Cell
-
 KAIST Predicts Diseases by Early Detection of Aging Signals in Liver Tissue
- KAIST-KRIBB Develops ‘FiNi-seq’ Technology to Capture Characteristics of Fibrotic Microenvironments Accumulated in Liver Tissue and Dynamic Changes of Early Aging Cells
- Elucidation of the Spatial Ecosystem of Aged Liver Tissue, where Reprogramming of Senescent Cells and Immune Exhaustion Progresses, at the Single-Cell Genome and Epigenome Levels
< (From left) Professor Jong-Eun Park of KAIST Graduate School of Medical Science and Engineering (GSMSE), Dr. Chuna Kim of KRIBB, Dr. Kwon Yong Tak of KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, Ph.D. Candidate Myungsun Park of KAIST GSMSE >
Aging and chronic diseases involve the gradual accumulation of subtle tissue changes over a long period. Therefore, there are still limitations in quantitatively understanding these changes within organs and linking them to early signs of disease onset. In response, Korean researchers have successfully developed a platform technology that accurately captures localized changes that first occur within tissue, significantly aiding in faster disease discovery and prediction, and in setting personalized treatment targets.
KAIST (President Kwang Hyung Lee) announced on June 12th that a joint research team led by Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering at KAIST and Dr. Chuna Kim of the Aging Convergence Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB, President Seok-Yoon Kwon) has developed ‘FiNi-seq (Fibrotic Niche enrichment sequencing)’ technology. This technology captures fibrotic microenvironments locally occurring in aged liver tissue and enables precise analysis at the single-cell transcriptome level*.
*Single-cell transcriptome analysis: A method to measure how actively each cell uses which genes, allowing identification and function of individual diseased cells.
The researchers developed a method to selectively enrich early aging microenvironments where regeneration is delayed and fibrosis accumulates, by physically selecting regions with high tissue degradation resistance in aged liver tissue.
In this process, high-resolution identification of fibrosis-related endothelial cells, fibroblasts interacting with the immune system, and immune-exhausted cells such as PD-1 highly expressing CD8 T cells, which were difficult to capture with existing single-cell analysis technologies, was possible.
In particular, the research team confirmed through ‘FiNi-seq’ technology that specific cells observed in fibrotic areas within aged liver tissue secondarily age the surrounding environment through secreted factors, and that this leads to the expansion of the aged environment.
Furthermore, they also elucidated the mechanism by which endothelial cells lose their tissue-specific identity and induce innate immune responses, promoting immune cell infiltration. Through spatial transcriptome analysis, the spatial distribution of fibroblasts interacting with immune cells was quantified, revealing their involvement in tissue regeneration, induction of inflammatory responses, and progression to chronic fibrosis.
The research team performed integrated analysis of multi-omics\* data to obtain transcriptome and epigenome information, precisely interpreting the microenvironment of aged liver tissue and its spatial heterogeneity, and confirming how these changes are connected to the intrahepatic vascular structure.
*Multi-omics: An integrated analysis method for various biological information within an organism, such as genes, proteins, metabolites, and cell information.
The newly developed ‘FiNi-seq’ technology is expected to be a useful platform for high-resolution capture of pathophysiological signals in most chronic liver diseases, including the aging process that causes fibrosis.
< Figure 1. Isolation of fibrotic regions from aged liver tissue, followed by single-cell transcriptome analysis and validation in a fibrosis model. >
The first author, Dr. Kwon Yong Tak of KAIST Graduate School of Medical Science and Engineering (GSMSE), a hepatologist at Seoul St. Mary's Hospital, designed this study to lay the groundwork for early diagnosis and treatment of fibrosis progression, the most important clinical prognostic indicator in chronic liver disease, while pursuing his Ph.D. at KAIST KAIST GSMSE with support from the physician-scientist training program. Co-first author Myungsun Park, a Ph.D. candidate at KAIST KAIST GSMSE, was responsible for the technical implementation of FiNi-seq technology, and Juyeon Kim, a Ph.D. candidate at KRIBB's Aging Convergence Research Center, was responsible for imaging analysis of aged tissue, playing a key role in the research.
Dr. Chuna Kim of KRIBB stated, “Through this study, we were able to precisely elucidate the cellular composition and spatial characteristics of the fibrotic microenvironment observed in aged liver tissue at the single-cell level.”
< Figure 2. Spatially defined stepwise progression patterns of aging-related regions within the liver and identification of regulatory factors inducing them. >
Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering said, “As an analytical technology that can capture subtle changes occurring in the early stages of aging and chronic diseases, it is expected to play a significant role in finding effective treatment targets in the future. Also, we plan to expand this research to chronic diseases in other organs such as the lungs and kidneys, as well as various liver disease models.”
This research was published in the international journal ‘Nature Aging’ on May 5, 2025, with Dr. Kwon Yong Tak of KAIST KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, and Ph.D. Candidate Myungsun Park of KAIST as co-first authors.
*Paper Title: Quasi-spatial single-cell transcriptome based on physical tissue properties defines early aging associated niche in liver
*DOI: https://doi.org/10.1038/s43587-025-00857-7
This research was supported by several domestic institutions, including the National Research Foundation of Korea, the Korea Health Industry Development Institute (KHIDI), the Korea Research Institute of Bioscience and Biotechnology (KRIBB), KIST, POSCO Science Fellowship, and the Convergence Medical Scientist Training Program.
2025.06.12 View 1959
KAIST Predicts Diseases by Early Detection of Aging Signals in Liver Tissue
- KAIST-KRIBB Develops ‘FiNi-seq’ Technology to Capture Characteristics of Fibrotic Microenvironments Accumulated in Liver Tissue and Dynamic Changes of Early Aging Cells
- Elucidation of the Spatial Ecosystem of Aged Liver Tissue, where Reprogramming of Senescent Cells and Immune Exhaustion Progresses, at the Single-Cell Genome and Epigenome Levels
< (From left) Professor Jong-Eun Park of KAIST Graduate School of Medical Science and Engineering (GSMSE), Dr. Chuna Kim of KRIBB, Dr. Kwon Yong Tak of KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, Ph.D. Candidate Myungsun Park of KAIST GSMSE >
Aging and chronic diseases involve the gradual accumulation of subtle tissue changes over a long period. Therefore, there are still limitations in quantitatively understanding these changes within organs and linking them to early signs of disease onset. In response, Korean researchers have successfully developed a platform technology that accurately captures localized changes that first occur within tissue, significantly aiding in faster disease discovery and prediction, and in setting personalized treatment targets.
