DNA
-
 Genome Sequencing Unveils Mutational Impacts of Radiation on Mammalian Cells
Recent release of the waste water from Japan's Fukushima nuclear disaster stirred apprehension regarding the health implications of radiation exposure. Classified as a Group 1 carcinogen, ionizing radiation has long been associated with various cancers and genetic disorders, as evidenced by survivors and descendants of atomic bombings and the Chernobyl disaster. Despite much smaller amount, we remain consistently exposed to low levels of radiation in everyday life and medical procedures.
Radiation, whether in the form of high-energy particles or electromagnetic waves, is conventionally known to break our cellular DNA, leading to cancer and genetic disorders. Yet, our understanding of the quantitative and qualitative mutational impacts of ionizing radiation has been incomplete.
On the 14th, Professor Young Seok Ju and his research team from KAIST, in collaboration with Dr. Tae Gen Son from the Dongnam Institute of Radiological and Medical Science, and Professors Kyung Su Kim and Ji Hyun Chang from Seoul National University, unveiled a breakthrough. Their study, led by joint first authors Drs. Jeonghwan Youk, Hyun Woo Kwon, Joonoh Lim, Eunji Kim and Tae-Woo Kim, titled "Quantitative and qualitative mutational impact of ionizing radiation on normal cells," was published in Cell Genomics.
Employing meticulous techniques, the research team comprehensively analyzed the whole-genome sequences of cells pre- and post-radiation exposure, pinpointing radiation-induced DNA mutations. Experiments involving cells from different organs of humans and mice exposed to varying radiation doses revealed mutation patterns correlating with exposure levels. (Figure 1)
Notably, exposure to 1 Gray (Gy) of radiation resulted in on average 14 mutations in every post-exposure cell. (Figure 2) Unlike other carcinogens, radiation-induced mutations primarily comprised short base deletions and a set of structural variations including inversions, translocations, and various complex genomic rearrangements. (Figure 3) Interestingly, experiments subjecting cells to low radiation dose rate over 100 days demonstrated that mutation quantities, under equivalent total radiation doses, mirrored those of high-dose exposure.
"Through this study, we have clearly elucidated the effects of radiation on cells at the molecular level," said Prof. Ju at KAIST. "Now we understand better how radiation changes the DNA of our cells," he added.
Dr. Son from the Dongnam Institute of Radiological and Medical Science stated, "Based on this study, we will continue to research the effects of very low and very high doses of radiation on the human body," and further remarked, "We will advance the development of safe and effective radiation therapy techniques."
Professors Kim and Chang from Seoul National University College of Medicine expressed their views, saying, "Through this study, we believe we now have a tool to accurately understand the impact of radiation on human DNA," and added, "We hope that many subsequent studies will emerge using the research methodologies employed in this study."
This research represents a significant leap forward in radiation studies, made possible through collaborative efforts and interdisciplinary approaches. This pioneering research engaged scholars from diverse backgrounds, spanning from the Genetic Engineering Research Institute at Seoul National University, the Cambridge Stem Cell Institute in the UK, the Institute for Molecular Biotechnology in Austria (IMBA), and the Genome Insight Inc. (a KAIST spin-off start-up). This study was supported by various institutions including the National Research Foundation of Korea, Dongnam Institute of Radiological and Medical Science (supported by Ministry of Science and ICT, the government of South Korea), the Suh Kyungbae Foundation, the Human Frontier Science Program (HFSP), and the Korea University Anam Hospital Korea Foundation for the Advancement of Science and Creativity, the Ministry of Science and ICT, and the National R&D Program.
2024.02.15 View 8840
Genome Sequencing Unveils Mutational Impacts of Radiation on Mammalian Cells
Recent release of the waste water from Japan's Fukushima nuclear disaster stirred apprehension regarding the health implications of radiation exposure. Classified as a Group 1 carcinogen, ionizing radiation has long been associated with various cancers and genetic disorders, as evidenced by survivors and descendants of atomic bombings and the Chernobyl disaster. Despite much smaller amount, we remain consistently exposed to low levels of radiation in everyday life and medical procedures.
Radiation, whether in the form of high-energy particles or electromagnetic waves, is conventionally known to break our cellular DNA, leading to cancer and genetic disorders. Yet, our understanding of the quantitative and qualitative mutational impacts of ionizing radiation has been incomplete.
On the 14th, Professor Young Seok Ju and his research team from KAIST, in collaboration with Dr. Tae Gen Son from the Dongnam Institute of Radiological and Medical Science, and Professors Kyung Su Kim and Ji Hyun Chang from Seoul National University, unveiled a breakthrough. Their study, led by joint first authors Drs. Jeonghwan Youk, Hyun Woo Kwon, Joonoh Lim, Eunji Kim and Tae-Woo Kim, titled "Quantitative and qualitative mutational impact of ionizing radiation on normal cells," was published in Cell Genomics.
Employing meticulous techniques, the research team comprehensively analyzed the whole-genome sequences of cells pre- and post-radiation exposure, pinpointing radiation-induced DNA mutations. Experiments involving cells from different organs of humans and mice exposed to varying radiation doses revealed mutation patterns correlating with exposure levels. (Figure 1)
Notably, exposure to 1 Gray (Gy) of radiation resulted in on average 14 mutations in every post-exposure cell. (Figure 2) Unlike other carcinogens, radiation-induced mutations primarily comprised short base deletions and a set of structural variations including inversions, translocations, and various complex genomic rearrangements. (Figure 3) Interestingly, experiments subjecting cells to low radiation dose rate over 100 days demonstrated that mutation quantities, under equivalent total radiation doses, mirrored those of high-dose exposure.
"Through this study, we have clearly elucidated the effects of radiation on cells at the molecular level," said Prof. Ju at KAIST. "Now we understand better how radiation changes the DNA of our cells," he added.
Dr. Son from the Dongnam Institute of Radiological and Medical Science stated, "Based on this study, we will continue to research the effects of very low and very high doses of radiation on the human body," and further remarked, "We will advance the development of safe and effective radiation therapy techniques."
Professors Kim and Chang from Seoul National University College of Medicine expressed their views, saying, "Through this study, we believe we now have a tool to accurately understand the impact of radiation on human DNA," and added, "We hope that many subsequent studies will emerge using the research methodologies employed in this study."
This research represents a significant leap forward in radiation studies, made possible through collaborative efforts and interdisciplinary approaches. This pioneering research engaged scholars from diverse backgrounds, spanning from the Genetic Engineering Research Institute at Seoul National University, the Cambridge Stem Cell Institute in the UK, the Institute for Molecular Biotechnology in Austria (IMBA), and the Genome Insight Inc. (a KAIST spin-off start-up). This study was supported by various institutions including the National Research Foundation of Korea, Dongnam Institute of Radiological and Medical Science (supported by Ministry of Science and ICT, the government of South Korea), the Suh Kyungbae Foundation, the Human Frontier Science Program (HFSP), and the Korea University Anam Hospital Korea Foundation for the Advancement of Science and Creativity, the Ministry of Science and ICT, and the National R&D Program.
