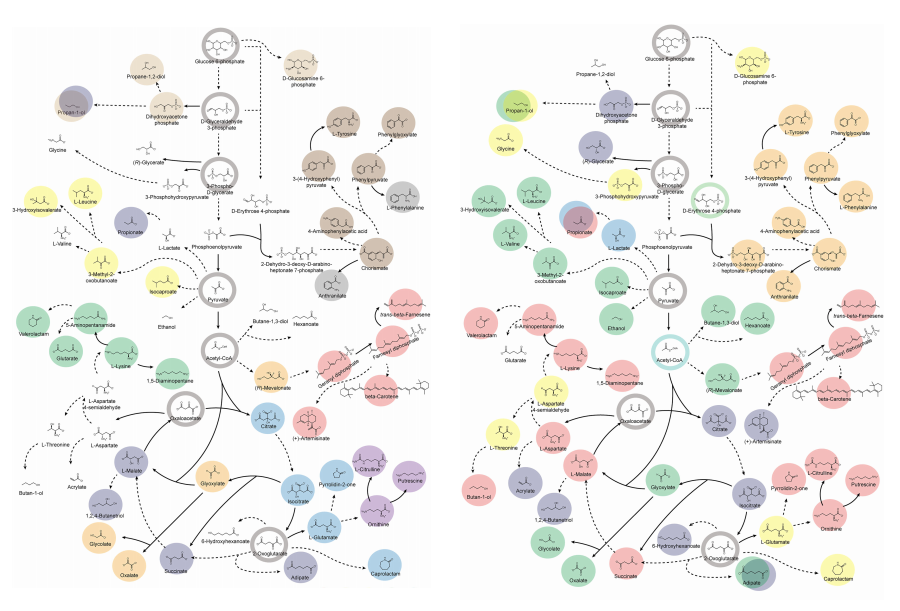
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3721
KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3721 -
 KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 4918
KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 4918 -
 Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 15274
Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 15274 -
 Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 11222
Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 11222 -
 Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 11471
Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 11471 -
 Natural Rainbow Colorants Microbially Produced
Integrated strategies of systems metabolic engineering and membrane engineering led to the production of natural rainbow colorants comprising seven natural colorants from bacteria for the first time
A research group at KAIST has engineered bacterial strains capable of producing three carotenoids and four violacein derivatives, completing the seven colors in the rainbow spectrum. The research team integrated systems metabolic engineering and membrane engineering strategies for the production of seven natural rainbow colorants in engineered Escherichia coli strains. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Colorants are widely used in our lives and are directly related to human health when we eat food additives and wear cosmetics. However, most of these colorants are made from petroleum, causing unexpected side effects and health problems. Furthermore, they raise environmental concerns such as water pollution from dyeing fabric in the textiles industry. For these reasons, the demand for the production of natural colorants using microorganisms has increased, but could not be readily realized due to the high cost and low yield of the bioprocesses.
These challenges inspired the metabolic engineers at KAIST including researchers Dr. Dongsoo Yang and Dr. Seon Young Park, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team reported the study entitled “Production of rainbow colorants by metabolically engineered Escherichia coli” in Advanced Science online on May 5. It was selected as the journal cover of the July 7 issue.
This research reports for the first time the production of rainbow colorants comprising three carotenoids and four violacein derivatives from glucose or glycerol via systems metabolic engineering and membrane engineering. The research group focused on the production of hydrophobic natural colorants useful for lipophilic food and dyeing garments. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, three carotenoids comprising astaxanthin (red), -carotene (orange), and zeaxanthin (yellow), and four violacein derivatives comprising proviolacein (green), prodeoxyviolacein (blue), violacein (navy), and deoxyviolacein (purple) could be produced. Thus, the production of natural colorants covering the complete rainbow spectrum was achieved.
When hydrophobic colorants are produced from microorganisms, the colorants are accumulated inside the cell. As the accumulation capacity is limited, the hydrophobic colorants could not be produced with concentrations higher than the limit. In this regard, the researchers engineered the cell morphology and generated inner-membrane vesicles (spherical membranous structures) to increase the intracellular capacity for accumulating the natural colorants. To further promote production, the researchers generated outer-membrane vesicles to secrete the natural colorants, thus succeeding in efficiently producing all of seven rainbow colorants. It was even more impressive that the production of natural green and navy colorants was achieved for the first time.
“The production of the seven natural rainbow colorants that can replace the current petroleum-based synthetic colorants was achieved for the first time,” said Dr. Dongsoo Yang. He explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals. “As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Lee.
This work was supported by the "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01550602)" Rural Development Administration, Republic of Korea.
-Publication:Dongsoo Yang, Seon Young Park, and Sang Yup Lee. Production of rainbow colorants by metabolically engineered Escherichia coli. Advanced Science, 2100743.