KAIST (President Kwang Hyung Lee) announced on June 12th that a joint research team led by Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering at KAIST and Dr. Chuna Kim of the Aging Convergence Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB, President Seok-Yoon Kwon) has developed ‘FiNi-seq (Fibrotic Niche enrichment sequencing)’ technology. This technology captures fibrotic microenvironments locally occurring in aged liver tissue and enables precise analysis at the single-cell transcriptome level*.
*Single-cell transcriptome analysis: A method to measure how actively each cell uses which genes, allowing identification and function of individual diseased cells.
The researchers developed a method to selectively enrich early aging microenvironments where regeneration is delayed and fibrosis accumulates, by physically selecting regions with high tissue degradation resistance in aged liver tissue.
In this process, high-resolution identification of fibrosis-related endothelial cells, fibroblasts interacting with the immune system, and immune-exhausted cells such as PD-1 highly expressing CD8 T cells, which were difficult to capture with existing single-cell analysis technologies, was possible.
In particular, the research team confirmed through ‘FiNi-seq’ technology that specific cells observed in fibrotic areas within aged liver tissue secondarily age the surrounding environment through secreted factors, and that this leads to the expansion of the aged environment.
Furthermore, they also elucidated the mechanism by which endothelial cells lose their tissue-specific identity and induce innate immune responses, promoting immune cell infiltration. Through spatial transcriptome analysis, the spatial distribution of fibroblasts interacting with immune cells was quantified, revealing their involvement in tissue regeneration, induction of inflammatory responses, and progression to chronic fibrosis.
The research team performed integrated analysis of multi-omics\* data to obtain transcriptome and epigenome information, precisely interpreting the microenvironment of aged liver tissue and its spatial heterogeneity, and confirming how these changes are connected to the intrahepatic vascular structure.
*Multi-omics: An integrated analysis method for various biological information within an organism, such as genes, proteins, metabolites, and cell information.
The newly developed ‘FiNi-seq’ technology is expected to be a useful platform for high-resolution capture of pathophysiological signals in most chronic liver diseases, including the aging process that causes fibrosis.
< Figure 1. Isolation of fibrotic regions from aged liver tissue, followed by single-cell transcriptome analysis and validation in a fibrosis model. >
The first author, Dr. Kwon Yong Tak of KAIST Graduate School of Medical Science and Engineering (GSMSE), a hepatologist at Seoul St. Mary's Hospital, designed this study to lay the groundwork for early diagnosis and treatment of fibrosis progression, the most important clinical prognostic indicator in chronic liver disease, while pursuing his Ph.D. at KAIST KAIST GSMSE with support from the physician-scientist training program. Co-first author Myungsun Park, a Ph.D. candidate at KAIST KAIST GSMSE, was responsible for the technical implementation of FiNi-seq technology, and Juyeon Kim, a Ph.D. candidate at KRIBB's Aging Convergence Research Center, was responsible for imaging analysis of aged tissue, playing a key role in the research.
Dr. Chuna Kim of KRIBB stated, “Through this study, we were able to precisely elucidate the cellular composition and spatial characteristics of the fibrotic microenvironment observed in aged liver tissue at the single-cell level.”
< Figure 2. Spatially defined stepwise progression patterns of aging-related regions within the liver and identification of regulatory factors inducing them. >
Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering said, “As an analytical technology that can capture subtle changes occurring in the early stages of aging and chronic diseases, it is expected to play a significant role in finding effective treatment targets in the future. Also, we plan to expand this research to chronic diseases in other organs such as the lungs and kidneys, as well as various liver disease models.”
This research was published in the international journal ‘Nature Aging’ on May 5, 2025, with Dr. Kwon Yong Tak of KAIST KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, and Ph.D. Candidate Myungsun Park of KAIST as co-first authors.
*Paper Title: Quasi-spatial single-cell transcriptome based on physical tissue properties defines early aging associated niche in liver
*DOI: https://doi.org/10.1038/s43587-025-00857-7
This research was supported by several domestic institutions, including the National Research Foundation of Korea, the Korea Health Industry Development Institute (KHIDI), the Korea Research Institute of Bioscience and Biotechnology (KRIBB), KIST, POSCO Science Fellowship, and the Convergence Medical Scientist Training Program.
2025.06.12 View 1959 -
 KAIST Develops Retinal Therapy to Restore Lost Vision
Vision is one of the most crucial human senses, yet over 300 million people worldwide are at risk of vision loss due to various retinal diseases. While recent advancements in retinal disease treatments have successfully slowed disease progression, no effective therapy has been developed to restore already lost vision—until now. KAIST researchers have successfully developed a novel drug to restore vision.
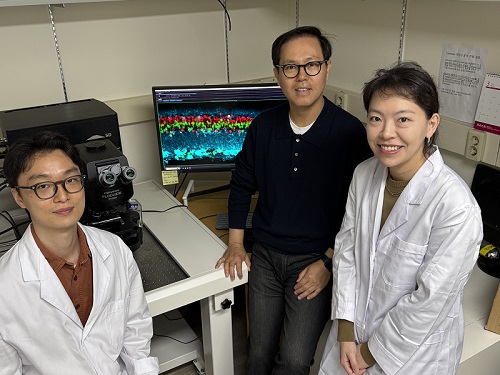
< Photo 1. (From left) Ph.D. candidate Museong Kim, Professor Jin Woo Kim, and Dr. Eun Jung Lee of KAIST Department of Biological Sciences >
KAIST (represented by President Kwang Hyung Lee) announced on the 30th of March that a research team led by Professor Jin Woo Kim from the Department of Biological Sciences has developed a treatment method that restores vision through retinal nerve regeneration.
The research team successfully induced retinal regeneration and vision recovery in a disease-model mouse by administering a compound that blocks the PROX1 (prospero homeobox 1) protein, which suppresses retinal regeneration. Furthermore, the effect lasted for more than six months.
This study marks the first successful induction of long-term neural regeneration in mammalian retinas, offering new hope to patients with degenerative retinal diseases who previously had no treatment options.
As the global population continues to age, the number of retinal disease patients is steadily increasing. However, no treatments exist to restore damaged retinas and vision. The primary reason for this is the mammalian retina's inability to regenerate once damaged.
Studies on cold-blooded animals, such as fish—known for their robust retinal regeneration—have shown that retinal injuries trigger Müller glia cells to dedifferentiate into retinal progenitor cells, which then generate new neurons. However, in mammals, this process is impaired, leading to permanent retinal damage.