2024.02.15 View 8840 -
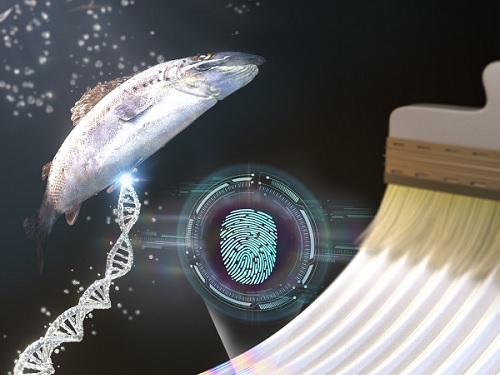 KAIST research team develops a forgery prevention technique using salmon DNA
The authenticity scandal that plagued the artwork “Beautiful Woman” by Kyung-ja Chun for 30 years shows how concerns about replicas can become a burden to artists, as most of them are not experts in the field of anti-counterfeiting. To solve this problem, artist-friendly physical unclonable functions (PUFs) based on optical techniques instead of electronic ones, which can be applied immediately onto artwork through brushstrokes are needed.
On May 23, a KAIST research team led by Professor Dong Ki Yoon in the Department of Chemistry revealed the development of a proprietary technology for security and certification using random patterns that occur during the self-assembly of soft materials.
With the development of the Internet of Things in recent years, various electronic devices and services can now be connected to the internet and carry out new innovative functions. However, counterfeiting technologies that infringe on individuals’ privacy have also entered the marketplace.
The technique developed by the research team involves random and spontaneous patterns that naturally occur during the self-assembly of two different types of soft materials, which can be used in the same way as human fingerprints for non-replicable security. This is very significant in that even non-experts in the field of security can construct anti-counterfeiting systems through simple actions like drawing a picture.
The team developed two unique methods. The first method uses liquid crystals. When liquid crystals become trapped in patterned substrates, they induce the symmetrical destruction of the structure and create a maze-like topology (Figure 1). The research team defined the pathways open to the right as 0 (blue), and those open to the left as 1 (red), and confirmed that the structure could be converted into a digital code composed of 0’s and 1’s that can serve as a type of fingerprint through object recognition using machine learning. This groundbreaking technique can be utilized by non-experts, as it does not require complex semiconductor patterns that are required by existing technology, and can be observed through the level of resolution of a smartphone camera. In particular, this technique can reconstruct information more easily than conventional methods that use semiconductor chips.
< Figure 1. Security technology using the maze made up of magnetically-assembled structures formed on a substrate patterned with liquid crystal materials. >
The second method uses DNA extracted from salmon. The DNA can be dissolved in water and applied with a brush to induce bulking instability, which forms random patterns similar to a zebra’s stripes. Here, the patterns create ridge endings and bifurcation, which are characteristics in fingerprints, and these can also be digitalized into 0’s and 1’s through machine learning. The research team applied conventional fingerprint recognition technology to this patterning technique and demonstrated its use as an artificial fingerprint. This method can be easily carried out using a brush, and the solution can be mixed into various colors and used as a new security ink.
< Figure 2. Technology to produce security ink using DNA polymers extracted from salmon >
This new security technology developed by the research team uses only simple organic materials and requires basic manufacturing processes, making it possible to enhance security at a low cost. In addition, users can produce patterns in the shapes and sizes they want, and even if the patterns are made in the same way, their randomness makes each individual pattern different. This provides high levels of security and gives the technique enhanced marketability.
Professor Dong Ki Yoon said, “These studies have taken the randomness that naturally occurs during self-assembly to create non-replicable patterns that can act like human fingerprints.” He added, “These ideas will be the cornerstone of technology that applies the many randomities that exist in nature to security systems.”
The two studies were published in the journal Advanced Materials under the titles “1Planar Spin Glass with Topologically-Protected Mazes in the Liquid Crystal Targeting for Reconfigurable Micro Security Media” and “2Paintable Physical Unclonable Function Using DNA” on May 6 and 5, respectively.
Author Information: 1Geonhyeong Park, Yun-Seok Choi, S. Joon Kwon*, and Dong Ki Yoon*/ 2Soon Mo Park†, Geonhyeong Park†, Dong Ki Yoon*: †co-first authors, *corresponding author
This research was funded by the Center for Multiscale Chiral Architectures and supported by the Ministry of Science and ICT-Korea Research Foundation, BRIDGE Convergent Research and Development Program, the Running Together Project, and the Samsung Future Technology Development Program.
< Figure 1-1. A scene from the schematic animation of the process of Blues (0) and Reds (1) forming the PUF by exploring the maze. From "Planar Spin Glass with Topologically-Protected Mazes in the Liquid Crystal Targeting for Reconfigurable Micro Security Media" by Geonhyeong Park, Yun-Seok Choi, S. Joon Kwon, Dong Ki Yoon. https://doi.org/10.1002/adma.202303077 >
< Figure 2-1. A schematic diagram of the formation of digital fingerprints formed using the DNA ink. From "Paintable Physical Unclonable Function Using DNA" by Soon Mo Park, Geonhyeong Park, Dong Ki Yoon. https://doi.org/10.1002/adma.202302135 >
2023.06.08 View 8545
KAIST research team develops a forgery prevention technique using salmon DNA
The authenticity scandal that plagued the artwork “Beautiful Woman” by Kyung-ja Chun for 30 years shows how concerns about replicas can become a burden to artists, as most of them are not experts in the field of anti-counterfeiting. To solve this problem, artist-friendly physical unclonable functions (PUFs) based on optical techniques instead of electronic ones, which can be applied immediately onto artwork through brushstrokes are needed.
On May 23, a KAIST research team led by Professor Dong Ki Yoon in the Department of Chemistry revealed the development of a proprietary technology for security and certification using random patterns that occur during the self-assembly of soft materials.
With the development of the Internet of Things in recent years, various electronic devices and services can now be connected to the internet and carry out new innovative functions. However, counterfeiting technologies that infringe on individuals’ privacy have also entered the marketplace.
The technique developed by the research team involves random and spontaneous patterns that naturally occur during the self-assembly of two different types of soft materials, which can be used in the same way as human fingerprints for non-replicable security. This is very significant in that even non-experts in the field of security can construct anti-counterfeiting systems through simple actions like drawing a picture.