-Profile Distinguished Professor Sang Yup LeeMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2021.06.09 View 12443
Natural Rainbow Colorants Microbially Produced
Integrated strategies of systems metabolic engineering and membrane engineering led to the production of natural rainbow colorants comprising seven natural colorants from bacteria for the first time
A research group at KAIST has engineered bacterial strains capable of producing three carotenoids and four violacein derivatives, completing the seven colors in the rainbow spectrum. The research team integrated systems metabolic engineering and membrane engineering strategies for the production of seven natural rainbow colorants in engineered Escherichia coli strains. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Colorants are widely used in our lives and are directly related to human health when we eat food additives and wear cosmetics. However, most of these colorants are made from petroleum, causing unexpected side effects and health problems. Furthermore, they raise environmental concerns such as water pollution from dyeing fabric in the textiles industry. For these reasons, the demand for the production of natural colorants using microorganisms has increased, but could not be readily realized due to the high cost and low yield of the bioprocesses.
These challenges inspired the metabolic engineers at KAIST including researchers Dr. Dongsoo Yang and Dr. Seon Young Park, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team reported the study entitled “Production of rainbow colorants by metabolically engineered Escherichia coli” in Advanced Science online on May 5. It was selected as the journal cover of the July 7 issue.
This research reports for the first time the production of rainbow colorants comprising three carotenoids and four violacein derivatives from glucose or glycerol via systems metabolic engineering and membrane engineering. The research group focused on the production of hydrophobic natural colorants useful for lipophilic food and dyeing garments. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, three carotenoids comprising astaxanthin (red), -carotene (orange), and zeaxanthin (yellow), and four violacein derivatives comprising proviolacein (green), prodeoxyviolacein (blue), violacein (navy), and deoxyviolacein (purple) could be produced. Thus, the production of natural colorants covering the complete rainbow spectrum was achieved.
When hydrophobic colorants are produced from microorganisms, the colorants are accumulated inside the cell. As the accumulation capacity is limited, the hydrophobic colorants could not be produced with concentrations higher than the limit. In this regard, the researchers engineered the cell morphology and generated inner-membrane vesicles (spherical membranous structures) to increase the intracellular capacity for accumulating the natural colorants. To further promote production, the researchers generated outer-membrane vesicles to secrete the natural colorants, thus succeeding in efficiently producing all of seven rainbow colorants. It was even more impressive that the production of natural green and navy colorants was achieved for the first time.
“The production of the seven natural rainbow colorants that can replace the current petroleum-based synthetic colorants was achieved for the first time,” said Dr. Dongsoo Yang. He explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals. “As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Lee.
This work was supported by the "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01550602)" Rural Development Administration, Republic of Korea.
-Publication:Dongsoo Yang, Seon Young Park, and Sang Yup Lee. Production of rainbow colorants by metabolically engineered Escherichia coli. Advanced Science, 2100743.
-Profile Distinguished Professor Sang Yup LeeMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2021.06.09 View 12443 -
 Microbial Production of a Natural Red Colorant Carminic Acid
Metabolic engineering and computer-simulated enzyme engineering led to the production of carminic acid, a natural red colorant, from bacteria for the first time
A research group at KAIST has engineered a bacterium capable of producing a natural red colorant, carminic acid, which is widely used for food and cosmetics. The research team reported the complete biosynthesis of carminic acid from glucose in engineered Escherichia coli. The strategies will be useful for the design and construction of biosynthetic pathways involving unknown enzymes and consequently the production of diverse industrially important natural products for the food, pharmaceutical, and cosmetic industries.
Carminic acid is a natural red colorant widely being used for products such as strawberry milk and lipstick. However, carminic acid has been produced by farming cochineals, a scale insect which only grows in the region around Peru and Canary Islands, followed by complicated multi-step purification processes. Moreover, carminic acid often contains protein contaminants that cause allergies so many people are unwilling to consume products made of insect-driven colorants. On that account, manufacturers around the world are using alternative red colorants despite the fact that carminic acid is one of the most stable natural red colorants.
These challenges inspired the metabolic engineering research group at KAIST to address this issue. Its members include postdoctoral researchers Dongsoo Yang and Woo Dae Jang, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering. This study entitled “Production of carminic acid by metabolically engineered Escherichia coli” was published online in the Journal of the American Chemical Society (JACS) on April 2.
This research reports for the first time the development of a bacterial strain capable of producing carminic acid from glucose via metabolic engineering and computer simulation-assisted enzyme engineering. The research group optimized the type II polyketide synthase machinery to efficiently produce the precursor of carminic acid, flavokermesic acid.
Since the enzymes responsible for the remaining two reactions were neither discovered nor functional, biochemical reaction analysis was performed to identify enzymes that can convert flavokermesic acid into carminic acid. Then, homology modeling and docking simulations were performed to enhance the activities of the two identified enzymes. The team could confirm that the final engineered strain could produce carminic acid directly from glucose. The C-glucosyltransferase developed in this study was found to be generally applicable for other natural products as showcased by the successful production of an additional product, aloesin, which is found in aloe leaves.