< Figure 1. Schematic diagram of the mechanism of retinal regeneration through inhibition of PROX1 migration. PROX1 protein secreted from retinal damaged retinal neurons transfers to Müllerglia and inhibits dedifferentiation into neural progenitor cells and neural regeneration. When PROX1 is captured outside the cells by an antibody against PROX1 and its transfer to Müllerglia is interfered, dedifferentiation of Müllerglia cells and retinal regeneration processes are resumed, restoring visual function. >
Through this study, the research team identified the PROX1 protein as a key inhibitor of Müller glia dedifferentiation in mammals. PROX1 is a protein found in neurons of the retina, hippocampus, and spinal cord, where it suppresses neural stem cell proliferation and promotes differentiation into neurons.
The researchers discovered that PROX1 accumulates in damaged mouse retinal Müller glia, but is absent in the highly regenerative Müller glia of fish. Furthermore, they demonstrated that the PROX1 found in Müller glia is not synthesized internally but rather taken up from surrounding neurons, which fail to degrade and instead secrete the protein.
Based on this finding, the team developed a method to restore Müller glia’s regenerative ability by eliminating extracellular PROX1 before it reaches these cells.
< Figure 2. Retinal regeneration and visual recovery in a retinitis pigmentosa model mouse through Anti-PROX1 gene therapy. After administration of adeno-associated virus expressing PROX1 neutralizing antibodies (AAV2-Anti-PROX1) to the eyes of RP1 retinitis pigmentosa model mice with vision loss, the photoreceptor cell layer of the retina is restored (A) and vision is restored (B). >
This approach involves using an antibody that binds to PROX1, developed by Celliaz Inc., a biotech startup founded by Professor Jin Woo Kim’s research lab. When administered to disease-model mouse retinas, this antibody significantly promoted neural regeneration. Additionally, when delivered, the antibody gene to the retinas of retinitis pigmentosa disease model mice, it enabled sustained retinal regeneration and vision restoration for over six months.
The retinal regeneration-inducing therapy is currently being developed by Celliaz Inc. for application in various degenerative retinal diseases that currently lack effective treatments. The company aims to begin clinical trials by 2028.
This study was co-authored by Dr. Eun Jung Lee of Celliaz Inc. and Museong Kim, a Ph.D. candidate at KAIST, as joint first authors. The findings were published online on March 26 in the international journal Nature Communications. (Paper Title: Restoration of retinal regenerative potential of Müller glia by disrupting intercellular Prox1 transfer | DOI: 10.1038/s41467-025-58290-8)
Dr. Eun Jung Lee stated, "We are about completing the optimization of the PROX1-neutralizing antibody (CLZ001) and move to preclinical studies before administering it to retinal disease patients. Our goal is to provide a solution for patients at risk of blindness who currently lack proper treatment options."
This research was supported by research funds from Korean National Research Foundation (NRF) and the Korea Drug Development Foundation (KDDF).
2025.03.31 View 14242
KAIST Develops Retinal Therapy to Restore Lost Vision
Vision is one of the most crucial human senses, yet over 300 million people worldwide are at risk of vision loss due to various retinal diseases. While recent advancements in retinal disease treatments have successfully slowed disease progression, no effective therapy has been developed to restore already lost vision—until now. KAIST researchers have successfully developed a novel drug to restore vision.
< Photo 1. (From left) Ph.D. candidate Museong Kim, Professor Jin Woo Kim, and Dr. Eun Jung Lee of KAIST Department of Biological Sciences >
KAIST (represented by President Kwang Hyung Lee) announced on the 30th of March that a research team led by Professor Jin Woo Kim from the Department of Biological Sciences has developed a treatment method that restores vision through retinal nerve regeneration.
The research team successfully induced retinal regeneration and vision recovery in a disease-model mouse by administering a compound that blocks the PROX1 (prospero homeobox 1) protein, which suppresses retinal regeneration. Furthermore, the effect lasted for more than six months.
This study marks the first successful induction of long-term neural regeneration in mammalian retinas, offering new hope to patients with degenerative retinal diseases who previously had no treatment options.
As the global population continues to age, the number of retinal disease patients is steadily increasing. However, no treatments exist to restore damaged retinas and vision. The primary reason for this is the mammalian retina's inability to regenerate once damaged.
Studies on cold-blooded animals, such as fish—known for their robust retinal regeneration—have shown that retinal injuries trigger Müller glia cells to dedifferentiate into retinal progenitor cells, which then generate new neurons. However, in mammals, this process is impaired, leading to permanent retinal damage.
< Figure 1. Schematic diagram of the mechanism of retinal regeneration through inhibition of PROX1 migration. PROX1 protein secreted from retinal damaged retinal neurons transfers to Müllerglia and inhibits dedifferentiation into neural progenitor cells and neural regeneration. When PROX1 is captured outside the cells by an antibody against PROX1 and its transfer to Müllerglia is interfered, dedifferentiation of Müllerglia cells and retinal regeneration processes are resumed, restoring visual function. >
Through this study, the research team identified the PROX1 protein as a key inhibitor of Müller glia dedifferentiation in mammals. PROX1 is a protein found in neurons of the retina, hippocampus, and spinal cord, where it suppresses neural stem cell proliferation and promotes differentiation into neurons.
The researchers discovered that PROX1 accumulates in damaged mouse retinal Müller glia, but is absent in the highly regenerative Müller glia of fish. Furthermore, they demonstrated that the PROX1 found in Müller glia is not synthesized internally but rather taken up from surrounding neurons, which fail to degrade and instead secrete the protein.
Based on this finding, the team developed a method to restore Müller glia’s regenerative ability by eliminating extracellular PROX1 before it reaches these cells.
< Figure 2. Retinal regeneration and visual recovery in a retinitis pigmentosa model mouse through Anti-PROX1 gene therapy. After administration of adeno-associated virus expressing PROX1 neutralizing antibodies (AAV2-Anti-PROX1) to the eyes of RP1 retinitis pigmentosa model mice with vision loss, the photoreceptor cell layer of the retina is restored (A) and vision is restored (B). >
This approach involves using an antibody that binds to PROX1, developed by Celliaz Inc., a biotech startup founded by Professor Jin Woo Kim’s research lab. When administered to disease-model mouse retinas, this antibody significantly promoted neural regeneration. Additionally, when delivered, the antibody gene to the retinas of retinitis pigmentosa disease model mice, it enabled sustained retinal regeneration and vision restoration for over six months.