The team developed two unique methods. The first method uses liquid crystals. When liquid crystals become trapped in patterned substrates, they induce the symmetrical destruction of the structure and create a maze-like topology (Figure 1). The research team defined the pathways open to the right as 0 (blue), and those open to the left as 1 (red), and confirmed that the structure could be converted into a digital code composed of 0’s and 1’s that can serve as a type of fingerprint through object recognition using machine learning. This groundbreaking technique can be utilized by non-experts, as it does not require complex semiconductor patterns that are required by existing technology, and can be observed through the level of resolution of a smartphone camera. In particular, this technique can reconstruct information more easily than conventional methods that use semiconductor chips.
< Figure 1. Security technology using the maze made up of magnetically-assembled structures formed on a substrate patterned with liquid crystal materials. >
The second method uses DNA extracted from salmon. The DNA can be dissolved in water and applied with a brush to induce bulking instability, which forms random patterns similar to a zebra’s stripes. Here, the patterns create ridge endings and bifurcation, which are characteristics in fingerprints, and these can also be digitalized into 0’s and 1’s through machine learning. The research team applied conventional fingerprint recognition technology to this patterning technique and demonstrated its use as an artificial fingerprint. This method can be easily carried out using a brush, and the solution can be mixed into various colors and used as a new security ink.
< Figure 2. Technology to produce security ink using DNA polymers extracted from salmon >
This new security technology developed by the research team uses only simple organic materials and requires basic manufacturing processes, making it possible to enhance security at a low cost. In addition, users can produce patterns in the shapes and sizes they want, and even if the patterns are made in the same way, their randomness makes each individual pattern different. This provides high levels of security and gives the technique enhanced marketability.
Professor Dong Ki Yoon said, “These studies have taken the randomness that naturally occurs during self-assembly to create non-replicable patterns that can act like human fingerprints.” He added, “These ideas will be the cornerstone of technology that applies the many randomities that exist in nature to security systems.”
The two studies were published in the journal Advanced Materials under the titles “1Planar Spin Glass with Topologically-Protected Mazes in the Liquid Crystal Targeting for Reconfigurable Micro Security Media” and “2Paintable Physical Unclonable Function Using DNA” on May 6 and 5, respectively.
Author Information: 1Geonhyeong Park, Yun-Seok Choi, S. Joon Kwon*, and Dong Ki Yoon*/ 2Soon Mo Park†, Geonhyeong Park†, Dong Ki Yoon*: †co-first authors, *corresponding author
This research was funded by the Center for Multiscale Chiral Architectures and supported by the Ministry of Science and ICT-Korea Research Foundation, BRIDGE Convergent Research and Development Program, the Running Together Project, and the Samsung Future Technology Development Program.
< Figure 1-1. A scene from the schematic animation of the process of Blues (0) and Reds (1) forming the PUF by exploring the maze. From "Planar Spin Glass with Topologically-Protected Mazes in the Liquid Crystal Targeting for Reconfigurable Micro Security Media" by Geonhyeong Park, Yun-Seok Choi, S. Joon Kwon, Dong Ki Yoon. https://doi.org/10.1002/adma.202303077 >
< Figure 2-1. A schematic diagram of the formation of digital fingerprints formed using the DNA ink. From "Paintable Physical Unclonable Function Using DNA" by Soon Mo Park, Geonhyeong Park, Dong Ki Yoon. https://doi.org/10.1002/adma.202302135 >
2023.06.08 View 8545 -
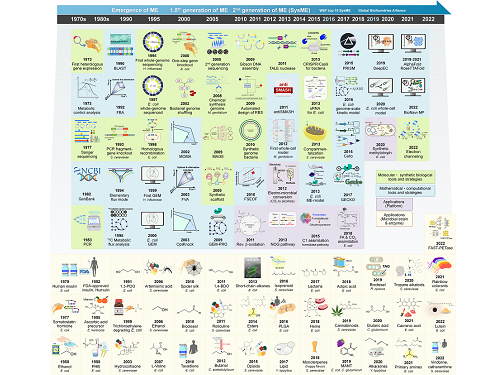 Overview of the 30-year history of metabolic engineering
< Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST >
A research team comprised of Gi Bae Kim, Dr. So Young Choi, Dr. In Jin Cho, Da-Hee Ahn, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST reported the 30-year history of metabolic engineering, highlighting examples of recent progress in the field and contributions to sustainability and health. Their paper “Metabolic engineering for sustainability and health” was published online in the 40th anniversary special issue of Trends in Biotechnology on January 10, 2023.
Metabolic engineering, a discipline of engineering that modifies cell phenotypes through molecular and genetic-level manipulations to improve cellular activities, has been studied since the early 1990s, and has progressed significantly over the past 30 years. In particular, metabolic engineering has enabled the engineering of microorganisms for the development of microbial cell factories capable of efficiently producing chemicals and materials as well as degrading recalcitrant contaminants.
This review article revisited how metabolic engineering has advanced over the past 30 years, from the advent of genetic engineering techniques such as recombinant DNA technologies to recent breakthroughs in systems metabolic engineering and data science aided by artificial intelligence. The research team highlighted momentous events and achievements in metabolic engineering, providing both trends and future directions in the field. Metabolic engineering’s contributions to bio-based sustainable chemicals and clean energy, health, and bioremediation were also reviewed. Finally, the research team shared their perspectives on the future challenges impacting metabolic engineering than must be overcome in order to achieve advancements in sustainability and health.
Distinguished Professor Sang Yup Lee said, “Replacing fossil resource-based chemical processes with bio-based sustainable processes for the production of chemicals, fuels, and materials using metabolic engineering has become our essential task for the future. By looking back on the 30+ years of metabolic engineering, we aimed to highlight the contributions of metabolic engineering to achieve sustainability and good health.” He added, “Metabolic engineering will play an increasingly important role as a key solution to the climate crisis, environmental pollution, food and energy shortages, and health problems in aging societies.”
< Figure: Metabolic Engineering Timeline >
2023.01.25 View 11568
Overview of the 30-year history of metabolic engineering
< Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST >
A research team comprised of Gi Bae Kim, Dr. So Young Choi, Dr. In Jin Cho, Da-Hee Ahn, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST reported the 30-year history of metabolic engineering, highlighting examples of recent progress in the field and contributions to sustainability and health. Their paper “Metabolic engineering for sustainability and health” was published online in the 40th anniversary special issue of Trends in Biotechnology on January 10, 2023.
Metabolic engineering, a discipline of engineering that modifies cell phenotypes through molecular and genetic-level manipulations to improve cellular activities, has been studied since the early 1990s, and has progressed significantly over the past 30 years. In particular, metabolic engineering has enabled the engineering of microorganisms for the development of microbial cell factories capable of efficiently producing chemicals and materials as well as degrading recalcitrant contaminants.