“The most important part of this research is that unknown enzymes for the production of target natural products were identified and improved by biochemical reaction analyses and computer simulation-assisted enzyme engineering,” says Dr. Dongsoo Yang. He explained the development of a generally applicable C-glucosyltransferase is also useful since C-glucosylation is a relatively unexplored reaction in bacteria including Escherichia coli. Using the C-glucosyltransferase developed in this study, both carminic acid and aloesin were successfully produced from glucose.
“A sustainable and insect-free method of producing carminic acid was achieved for the first time in this study. Unknown or inefficient enzymes have always been a major problem in natural product biosynthesis, and here we suggest one effective solution for solving this problem. As maintaining good health in the aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” said Distinguished Professor Sang Yup Lee.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries of the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea and the KAIST Cross-Generation Collaborative Lab project; Sang Yup Lee and Dongsoo Yang were also supported by Novo Nordisk Foundation in Denmark.
Publication:
Dongsoo Yang, Woo Dae Jang, and Sang Yup Lee. Production of carminic acid by metabolically engineered Escherichia coli. at the Journal of the American Chemical Society. https://doi.org.10.1021/jacs.0c12406
Profile:
Sang Yup Lee, PhD
Distinguished Professor
leesy@kaist.ac.kr
http://mbel.kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
Department of Chemical and Biomolecular Engineering
KAIST
2021.04.06 View 13749
Microbial Production of a Natural Red Colorant Carminic Acid
Metabolic engineering and computer-simulated enzyme engineering led to the production of carminic acid, a natural red colorant, from bacteria for the first time
A research group at KAIST has engineered a bacterium capable of producing a natural red colorant, carminic acid, which is widely used for food and cosmetics. The research team reported the complete biosynthesis of carminic acid from glucose in engineered Escherichia coli. The strategies will be useful for the design and construction of biosynthetic pathways involving unknown enzymes and consequently the production of diverse industrially important natural products for the food, pharmaceutical, and cosmetic industries.
Carminic acid is a natural red colorant widely being used for products such as strawberry milk and lipstick. However, carminic acid has been produced by farming cochineals, a scale insect which only grows in the region around Peru and Canary Islands, followed by complicated multi-step purification processes. Moreover, carminic acid often contains protein contaminants that cause allergies so many people are unwilling to consume products made of insect-driven colorants. On that account, manufacturers around the world are using alternative red colorants despite the fact that carminic acid is one of the most stable natural red colorants.
These challenges inspired the metabolic engineering research group at KAIST to address this issue. Its members include postdoctoral researchers Dongsoo Yang and Woo Dae Jang, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering. This study entitled “Production of carminic acid by metabolically engineered Escherichia coli” was published online in the Journal of the American Chemical Society (JACS) on April 2.
This research reports for the first time the development of a bacterial strain capable of producing carminic acid from glucose via metabolic engineering and computer simulation-assisted enzyme engineering. The research group optimized the type II polyketide synthase machinery to efficiently produce the precursor of carminic acid, flavokermesic acid.
Since the enzymes responsible for the remaining two reactions were neither discovered nor functional, biochemical reaction analysis was performed to identify enzymes that can convert flavokermesic acid into carminic acid. Then, homology modeling and docking simulations were performed to enhance the activities of the two identified enzymes. The team could confirm that the final engineered strain could produce carminic acid directly from glucose. The C-glucosyltransferase developed in this study was found to be generally applicable for other natural products as showcased by the successful production of an additional product, aloesin, which is found in aloe leaves.
“The most important part of this research is that unknown enzymes for the production of target natural products were identified and improved by biochemical reaction analyses and computer simulation-assisted enzyme engineering,” says Dr. Dongsoo Yang. He explained the development of a generally applicable C-glucosyltransferase is also useful since C-glucosylation is a relatively unexplored reaction in bacteria including Escherichia coli. Using the C-glucosyltransferase developed in this study, both carminic acid and aloesin were successfully produced from glucose.
“A sustainable and insect-free method of producing carminic acid was achieved for the first time in this study. Unknown or inefficient enzymes have always been a major problem in natural product biosynthesis, and here we suggest one effective solution for solving this problem. As maintaining good health in the aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” said Distinguished Professor Sang Yup Lee.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries of the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea and the KAIST Cross-Generation Collaborative Lab project; Sang Yup Lee and Dongsoo Yang were also supported by Novo Nordisk Foundation in Denmark.
Publication:
Dongsoo Yang, Woo Dae Jang, and Sang Yup Lee. Production of carminic acid by metabolically engineered Escherichia coli. at the Journal of the American Chemical Society. https://doi.org.10.1021/jacs.0c12406
Profile:
Sang Yup Lee, PhD
Distinguished Professor
leesy@kaist.ac.kr
http://mbel.kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
Department of Chemical and Biomolecular Engineering
KAIST
2021.04.06 View 13749