The retinal regeneration-inducing therapy is currently being developed by Celliaz Inc. for application in various degenerative retinal diseases that currently lack effective treatments. The company aims to begin clinical trials by 2028.
This study was co-authored by Dr. Eun Jung Lee of Celliaz Inc. and Museong Kim, a Ph.D. candidate at KAIST, as joint first authors. The findings were published online on March 26 in the international journal Nature Communications. (Paper Title: Restoration of retinal regenerative potential of Müller glia by disrupting intercellular Prox1 transfer | DOI: 10.1038/s41467-025-58290-8)
Dr. Eun Jung Lee stated, "We are about completing the optimization of the PROX1-neutralizing antibody (CLZ001) and move to preclinical studies before administering it to retinal disease patients. Our goal is to provide a solution for patients at risk of blindness who currently lack proper treatment options."
This research was supported by research funds from Korean National Research Foundation (NRF) and the Korea Drug Development Foundation (KDDF).
2025.03.31 View 14242 -
 KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 28518
KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 28518 -
 KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures
- Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS)
- Expected to have price competitiveness over bio-templating method as it expands the range of biological samples
- Expected to expand the range of application of nanostructure synthesis technology by utilizing various biological structures
< Photo 1. (From left) Professor Yeon Sik Jung, Ph.D. candidate Dae-Hyeon Song, Professor Jae-Byum Chang, and (from top right) Dr. Chang Woo Song and Dr. Seunghee H. Cho of the Department of Materials Science and Engineering >
Biological structures have complex characteristics that are difficult to replicate artificially, but biotemplating methods* that directly utilize these biological structures have been used in various fields of application. The KAIST research team succeeded in utilizing previously unusable biological structures and expanding the areas in which biotemplate methods can be applied.
*Biotemplating: A method of using biotemplates as a mold to create functional structural materials, utilizing the functions of these biological structures, from viruses to the tissues and organs that make up our bodies
KAIST (President Kwang Hyung Lee) announced on the 10th that a joint research team of Professors Jae-Byum Chang and Professor Yeon Sik Jung of the Department of Materials Science and Engineering developed a biotemplating method that utilizes specific intracellular proteins in biological samples and has high tunability.
Existing biotemplate methods mainly utilize only the external surface of biological samples or have limitations in utilizing the structure-function correlation of various biological structures due to limited dimensions and sample sizes, making it difficult to create functional nanostructures.
To solve this problem, the research team studied a way to utilize various biological structures within the cells while retaining high tunability.
< Figure 1. CamBio utilizing microtubules, a intracellular protein structure. The silver nanoparticle chains synthesized along the microtubules that span the entire cell interior can be observed through an electron microscope, and it is shown that this can be used as a successful SERS substrate. >
As a result of the research, the team developed the “Conversion to advanced materials via labeled Biostructure”, shortened as “CamBio”, which enables the selective synthesis of nanostructures with various characteristics and sizes from specific protein structures composed of diverse proteins within biological specimens.
The CamBio method secures high tunability of functional nanostructures that can be manufactured from biological samples by merging various manufacturing and biological technologies.
Through the technology of repeatedly attaching antibodies, arranging cells in a certain shape, and thinly slicing tissue, the functional nanostructures made with CamBio showed improved performance on the surface-enhanced Raman spectroscopy (SERS)* substrate used for material detection.
*Surface-enhanced Raman spectroscopy (SERS): A technology that can detect very small amounts of substances using light, based on the principle that specific substances react to light and amplifies signals on surfaces of metals such as gold or silver.
The research team found that the nanoparticle chains made using the intracellular protein structures through the process of repeated labeling with antibodies allowed easier control, and improved SERS performance by up to 230%.
In addition, the research team expanded from utilizing the structures inside cells to obtaining samples of muscle tissues inside meat using a cryostat and successfully producing a substrate with periodic bands made of metal particles by performing the CamBio process. This method of producing a substrate not only allows large-scale production using biological samples, but also shows that it is a cost-effective method.
< Figure 2. A method for securing tunability using CamBio at the cell level. Examples of controlling characteristics by integrating iterative labeling and cell pattering techniques with CamBio are shown. >
The CamBio developed by the research team is expected to be used as a way to solve problems faced by various research fields as it is to expand the range of bio-samples that can be produced for various usage.
The first author, Dae-Hyeon Song, a Ph.D. candidate of KAIST Department of Materials Science and Engineering said, “Through CamBio, we have comprehensively accumulated biotemplating methods that can utilize more diverse protein structures,” and “If combined with the state-of-the-art biological technologies such as gene editing and 3D bioprinting and new material synthesis technologies, biostructures can be utilized in various fields of application.”
< Figure 3. A method for securing tunability using CamBio at the tissue level. In order to utilize proteins inside muscle tissue, the frozen tissue sectioning technology is combined, and through this, a substrate with a periodic nanoparticle band pattern is successfully produced, and it is shown that large-area acquisition of samples and price competitiveness can be achieved. >
This study, in which the Ph.D. candidate Dae-Hyeon Song along with Dr. Chang Woo Song, and Dr. Seunghee H. Cho of the same department participated as the first authors, was published online in the international academic journal, Advanced Science, on November 13th, 2024.
(Paper title: Highly Tunable, Nanomaterial-Functionalized Structural Templating of Intracellular Protein Structures Within Biological Species) https://doi.org/10.1002/advs.202406492
This study was conducted with a combination of support from various programs including the National Convergence Research of Scientific Challenges (National Research Foundation of Korea (NRF) 2024), Engineering Reseach Center (ERC) (Wearable Platform Materials Technology Center, NRF 2023), ERC (Global Bio-integrated Materials Center, NRF 2024), and the National Advanced Program for Biological Research Resources (Bioimaging Data Curation Center, NRF 2024) funded by Ministry of Science and ICT.
2025.01.10 View 6091
KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures
- Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS)
- Expected to have price competitiveness over bio-templating method as it expands the range of biological samples
- Expected to expand the range of application of nanostructure synthesis technology by utilizing various biological structures
< Photo 1. (From left) Professor Yeon Sik Jung, Ph.D. candidate Dae-Hyeon Song, Professor Jae-Byum Chang, and (from top right) Dr. Chang Woo Song and Dr. Seunghee H. Cho of the Department of Materials Science and Engineering >
Biological structures have complex characteristics that are difficult to replicate artificially, but biotemplating methods* that directly utilize these biological structures have been used in various fields of application. The KAIST research team succeeded in utilizing previously unusable biological structures and expanding the areas in which biotemplate methods can be applied.