This review article revisited how metabolic engineering has advanced over the past 30 years, from the advent of genetic engineering techniques such as recombinant DNA technologies to recent breakthroughs in systems metabolic engineering and data science aided by artificial intelligence. The research team highlighted momentous events and achievements in metabolic engineering, providing both trends and future directions in the field. Metabolic engineering’s contributions to bio-based sustainable chemicals and clean energy, health, and bioremediation were also reviewed. Finally, the research team shared their perspectives on the future challenges impacting metabolic engineering than must be overcome in order to achieve advancements in sustainability and health.
Distinguished Professor Sang Yup Lee said, “Replacing fossil resource-based chemical processes with bio-based sustainable processes for the production of chemicals, fuels, and materials using metabolic engineering has become our essential task for the future. By looking back on the 30+ years of metabolic engineering, we aimed to highlight the contributions of metabolic engineering to achieve sustainability and good health.” He added, “Metabolic engineering will play an increasingly important role as a key solution to the climate crisis, environmental pollution, food and energy shortages, and health problems in aging societies.”
< Figure: Metabolic Engineering Timeline >
2023.01.25 View 11568 -
 Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 15186
Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 15186 -
 Now You Can See Floral Scents!
Optical interferometry visualizes how often lilies emit volatile organic compounds
Have you ever thought about when flowers emit their scents?
KAIST mechanical engineers and biological scientists directly visualized how often a lily releases a floral scent using a laser interferometry method. These measurement results can provide new insights for understanding and further exploring the biosynthesis and emission mechanisms of floral volatiles.
Why is it important to know this? It is well known that the fragrance of flowers affects their interactions with pollinators, microorganisms, and florivores. For instance, many flowering plants can tune their scent emission rates when pollinators are active for pollination. Petunias and the wild tobacco Nicotiana attenuata emit floral scents at night to attract night-active pollinators. Thus, visualizing scent emissions can help us understand the ecological evolution of plant-pollinator interactions.
Many groups have been trying to develop methods for scent analysis. Mass spectrometry has been one widely used method for investigating the fragrance of flowers. Although mass spectrometry reveals the quality and quantity of floral scents, it is impossible to directly measure the releasing frequency. A laser-based gas detection system and a smartphone-based detection system using chemo-responsive dyes have also been used to measure volatile organic compounds (VOCs) in real-time, but it is still hard to measure the time-dependent emission rate of floral scents.
However, the KAIST research team co-led by Professor Hyoungsoo Kim from the Department of Mechanical Engineering and Professor Sang-Gyu Kim from the Department of Biological Sciences measured a refractive index difference between the vapor of the VOCs of lilies and the air to measure the emission frequency. The floral scent vapor was detected and the refractive index of air was 1.0 while that of the major floral scent of a linalool lily was 1.46.
Professor Hyoungsoo Kim said, “We expect this technology to be further applicable to various industrial sectors such as developing it to detect hazardous substances in a space.” The research team also plans to identify the DNA mechanism that controls floral scent secretion.
The current work entitled “Real-time visualization of scent accumulation reveals the frequency of floral scent emissions” was published in ‘Frontiers in Plant Science’ on April 18, 2022. (https://doi.org/10.3389/fpls.2022.835305).
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2021R1A2C2007835), the Rural Development Administration (PJ016403), and the KAIST-funded Global Singularity Research PREP-Program.
-Publication:H. Kim, G. Lee, J. Song, and S.-G. Kim, "Real-time visualization of scent accumulation reveals the frequency of floral scent emissions," Frontiers in Plant Science 18, 835305 (2022) (https://doi.org/10.3389/fpls.2022.835305)
-Profile:Professor Hyoungsoo Kimhttp://fil.kaist.ac.kr
@MadeInH on TwitterDepartment of Mechanical EngineeringKAIST
Professor Sang-Gyu Kimhttps://sites.google.com/view/kimlab/home Department of Biological SciencesKAIST
2022.05.25 View 10768
Now You Can See Floral Scents!
Optical interferometry visualizes how often lilies emit volatile organic compounds
Have you ever thought about when flowers emit their scents?
KAIST mechanical engineers and biological scientists directly visualized how often a lily releases a floral scent using a laser interferometry method. These measurement results can provide new insights for understanding and further exploring the biosynthesis and emission mechanisms of floral volatiles.
Why is it important to know this? It is well known that the fragrance of flowers affects their interactions with pollinators, microorganisms, and florivores. For instance, many flowering plants can tune their scent emission rates when pollinators are active for pollination. Petunias and the wild tobacco Nicotiana attenuata emit floral scents at night to attract night-active pollinators. Thus, visualizing scent emissions can help us understand the ecological evolution of plant-pollinator interactions.
Many groups have been trying to develop methods for scent analysis. Mass spectrometry has been one widely used method for investigating the fragrance of flowers. Although mass spectrometry reveals the quality and quantity of floral scents, it is impossible to directly measure the releasing frequency. A laser-based gas detection system and a smartphone-based detection system using chemo-responsive dyes have also been used to measure volatile organic compounds (VOCs) in real-time, but it is still hard to measure the time-dependent emission rate of floral scents.
However, the KAIST research team co-led by Professor Hyoungsoo Kim from the Department of Mechanical Engineering and Professor Sang-Gyu Kim from the Department of Biological Sciences measured a refractive index difference between the vapor of the VOCs of lilies and the air to measure the emission frequency. The floral scent vapor was detected and the refractive index of air was 1.0 while that of the major floral scent of a linalool lily was 1.46.
Professor Hyoungsoo Kim said, “We expect this technology to be further applicable to various industrial sectors such as developing it to detect hazardous substances in a space.” The research team also plans to identify the DNA mechanism that controls floral scent secretion.
The current work entitled “Real-time visualization of scent accumulation reveals the frequency of floral scent emissions” was published in ‘Frontiers in Plant Science’ on April 18, 2022. (https://doi.org/10.3389/fpls.2022.835305).
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2021R1A2C2007835), the Rural Development Administration (PJ016403), and the KAIST-funded Global Singularity Research PREP-Program.
-Publication:H. Kim, G. Lee, J. Song, and S.-G. Kim, "Real-time visualization of scent accumulation reveals the frequency of floral scent emissions," Frontiers in Plant Science 18, 835305 (2022) (https://doi.org/10.3389/fpls.2022.835305)
-Profile:Professor Hyoungsoo Kimhttp://fil.kaist.ac.kr
@MadeInH on TwitterDepartment of Mechanical EngineeringKAIST
Professor Sang-Gyu Kimhttps://sites.google.com/view/kimlab/home Department of Biological SciencesKAIST
2022.05.25 View 10768 -
 Machine Learning-Based Algorithm to Speed up DNA Sequencing
The algorithm presents the first full-fledged, short-read alignment software that leverages learned indices for solving the exact match search problem for efficient seeding
The human genome consists of a complete set of DNA, which is about 6.4 billion letters long. Because of its size, reading the whole genome sequence at once is challenging. So scientists use DNA sequencers to produce hundreds of millions of DNA sequence fragments, or short reads, up to 300 letters long. Then the DNA sequencer assembles all the short reads like a giant jigsaw puzzle to reconstruct the entire genome sequence. Even with very fast computers, this job can take hours to complete.