*Biotemplating: A method of using biotemplates as a mold to create functional structural materials, utilizing the functions of these biological structures, from viruses to the tissues and organs that make up our bodies
KAIST (President Kwang Hyung Lee) announced on the 10th that a joint research team of Professors Jae-Byum Chang and Professor Yeon Sik Jung of the Department of Materials Science and Engineering developed a biotemplating method that utilizes specific intracellular proteins in biological samples and has high tunability.
Existing biotemplate methods mainly utilize only the external surface of biological samples or have limitations in utilizing the structure-function correlation of various biological structures due to limited dimensions and sample sizes, making it difficult to create functional nanostructures.
To solve this problem, the research team studied a way to utilize various biological structures within the cells while retaining high tunability.
< Figure 1. CamBio utilizing microtubules, a intracellular protein structure. The silver nanoparticle chains synthesized along the microtubules that span the entire cell interior can be observed through an electron microscope, and it is shown that this can be used as a successful SERS substrate. >
As a result of the research, the team developed the “Conversion to advanced materials via labeled Biostructure”, shortened as “CamBio”, which enables the selective synthesis of nanostructures with various characteristics and sizes from specific protein structures composed of diverse proteins within biological specimens.
The CamBio method secures high tunability of functional nanostructures that can be manufactured from biological samples by merging various manufacturing and biological technologies.
Through the technology of repeatedly attaching antibodies, arranging cells in a certain shape, and thinly slicing tissue, the functional nanostructures made with CamBio showed improved performance on the surface-enhanced Raman spectroscopy (SERS)* substrate used for material detection.
*Surface-enhanced Raman spectroscopy (SERS): A technology that can detect very small amounts of substances using light, based on the principle that specific substances react to light and amplifies signals on surfaces of metals such as gold or silver.
The research team found that the nanoparticle chains made using the intracellular protein structures through the process of repeated labeling with antibodies allowed easier control, and improved SERS performance by up to 230%.
In addition, the research team expanded from utilizing the structures inside cells to obtaining samples of muscle tissues inside meat using a cryostat and successfully producing a substrate with periodic bands made of metal particles by performing the CamBio process. This method of producing a substrate not only allows large-scale production using biological samples, but also shows that it is a cost-effective method.
< Figure 2. A method for securing tunability using CamBio at the cell level. Examples of controlling characteristics by integrating iterative labeling and cell pattering techniques with CamBio are shown. >
The CamBio developed by the research team is expected to be used as a way to solve problems faced by various research fields as it is to expand the range of bio-samples that can be produced for various usage.
The first author, Dae-Hyeon Song, a Ph.D. candidate of KAIST Department of Materials Science and Engineering said, “Through CamBio, we have comprehensively accumulated biotemplating methods that can utilize more diverse protein structures,” and “If combined with the state-of-the-art biological technologies such as gene editing and 3D bioprinting and new material synthesis technologies, biostructures can be utilized in various fields of application.”
< Figure 3. A method for securing tunability using CamBio at the tissue level. In order to utilize proteins inside muscle tissue, the frozen tissue sectioning technology is combined, and through this, a substrate with a periodic nanoparticle band pattern is successfully produced, and it is shown that large-area acquisition of samples and price competitiveness can be achieved. >
This study, in which the Ph.D. candidate Dae-Hyeon Song along with Dr. Chang Woo Song, and Dr. Seunghee H. Cho of the same department participated as the first authors, was published online in the international academic journal, Advanced Science, on November 13th, 2024.
(Paper title: Highly Tunable, Nanomaterial-Functionalized Structural Templating of Intracellular Protein Structures Within Biological Species) https://doi.org/10.1002/advs.202406492
This study was conducted with a combination of support from various programs including the National Convergence Research of Scientific Challenges (National Research Foundation of Korea (NRF) 2024), Engineering Reseach Center (ERC) (Wearable Platform Materials Technology Center, NRF 2023), ERC (Global Bio-integrated Materials Center, NRF 2024), and the National Advanced Program for Biological Research Resources (Bioimaging Data Curation Center, NRF 2024) funded by Ministry of Science and ICT.
2025.01.10 View 6091 -
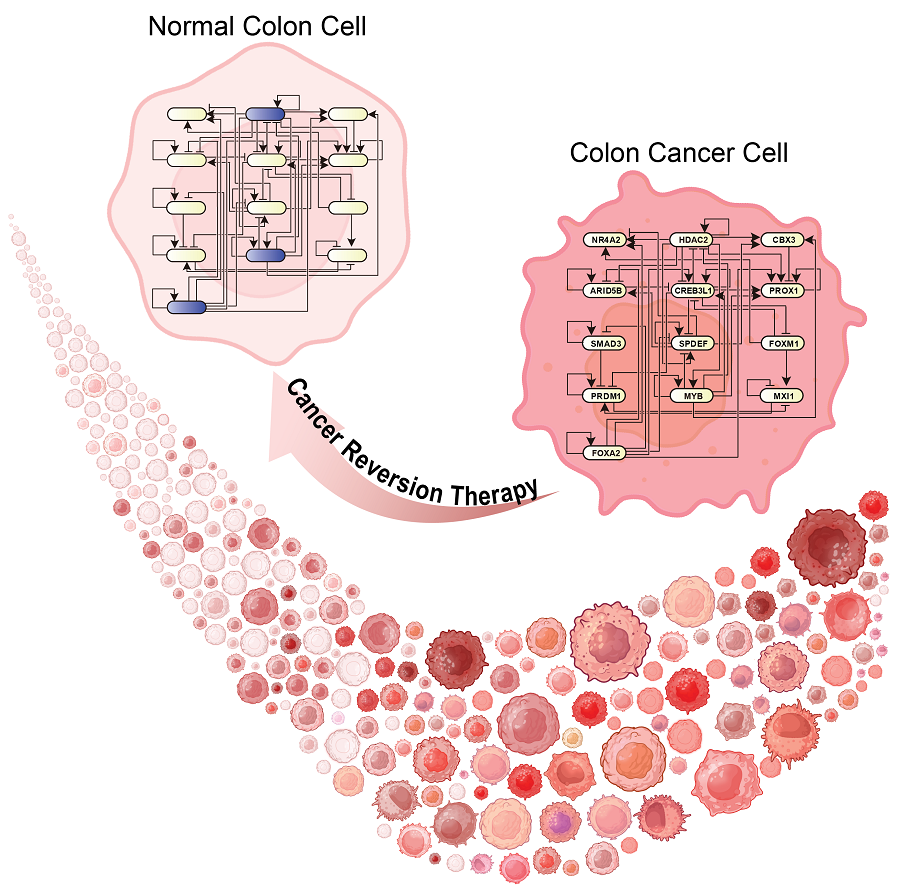 KAIST Develops Foundational Technology to Revert Cancer Cells to Normal Cells
Despite the development of numerous cancer treatment technologies, the common goal of current cancer therapies is to eliminate cancer cells. This approach, however, faces fundamental limitations, including cancer cells developing resistance and returning, as well as severe side effects from the destruction of healthy cells.
< (From top left) Bio and Brain Engineering PhD candidates Juhee Kim, Jeong-Ryeol Gong, Chun-Kyung Lee, and Hoon-Min Kim posed for a group photo with Professor Kwang-Hyun Cho >
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of December that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering has developed a groundbreaking technology that can treat colon cancer by converting cancer cells into a state resembling normal colon cells without killing them, thus avoiding side effects.
The research team focused on the observation that during the oncogenesis process, normal cells regress along their differentiation trajectory. Building on this insight, they developed a technology to create a digital twin of the gene network associated with the differentiation trajectory of normal cells.
< Figure 1. Technology for creating a digital twin of a gene network from single-cell transcriptome data of a normal cell differentiation trajectory. Professor Kwang-Hyun Cho's research team developed a digital twin creation technology that precisely observes the dynamics of gene regulatory relationships during the process of normal cells differentiating along a differentiation trajectory and analyzes the relationships among key genes to build a mathematical model that can be simulated (A-F). In addition, they developed a technology to discover key regulatory factors that control the differentiation trajectory of normal cells by simulating and analyzing this digital twin. >
< Figure 2. Digital twin simulation simulating the differentiation trajectory of normal colon cells. The dynamics of single-cell transcriptome data for the differentiation trajectory of normal colon cells were analyzed (A) and a digital twin of the gene network was developed representing the regulatory relationships of key genes in this differentiation trajectory (B). The simulation results of the digital twin confirm that it readily reproduces the dynamics of single-cell transcriptome data (C, D). >
Through simulation analysis, the team systematically identified master molecular switches that induce normal cell differentiation. When these switches were applied to colon cancer cells, the cancer cells reverted to a normal-like state, a result confirmed through molecular and cellular experiments as well as animal studies.
< Figure 3. Discovery of top-level key control factors that induce differentiation of normal colon cells. By applying control factor discovery technology to the digital twin model, three genes, HDAC2, FOXA2, and MYB, were discovered as key control factors that induce differentiation of normal colon cells (A, B). The results of simulation analysis of the regulatory effects of the discovered control factors through the digital twin confirmed that they could induce complete differentiation of colon cells (C). >
< Figure 4. Verification of the effect of the key control factors discovered using colon cancer cells and animal experiments on the reversibility of colon cancer. The key control factors of the normal colon cell differentiation trajectory discovered through digital twin simulation analysis were applied to actual colon cancer cells and colon cancer mouse animal models to experimentally verify the effect of cancer reversibility. The key control factors significantly reduced the proliferation of three colon cancer cell lines (A), and this was confirmed in the same way in animal models (B-D). >
This research demonstrates that cancer cell reversion can be systematically achieved by analyzing and utilizing the digital twin of the cancer cell gene network, rather than relying on serendipitous discoveries. The findings hold significant promise for developing reversible cancer therapies that can be applied to various types of cancer.
< Figure 5. The change in overall gene expression was confirmed through the regulation of the identified key regulatory factors, which converted the state of colon cancer cells to that of normal colon cells. The transcriptomes of colon cancer tissues and normal colon tissues from more than 400 colon cancer patients were compared with the transcriptomes of colon cancer cell lines and reversible colon cancer cell lines, respectively. The comparison results confirmed that the regulation of the identified key regulatory factors converted all three colon cancer cell lines to a state similar to the transcriptome expression of normal colon tissues. >
Professor Kwang-Hyun Cho remarked, "The fact that cancer cells can be converted back to normal cells is an astonishing phenomenon. This study proves that such reversion can be systematically induced."
He further emphasized, "This research introduces the novel concept of reversible cancer therapy by reverting cancer cells to normal cells. It also develops foundational technology for identifying targets for cancer reversion through the systematic analysis of normal cell differentiation trajectories."
This research included contributions from Jeong-Ryeol Gong, Chun-Kyung Lee, Hoon-Min Kim, Juhee Kim, and Jaeog Jeon, and was published in the online edition of the international journal Advanced Science by Wiley on December 11. (Title: “Control of Cellular Differentiation Trajectories for Cancer Reversion”) DOI: https://doi.org/10.1002/advs.202402132
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed a source technology to systematically discover key control factors that can induce reversibility of colon cancer cells through a systems biology approach and a digital twin simulation analysis of the differentiation trajectory of normal colon cells, and verified the effects of reversion on actual colon cancer through molecular cell experiments and animal experiments. >
The study was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Researcher Program and Basic Research Laboratory Program. The research findings have been transferred to BioRevert Inc., where they will be used for the development of practical cancer reversion therapies.
2024.12.23 View 104139
KAIST Develops Foundational Technology to Revert Cancer Cells to Normal Cells
Despite the development of numerous cancer treatment technologies, the common goal of current cancer therapies is to eliminate cancer cells. This approach, however, faces fundamental limitations, including cancer cells developing resistance and returning, as well as severe side effects from the destruction of healthy cells.
< (From top left) Bio and Brain Engineering PhD candidates Juhee Kim, Jeong-Ryeol Gong, Chun-Kyung Lee, and Hoon-Min Kim posed for a group photo with Professor Kwang-Hyun Cho >
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of December that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering has developed a groundbreaking technology that can treat colon cancer by converting cancer cells into a state resembling normal colon cells without killing them, thus avoiding side effects.
The research team focused on the observation that during the oncogenesis process, normal cells regress along their differentiation trajectory. Building on this insight, they developed a technology to create a digital twin of the gene network associated with the differentiation trajectory of normal cells.