A research team at KAIST has achieved up to 3.45x faster speeds by developing the first short-read alignment software that uses a recent advance in machine-learning called a learned index.
The research team reported their findings on March 7, 2022 in the journal Bioinformatics. The software has been released as open source and can be found on github (https://github.com/kaist-ina/BWA-MEME).
Next-generation sequencing (NGS) is a state-of-the-art DNA sequencing method. Projects are underway with the goal of producing genome sequencing at population scale. Modern NGS hardware is capable of generating billions of short reads in a single run. Then the short reads have to be aligned with the reference DNA sequence. With large-scale DNA sequencing operations running hundreds of next-generation sequences, the need for an efficient short read alignment tool has become even more critical. Accelerating the DNA sequence alignment would be a step toward achieving the goal of population-scale sequencing. However, existing algorithms are limited in their performance because of their frequent memory accesses.
BWA-MEM2 is a popular short-read alignment software package currently used to sequence the DNA. However, it has its limitations. The state-of-the-art alignment has two phases – seeding and extending. During the seeding phase, searches find exact matches of short reads in the reference DNA sequence. During the extending phase, the short reads from the seeding phase are extended. In the current process, bottlenecks occur in the seeding phase. Finding the exact matches slows the process.
The researchers set out to solve the problem of accelerating the DNA sequence alignment. To speed the process, they applied machine learning techniques to create an algorithmic improvement. Their algorithm, BWA-MEME (BWA-MEM emulated) leverages learned indices to solve the exact match search problem. The original software compared one character at a time for an exact match search. The team’s new algorithm achieves up to 3.45x faster speeds in seeding throughput over BWA-MEM2 by reducing the number of instructions by 4.60x and memory accesses by 8.77x. “Through this study, it has been shown that full genome big data analysis can be performed faster and less costly than conventional methods by applying machine learning technology,” said Professor Dongsu Han from the School of Electrical Engineering at KAIST.
The researchers’ ultimate goal was to develop efficient software that scientists from academia and industry could use on a daily basis for analyzing big data in genomics. “With the recent advances in artificial intelligence and machine learning, we see so many opportunities for designing better software for genomic data analysis. The potential is there for accelerating existing analysis as well as enabling new types of analysis, and our goal is to develop such software,” added Han.
Whole genome sequencing has traditionally been used for discovering genomic mutations and identifying the root causes of diseases, which leads to the discovery and development of new drugs and cures. There could be many potential applications. Whole genome sequencing is used not only for research, but also for clinical purposes. “The science and technology for analyzing genomic data is making rapid progress to make it more accessible for scientists and patients. This will enhance our understanding about diseases and develop a better cure for patients of various diseases.”
The research was funded by the National Research Foundation of the Korean government’s Ministry of Science and ICT.
-PublicationYoungmok Jung, Dongsu Han, “BWA-MEME:BWA-MEM emulated with a machine learning approach,” Bioinformatics, Volume 38, Issue 9, May 2022
(https://doi.org/10.1093/bioinformatics/btac137)
-ProfileProfessor Dongsu HanSchool of Electrical EngineeringKAIST
2022.05.10 View 10405
Machine Learning-Based Algorithm to Speed up DNA Sequencing
The algorithm presents the first full-fledged, short-read alignment software that leverages learned indices for solving the exact match search problem for efficient seeding
The human genome consists of a complete set of DNA, which is about 6.4 billion letters long. Because of its size, reading the whole genome sequence at once is challenging. So scientists use DNA sequencers to produce hundreds of millions of DNA sequence fragments, or short reads, up to 300 letters long. Then the DNA sequencer assembles all the short reads like a giant jigsaw puzzle to reconstruct the entire genome sequence. Even with very fast computers, this job can take hours to complete.
A research team at KAIST has achieved up to 3.45x faster speeds by developing the first short-read alignment software that uses a recent advance in machine-learning called a learned index.
The research team reported their findings on March 7, 2022 in the journal Bioinformatics. The software has been released as open source and can be found on github (https://github.com/kaist-ina/BWA-MEME).
Next-generation sequencing (NGS) is a state-of-the-art DNA sequencing method. Projects are underway with the goal of producing genome sequencing at population scale. Modern NGS hardware is capable of generating billions of short reads in a single run. Then the short reads have to be aligned with the reference DNA sequence. With large-scale DNA sequencing operations running hundreds of next-generation sequences, the need for an efficient short read alignment tool has become even more critical. Accelerating the DNA sequence alignment would be a step toward achieving the goal of population-scale sequencing. However, existing algorithms are limited in their performance because of their frequent memory accesses.
BWA-MEM2 is a popular short-read alignment software package currently used to sequence the DNA. However, it has its limitations. The state-of-the-art alignment has two phases – seeding and extending. During the seeding phase, searches find exact matches of short reads in the reference DNA sequence. During the extending phase, the short reads from the seeding phase are extended. In the current process, bottlenecks occur in the seeding phase. Finding the exact matches slows the process.
The researchers set out to solve the problem of accelerating the DNA sequence alignment. To speed the process, they applied machine learning techniques to create an algorithmic improvement. Their algorithm, BWA-MEME (BWA-MEM emulated) leverages learned indices to solve the exact match search problem. The original software compared one character at a time for an exact match search. The team’s new algorithm achieves up to 3.45x faster speeds in seeding throughput over BWA-MEM2 by reducing the number of instructions by 4.60x and memory accesses by 8.77x. “Through this study, it has been shown that full genome big data analysis can be performed faster and less costly than conventional methods by applying machine learning technology,” said Professor Dongsu Han from the School of Electrical Engineering at KAIST.
The researchers’ ultimate goal was to develop efficient software that scientists from academia and industry could use on a daily basis for analyzing big data in genomics. “With the recent advances in artificial intelligence and machine learning, we see so many opportunities for designing better software for genomic data analysis. The potential is there for accelerating existing analysis as well as enabling new types of analysis, and our goal is to develop such software,” added Han.
Whole genome sequencing has traditionally been used for discovering genomic mutations and identifying the root causes of diseases, which leads to the discovery and development of new drugs and cures. There could be many potential applications. Whole genome sequencing is used not only for research, but also for clinical purposes. “The science and technology for analyzing genomic data is making rapid progress to make it more accessible for scientists and patients. This will enhance our understanding about diseases and develop a better cure for patients of various diseases.”
The research was funded by the National Research Foundation of the Korean government’s Ministry of Science and ICT.
-PublicationYoungmok Jung, Dongsu Han, “BWA-MEME:BWA-MEM emulated with a machine learning approach,” Bioinformatics, Volume 38, Issue 9, May 2022
(https://doi.org/10.1093/bioinformatics/btac137)
-ProfileProfessor Dongsu HanSchool of Electrical EngineeringKAIST
2022.05.10 View 10405 -
 Scientists Discover the Mechanism of DNA High-Order Structure Formation
(Molecular structures of Abo1 in different energy states (left), Demonstration of an Abo1-assisted histone loading onto DNA by the DNA curtain assay. )
The genetic material of our cells—DNA—exists in a high-order structure called “chromatin”. Chromatin consists of DNA wrapped around histone proteins and efficiently packs DNA into a small volume. Moreover, using a spool and thread analogy, chromatin allows DNA to be locally wound or unwound, thus enabling genes to be enclosed or exposed. The misregulation of chromatin structures results in aberrant gene expression and can ultimately lead to developmental disorders or cancers. Despite the importance of DNA high-order structures, the complexity of the underlying machinery has circumvented molecular dissection.
For the first time, molecular biologists have uncovered how one particular mechanism uses energy to ensure proper histone placement onto DNA to form chromatin. They published their results on Dec. 17 in Nature Communications.
The study focused on proteins called histone chaperones. Histone chaperones are responsible for adding and removing specific histones at specific times during the DNA packaging process. The wrong histone at the wrong time and place could result in the misregulation of gene expression or aberrant DNA replication. Thus, histone chaperones are key players in the assembly and disassembly of chromatin.
“In order to carefully control the assembly and disassembly of chromatin units, histone chaperones act as molecular escorts that prevent histone aggregation and undesired interactions,” said Professor Ji-Joon Song in the Department of Biological Sciences at KAIST. “We set out to understand how a unique histone chaperone uses chemical energy to assemble or disassemble chromatin.”
Song and his team looked to Abo1, the only known histone chaperone that utilizes cellular energy (ATP). While Abo1 is found in yeast, it has an analogous partner in other organisms, including humans, called ATAD2. Both use ATP, which is produced through a cellular process where enzymes break down a molecule’s phosphate bond. ATP energy is typically used to power other cellular processes, but it is a rare partner for histone chaperones.
“This was an interesting problem in the field because all other histone chaperones studied to date do not use ATP,” Song said.
By imaging Abo1 with a single-molecule fluorescence imaging technique known as the DNA curtain assay, the researchers could examine the protein interactions at the single-molecule level. The technique allows scientists to arrange the DNA molecules and proteins on a single layer of a microfluidic chamber and examine the layer with fluorescence microscopy.
The researchers found through real-time observation that Abo1 is ring-shaped and changes its structure to accommodate a specific histone and deposit it on DNA. Moreover, they found that the accommodating structural changes are powered by ADP.
“We discovered a mechanism by which Abo1 accommodates histone substrates, ultimately allowing it to function as a unique energy-dependent histone chaperone,” Song said. “We also found that despite looking like a protein disassembly machine, Abo1 actually loads histone substrates onto DNA to facilitate chromatin assembly.”
The researchers plan to continue exploring how energy-dependent histone chaperones bind and release histones, with the ultimate goal of developing therapeutics that can target cancer-causing misbehavior by Abo1’s analogous human counterpart, ATAD2.
-Profile
Professor Ji-Joon Song
Department of Biological Sciences KI for the BioCentury (https://kis.kaist.ac.kr/index.php?mid=KIB_O) KAIST
2020.01.07 View 11814
Scientists Discover the Mechanism of DNA High-Order Structure Formation
(Molecular structures of Abo1 in different energy states (left), Demonstration of an Abo1-assisted histone loading onto DNA by the DNA curtain assay. )
The genetic material of our cells—DNA—exists in a high-order structure called “chromatin”. Chromatin consists of DNA wrapped around histone proteins and efficiently packs DNA into a small volume. Moreover, using a spool and thread analogy, chromatin allows DNA to be locally wound or unwound, thus enabling genes to be enclosed or exposed. The misregulation of chromatin structures results in aberrant gene expression and can ultimately lead to developmental disorders or cancers. Despite the importance of DNA high-order structures, the complexity of the underlying machinery has circumvented molecular dissection.
For the first time, molecular biologists have uncovered how one particular mechanism uses energy to ensure proper histone placement onto DNA to form chromatin. They published their results on Dec. 17 in Nature Communications.
The study focused on proteins called histone chaperones. Histone chaperones are responsible for adding and removing specific histones at specific times during the DNA packaging process. The wrong histone at the wrong time and place could result in the misregulation of gene expression or aberrant DNA replication. Thus, histone chaperones are key players in the assembly and disassembly of chromatin.
“In order to carefully control the assembly and disassembly of chromatin units, histone chaperones act as molecular escorts that prevent histone aggregation and undesired interactions,” said Professor Ji-Joon Song in the Department of Biological Sciences at KAIST. “We set out to understand how a unique histone chaperone uses chemical energy to assemble or disassemble chromatin.”
Song and his team looked to Abo1, the only known histone chaperone that utilizes cellular energy (ATP). While Abo1 is found in yeast, it has an analogous partner in other organisms, including humans, called ATAD2. Both use ATP, which is produced through a cellular process where enzymes break down a molecule’s phosphate bond. ATP energy is typically used to power other cellular processes, but it is a rare partner for histone chaperones.
“This was an interesting problem in the field because all other histone chaperones studied to date do not use ATP,” Song said.
By imaging Abo1 with a single-molecule fluorescence imaging technique known as the DNA curtain assay, the researchers could examine the protein interactions at the single-molecule level. The technique allows scientists to arrange the DNA molecules and proteins on a single layer of a microfluidic chamber and examine the layer with fluorescence microscopy.
The researchers found through real-time observation that Abo1 is ring-shaped and changes its structure to accommodate a specific histone and deposit it on DNA. Moreover, they found that the accommodating structural changes are powered by ADP.
“We discovered a mechanism by which Abo1 accommodates histone substrates, ultimately allowing it to function as a unique energy-dependent histone chaperone,” Song said. “We also found that despite looking like a protein disassembly machine, Abo1 actually loads histone substrates onto DNA to facilitate chromatin assembly.”
The researchers plan to continue exploring how energy-dependent histone chaperones bind and release histones, with the ultimate goal of developing therapeutics that can target cancer-causing misbehavior by Abo1’s analogous human counterpart, ATAD2.