< Figure 1. Technology for creating a digital twin of a gene network from single-cell transcriptome data of a normal cell differentiation trajectory. Professor Kwang-Hyun Cho's research team developed a digital twin creation technology that precisely observes the dynamics of gene regulatory relationships during the process of normal cells differentiating along a differentiation trajectory and analyzes the relationships among key genes to build a mathematical model that can be simulated (A-F). In addition, they developed a technology to discover key regulatory factors that control the differentiation trajectory of normal cells by simulating and analyzing this digital twin. >
< Figure 2. Digital twin simulation simulating the differentiation trajectory of normal colon cells. The dynamics of single-cell transcriptome data for the differentiation trajectory of normal colon cells were analyzed (A) and a digital twin of the gene network was developed representing the regulatory relationships of key genes in this differentiation trajectory (B). The simulation results of the digital twin confirm that it readily reproduces the dynamics of single-cell transcriptome data (C, D). >
Through simulation analysis, the team systematically identified master molecular switches that induce normal cell differentiation. When these switches were applied to colon cancer cells, the cancer cells reverted to a normal-like state, a result confirmed through molecular and cellular experiments as well as animal studies.
< Figure 3. Discovery of top-level key control factors that induce differentiation of normal colon cells. By applying control factor discovery technology to the digital twin model, three genes, HDAC2, FOXA2, and MYB, were discovered as key control factors that induce differentiation of normal colon cells (A, B). The results of simulation analysis of the regulatory effects of the discovered control factors through the digital twin confirmed that they could induce complete differentiation of colon cells (C). >
< Figure 4. Verification of the effect of the key control factors discovered using colon cancer cells and animal experiments on the reversibility of colon cancer. The key control factors of the normal colon cell differentiation trajectory discovered through digital twin simulation analysis were applied to actual colon cancer cells and colon cancer mouse animal models to experimentally verify the effect of cancer reversibility. The key control factors significantly reduced the proliferation of three colon cancer cell lines (A), and this was confirmed in the same way in animal models (B-D). >
This research demonstrates that cancer cell reversion can be systematically achieved by analyzing and utilizing the digital twin of the cancer cell gene network, rather than relying on serendipitous discoveries. The findings hold significant promise for developing reversible cancer therapies that can be applied to various types of cancer.
< Figure 5. The change in overall gene expression was confirmed through the regulation of the identified key regulatory factors, which converted the state of colon cancer cells to that of normal colon cells. The transcriptomes of colon cancer tissues and normal colon tissues from more than 400 colon cancer patients were compared with the transcriptomes of colon cancer cell lines and reversible colon cancer cell lines, respectively. The comparison results confirmed that the regulation of the identified key regulatory factors converted all three colon cancer cell lines to a state similar to the transcriptome expression of normal colon tissues. >
Professor Kwang-Hyun Cho remarked, "The fact that cancer cells can be converted back to normal cells is an astonishing phenomenon. This study proves that such reversion can be systematically induced."
He further emphasized, "This research introduces the novel concept of reversible cancer therapy by reverting cancer cells to normal cells. It also develops foundational technology for identifying targets for cancer reversion through the systematic analysis of normal cell differentiation trajectories."
This research included contributions from Jeong-Ryeol Gong, Chun-Kyung Lee, Hoon-Min Kim, Juhee Kim, and Jaeog Jeon, and was published in the online edition of the international journal Advanced Science by Wiley on December 11. (Title: “Control of Cellular Differentiation Trajectories for Cancer Reversion”) DOI: https://doi.org/10.1002/advs.202402132
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed a source technology to systematically discover key control factors that can induce reversibility of colon cancer cells through a systems biology approach and a digital twin simulation analysis of the differentiation trajectory of normal colon cells, and verified the effects of reversion on actual colon cancer through molecular cell experiments and animal experiments. >
The study was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Researcher Program and Basic Research Laboratory Program. The research findings have been transferred to BioRevert Inc., where they will be used for the development of practical cancer reversion therapies.
2024.12.23 View 104139 -
 KAIST Unveils New Possibilities for Treating Intractable Brain Tumors
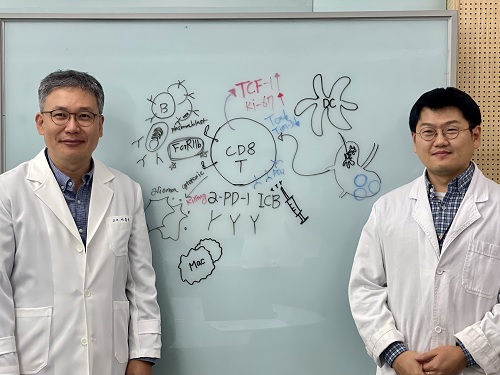
< Photo 1. (From left) Professor Heung Kyu Lee, KAIST Department of Biological Sciences, and Dr. Keun Bon Ku >
Immunotherapy, which enhances the immune system's T cell response to eliminate cancer cells, has emerged as a key approach in cancer treatment. However, in the case of glioblastoma, an aggressive and treatment-resistant brain tumor, numerous clinical trials have failed to confirm their efficacy. Korean researchers have recently analyzed the mechanisms that cause T cell exhaustion, which is characterized by a loss of function or a weakened response following prolonged exposure to antigens in such intractable cancers, identifying key control factors in T cell activation and clarifying the mechanisms that enhance therapeutic effectiveness.
KAIST (represented by President Kwang Hyung Lee) announced on the 6th of November that Professor Heung Kyu Lee’s team from the Department of Biological Sciences, in collaboration with the Korea Research Institute of Chemical Technology (represented by President Young Kuk Lee), has confirmed improved survival rates in a glioblastoma mouse model. By removing the inhibitory Fc gamma receptor (FcγRIIB), the research team was able to restore the responsiveness of cytotoxic T cells to immune checkpoint inhibitors, leading to enhanced anticancer activity.
The research team examined the effect of FcγRIIB, an inhibitory receptor recently found in cytotoxic T cells, on tumor-infiltrating T cells and the therapeutic effectiveness of the anti-PD-1 immune checkpoint inhibitor.
< Figure 1. Study results on improved survival rate due to increased antitumor activity of anti-PD-1 treatment in inhibitory Fc gamma receptor(Fcgr2b) ablation mice with murine glioblastoma. >
Their findings showed that deleting FcγRIIB induced the increase of tumor antigen-specific memory T cells, which helps to suppress exhaustion, enhances stem-like qualities, and reactivates T cell-mediated antitumor immunity, particularly in response to anti-PD-1 treatment. Furthermore, FcγRIIB deletion led to an increase in antigen-specific memory T cells that maintained continuous infiltration into the tumor tissue.
This study presents a new therapeutic target for tumors unresponsive to immune checkpoint inhibitors and demonstrates that combining FcγRIIB inhibition with anti-PD-1 treatment can produce synergistic effects, potentially improving therapeutic outcomes for tumors like glioblastoma, which typically show resistance to anti-PD-1 therapy.