-Profile
Professor Ji-Joon Song
Department of Biological Sciences KI for the BioCentury (https://kis.kaist.ac.kr/index.php?mid=KIB_O) KAIST
2020.01.07 View 11814 -
 Micropatch Made of DNA
Researchers reported the fabrication of microstructure arrays of DNA materials using topographic control. This method provides a platform for forming multiscale hierarchical orientations of soft and biomaterials using a process of simple shearing and controlled evaporation on a patterned substrate. This approach enables the potential of patterning applications using DNA or other anisotropic biomaterials.
DNA is one of the most abundant biomaterials found in all living organisms in nature. It has unique characteristics of fine feature size and liquid crystalline phase, enabling to create various kinds of microstructure DNA arrays. Based on these characteristics, DNA has been used as a building block for “origami” and textile art at the nanometer scale.
A KAIST research team led by Professors Dong Ki Yoon and Hyungsoo Kim fabricated a DNA-based micropatch using the “coffee ring effect” and its multi-angle control technology, which was published online in Nature Communications on June 7.
The research team used cheap DNA material extracted from salmon to realize the micropatch structure with well-aligned knit or ice cream cone shapes. When the DNA material in an aqueous solution is rubbed between two solid substrates while water is evaporating, DNA chains are unidirectionally aligned to make a thin film such as in LCD display devices. The DNA chains can make more complex microstructures such as knit or a texture with ice cream cone shapes when the same procedure is carried out in topographical patterns like microposts (Figure 1). This can be applied to make metamaterials by mixing with functionalized gold nanorods to show plasmonic color.
Plasmon resonance is a phenomenon in which electrons vibrate uniformly on the surface of a substrate made of metal, reacting only to light that matches a specific energy to enhance the clarity and expression of colors. For this, the most important factor is the orientation in which the gold nanorods align. That is, when the rods are aligned side by side in one direction, the optical and electrical characteristics are maximized. The research team focused on this point and made the DNA micropatch as a frame to orient the gold nanorods in a unique shape and fabricated a plasmonic color film (Figure 2).
Professor Yoon said this study is meaningful in that it deals with the evaporation phenomenon, which has not been studied much in the field of polymers and biopolymers in terms of basic science. He explained, “This will also help maximize the efficiency of polymeric materials that can be orientated in coating, 2D, and 3D printing applications. Furthermore, DNA that exists infinitely in nature can be expected to have industrial application value as a new material since it can easily form complexes with other materials as described in this study.”
(Figure 1. The DNA micropatch using topographic control. (a) The experimental scheme.
(b) Enlarged image of (e). (c-e) Different micropatches made of DNA using different shearing directions.)
(Figure 2. The knit-like structures made of DNA-gold nanorod complex. (a,b) Optical
and polarized optical microscopy images. (c-f) Plasmonic colors reflected from aligned DNA-gold nanorod complex depending on the sample rotation.)
2019.07.01 View 35556
Micropatch Made of DNA
Researchers reported the fabrication of microstructure arrays of DNA materials using topographic control. This method provides a platform for forming multiscale hierarchical orientations of soft and biomaterials using a process of simple shearing and controlled evaporation on a patterned substrate. This approach enables the potential of patterning applications using DNA or other anisotropic biomaterials.
DNA is one of the most abundant biomaterials found in all living organisms in nature. It has unique characteristics of fine feature size and liquid crystalline phase, enabling to create various kinds of microstructure DNA arrays. Based on these characteristics, DNA has been used as a building block for “origami” and textile art at the nanometer scale.
A KAIST research team led by Professors Dong Ki Yoon and Hyungsoo Kim fabricated a DNA-based micropatch using the “coffee ring effect” and its multi-angle control technology, which was published online in Nature Communications on June 7.
The research team used cheap DNA material extracted from salmon to realize the micropatch structure with well-aligned knit or ice cream cone shapes. When the DNA material in an aqueous solution is rubbed between two solid substrates while water is evaporating, DNA chains are unidirectionally aligned to make a thin film such as in LCD display devices. The DNA chains can make more complex microstructures such as knit or a texture with ice cream cone shapes when the same procedure is carried out in topographical patterns like microposts (Figure 1). This can be applied to make metamaterials by mixing with functionalized gold nanorods to show plasmonic color.
Plasmon resonance is a phenomenon in which electrons vibrate uniformly on the surface of a substrate made of metal, reacting only to light that matches a specific energy to enhance the clarity and expression of colors. For this, the most important factor is the orientation in which the gold nanorods align. That is, when the rods are aligned side by side in one direction, the optical and electrical characteristics are maximized. The research team focused on this point and made the DNA micropatch as a frame to orient the gold nanorods in a unique shape and fabricated a plasmonic color film (Figure 2).
Professor Yoon said this study is meaningful in that it deals with the evaporation phenomenon, which has not been studied much in the field of polymers and biopolymers in terms of basic science. He explained, “This will also help maximize the efficiency of polymeric materials that can be orientated in coating, 2D, and 3D printing applications. Furthermore, DNA that exists infinitely in nature can be expected to have industrial application value as a new material since it can easily form complexes with other materials as described in this study.”
(Figure 1. The DNA micropatch using topographic control. (a) The experimental scheme.
(b) Enlarged image of (e). (c-e) Different micropatches made of DNA using different shearing directions.)
(Figure 2. The knit-like structures made of DNA-gold nanorod complex. (a,b) Optical
and polarized optical microscopy images. (c-f) Plasmonic colors reflected from aligned DNA-gold nanorod complex depending on the sample rotation.)
2019.07.01 View 35556 -
 JETS Conference 2017
KAIST and four science and technology research universities in Korea co-hosted a technology start-up fair, the 2017 JETS (Job, Exhibition, Tech Forum, and Startup) Conference January 19 ~20 in the Ryu Geun-chul Sports Complex at KAIST.
Korea’s major science and technology research universities, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Gwangju Institute of Science and Technology (GIST), Pohang University of Science and Technology (Postech), and Ulsan National Institute of Science and Technology (UNIST), held the event in a collaborative effort to educate, inspire, and connect young entrepreneurs, especially those who will launch technology start-ups.
The conference brought entrepreneurs and innovators together who seek ways of working with and supporting start-ups and for their sustainable growth. It also drew aspiring young students and researchers from universities and the government-funded research institutions who are in the process of commercializing their technology. Students from each university’s industry-academia cooperation program who incubated their technology and ideas were key contributors.
At the Tech Forum, entrepreneurship and technology consultation specialists including Joe Jasin, managing director at DNA Investment Partners in the US, the founder of Cyworld Dong-Hyung Lee, and Professor Hawoong Jeong, a complex bio-network specialist from the Department of Physics of KAIST lectured on the ecosystem of start-ups and its trends and development.
The Dean of University-Industry Cooperation at KAIST Joongmyeon Bae said, "We organized this event in collaboration with four major research universities to further encourage technology start-ups from young students and help their ideas and technology bear fruit. We will continue to strive to create an ecosystem of start-ups which works efficiently.”
(Above photo: Founder of the Cyworld, Dong-Hyung Lee gives a lecture at the Tech Forum. Below photo: Students visit exhibition booth of each participating institution.)
2017.01.20 View 13734
JETS Conference 2017
KAIST and four science and technology research universities in Korea co-hosted a technology start-up fair, the 2017 JETS (Job, Exhibition, Tech Forum, and Startup) Conference January 19 ~20 in the Ryu Geun-chul Sports Complex at KAIST.
Korea’s major science and technology research universities, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Gwangju Institute of Science and Technology (GIST), Pohang University of Science and Technology (Postech), and Ulsan National Institute of Science and Technology (UNIST), held the event in a collaborative effort to educate, inspire, and connect young entrepreneurs, especially those who will launch technology start-ups.
The conference brought entrepreneurs and innovators together who seek ways of working with and supporting start-ups and for their sustainable growth. It also drew aspiring young students and researchers from universities and the government-funded research institutions who are in the process of commercializing their technology. Students from each university’s industry-academia cooperation program who incubated their technology and ideas were key contributors.
At the Tech Forum, entrepreneurship and technology consultation specialists including Joe Jasin, managing director at DNA Investment Partners in the US, the founder of Cyworld Dong-Hyung Lee, and Professor Hawoong Jeong, a complex bio-network specialist from the Department of Physics of KAIST lectured on the ecosystem of start-ups and its trends and development.
The Dean of University-Industry Cooperation at KAIST Joongmyeon Bae said, "We organized this event in collaboration with four major research universities to further encourage technology start-ups from young students and help their ideas and technology bear fruit. We will continue to strive to create an ecosystem of start-ups which works efficiently.”
(Above photo: Founder of the Cyworld, Dong-Hyung Lee gives a lecture at the Tech Forum. Below photo: Students visit exhibition booth of each participating institution.)
2017.01.20 View 13734 -
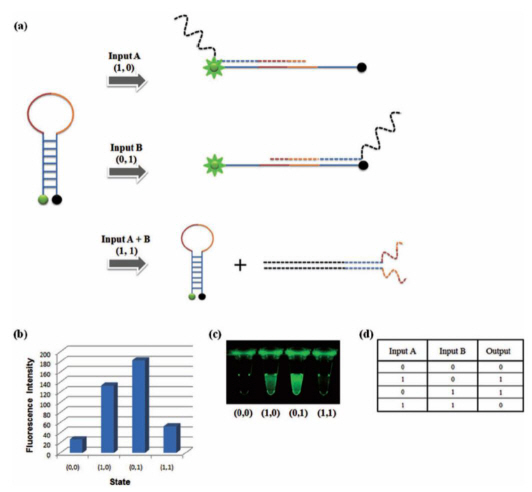 DNA based semiconductor technology developed
Professor Park Hyun Gyu’s research team from the Department of Chemical and Biomolecular Engineering at KAIST has successfully implemented all logic gates using DNA, a feat that led the research to be published as the cover paper for the international nanotechnology paper "Small".
Even with the latest technology, it was impossible to create a silicon based semiconductor smaller than 10nm, but because DNA has a thickness of only 2nm, this could lead to the creation of semiconductors with groundbreaking degrees of integration.
A 2 nm semiconductor will be able to store 10,000 HD movies within a size of a postage stamp, at least 100 times more than the current 20nm semiconductors.
DNAs are comprised of 4 bases which are continually connected: Adenine (A) with Thymine (T), and Guanine (G) with Cytosine (C).
For this research, the team used the specific binding properties of DNA, which forms its helix-shape, and a circular molecular beacon that has fluorescent signaling properties under structural changes.
The research team used input signals to open and close the circular DNA, the same principle that is applied to logic gates in digital circuits.
The output signal was measured using the increase and decrease of the fluorescent signal from the molecular beacon due to the opening and closing of the circular DNA respectively.
The team overcame the limited system problems of the existing logic gates and managed to implement all 8 logic gates (AND, OR, XOR, INHIBIT, NAND, NOR, XNOR, IMPlCATION). A multilevel circuit that connects different logic gates was also tested to show its regenerative properties.
Professor Park said that “cheap bio-electric devices with high degrees of integration will be made possible by this research” and that “there will be a large difference in the field of molecular level electronic research”
Mr. Park Gi Su, a doctoral candidate and the 1st author of this research, said that “a DNA sequence of 10 bases is only 3.4nm long and 2nm thick, which can be used to effectively increase the degree of integration of electronic devices” and that “a bio computer could materialize in the near future through DNA semiconductors with accurate logic gates”.
XOR Gate: The output signal 1 comes through the open circular DNA when either input DNA A or input DNA B is present. When both inputs are not present, the flourescent signal does not come through
2012.09.27 View 11049
DNA based semiconductor technology developed
Professor Park Hyun Gyu’s research team from the Department of Chemical and Biomolecular Engineering at KAIST has successfully implemented all logic gates using DNA, a feat that led the research to be published as the cover paper for the international nanotechnology paper "Small".
Even with the latest technology, it was impossible to create a silicon based semiconductor smaller than 10nm, but because DNA has a thickness of only 2nm, this could lead to the creation of semiconductors with groundbreaking degrees of integration.
A 2 nm semiconductor will be able to store 10,000 HD movies within a size of a postage stamp, at least 100 times more than the current 20nm semiconductors.
DNAs are comprised of 4 bases which are continually connected: Adenine (A) with Thymine (T), and Guanine (G) with Cytosine (C).
For this research, the team used the specific binding properties of DNA, which forms its helix-shape, and a circular molecular beacon that has fluorescent signaling properties under structural changes.
The research team used input signals to open and close the circular DNA, the same principle that is applied to logic gates in digital circuits.
The output signal was measured using the increase and decrease of the fluorescent signal from the molecular beacon due to the opening and closing of the circular DNA respectively.
The team overcame the limited system problems of the existing logic gates and managed to implement all 8 logic gates (AND, OR, XOR, INHIBIT, NAND, NOR, XNOR, IMPlCATION). A multilevel circuit that connects different logic gates was also tested to show its regenerative properties.
Professor Park said that “cheap bio-electric devices with high degrees of integration will be made possible by this research” and that “there will be a large difference in the field of molecular level electronic research”
Mr. Park Gi Su, a doctoral candidate and the 1st author of this research, said that “a DNA sequence of 10 bases is only 3.4nm long and 2nm thick, which can be used to effectively increase the degree of integration of electronic devices” and that “a bio computer could materialize in the near future through DNA semiconductors with accurate logic gates”.
XOR Gate: The output signal 1 comes through the open circular DNA when either input DNA A or input DNA B is present. When both inputs are not present, the flourescent signal does not come through
2012.09.27 View 11049