< Figure 2. Overview of the study on the enhanced response to anti-PD-1 therapy for glioblastoma brain tumors upon deletion of the inhibitory Fc gamma receptor (FcγRIIB) in tumor microenvironment. When the inhibitory Fc gamma receptor (FcγRIIB) of cytotoxic T cells is deleted, an increase in tumor-specific memory T cells (Ttsms) was observed. In addition, this T cell subset is identified as originating from the tumor-draining lymph nodes(TdLNs) and leads to persistent infiltration into the tumor tissue. Anti-PD-1 therapy leads to an increased anti-tumor immune response via Ttsms, which is confirmed by increased tumor cell toxicity and increased cell division and decreased cell de-migration indices. Ultimately, the increased cytotoxic T cell immune response leads to an increase in the survival rate of glioblastoma. >
Professor Heung Kyu Lee explained, "This study offers a way to overcome clinical failures in treating brain tumors with immune checkpoint therapy and opens possibilities for broader applications to other intractable cancers. It also highlights the potential of utilizing cytotoxic T cells for tumor cell therapy."
The study, led by Dr. Keun Bon Ku of KAIST (currently a senior researcher at the Korea Research Institute of Chemical Technology's Center for Infectious Disease Diagnosis and Prevention), along with Chae Won Kim, Yumin Kim, Byeong Hoon Kang, Jeongwoo La, In Kang, Won Hyung Park, Stephen Ahn, and Sung Ki Lee, was published online on October 26 in the Journal for ImmunoTherapy of Cancer, an international journal in tumor immunology and therapy from the Society for Immunotherapy of Cancer. (Paper title: “Inhibitory Fcγ receptor deletion enhances CD8 T cell stemness increasing anti-PD-1 therapy responsiveness against glioblastoma,” http://dx.doi.org/10.1136/jitc-2024-009449).
This research received support from the National Research Foundation of Korea, the Bio & Medical Technology Development Program, and the Samsung Science & Technology Foundation.
2024.11.15 View 6180
KAIST Unveils New Possibilities for Treating Intractable Brain Tumors
< Photo 1. (From left) Professor Heung Kyu Lee, KAIST Department of Biological Sciences, and Dr. Keun Bon Ku >
Immunotherapy, which enhances the immune system's T cell response to eliminate cancer cells, has emerged as a key approach in cancer treatment. However, in the case of glioblastoma, an aggressive and treatment-resistant brain tumor, numerous clinical trials have failed to confirm their efficacy. Korean researchers have recently analyzed the mechanisms that cause T cell exhaustion, which is characterized by a loss of function or a weakened response following prolonged exposure to antigens in such intractable cancers, identifying key control factors in T cell activation and clarifying the mechanisms that enhance therapeutic effectiveness.
KAIST (represented by President Kwang Hyung Lee) announced on the 6th of November that Professor Heung Kyu Lee’s team from the Department of Biological Sciences, in collaboration with the Korea Research Institute of Chemical Technology (represented by President Young Kuk Lee), has confirmed improved survival rates in a glioblastoma mouse model. By removing the inhibitory Fc gamma receptor (FcγRIIB), the research team was able to restore the responsiveness of cytotoxic T cells to immune checkpoint inhibitors, leading to enhanced anticancer activity.
The research team examined the effect of FcγRIIB, an inhibitory receptor recently found in cytotoxic T cells, on tumor-infiltrating T cells and the therapeutic effectiveness of the anti-PD-1 immune checkpoint inhibitor.
< Figure 1. Study results on improved survival rate due to increased antitumor activity of anti-PD-1 treatment in inhibitory Fc gamma receptor(Fcgr2b) ablation mice with murine glioblastoma. >
Their findings showed that deleting FcγRIIB induced the increase of tumor antigen-specific memory T cells, which helps to suppress exhaustion, enhances stem-like qualities, and reactivates T cell-mediated antitumor immunity, particularly in response to anti-PD-1 treatment. Furthermore, FcγRIIB deletion led to an increase in antigen-specific memory T cells that maintained continuous infiltration into the tumor tissue.
This study presents a new therapeutic target for tumors unresponsive to immune checkpoint inhibitors and demonstrates that combining FcγRIIB inhibition with anti-PD-1 treatment can produce synergistic effects, potentially improving therapeutic outcomes for tumors like glioblastoma, which typically show resistance to anti-PD-1 therapy.
< Figure 2. Overview of the study on the enhanced response to anti-PD-1 therapy for glioblastoma brain tumors upon deletion of the inhibitory Fc gamma receptor (FcγRIIB) in tumor microenvironment. When the inhibitory Fc gamma receptor (FcγRIIB) of cytotoxic T cells is deleted, an increase in tumor-specific memory T cells (Ttsms) was observed. In addition, this T cell subset is identified as originating from the tumor-draining lymph nodes(TdLNs) and leads to persistent infiltration into the tumor tissue. Anti-PD-1 therapy leads to an increased anti-tumor immune response via Ttsms, which is confirmed by increased tumor cell toxicity and increased cell division and decreased cell de-migration indices. Ultimately, the increased cytotoxic T cell immune response leads to an increase in the survival rate of glioblastoma. >
Professor Heung Kyu Lee explained, "This study offers a way to overcome clinical failures in treating brain tumors with immune checkpoint therapy and opens possibilities for broader applications to other intractable cancers. It also highlights the potential of utilizing cytotoxic T cells for tumor cell therapy."
The study, led by Dr. Keun Bon Ku of KAIST (currently a senior researcher at the Korea Research Institute of Chemical Technology's Center for Infectious Disease Diagnosis and Prevention), along with Chae Won Kim, Yumin Kim, Byeong Hoon Kang, Jeongwoo La, In Kang, Won Hyung Park, Stephen Ahn, and Sung Ki Lee, was published online on October 26 in the Journal for ImmunoTherapy of Cancer, an international journal in tumor immunology and therapy from the Society for Immunotherapy of Cancer. (Paper title: “Inhibitory Fcγ receptor deletion enhances CD8 T cell stemness increasing anti-PD-1 therapy responsiveness against glioblastoma,” http://dx.doi.org/10.1136/jitc-2024-009449).
This research received support from the National Research Foundation of Korea, the Bio & Medical Technology Development Program, and the Samsung Science & Technology Foundation.
2024.11.15 View 6180 -
 KAIST presents strategies for Holotomography in advanced bio research
Measuring and analyzing three-dimensional (3D) images of live cells and tissues is considered crucial in advanced fields of biology and medicine. Organoids, which are 3D structures that mimic organs, are particular examples that significantly benefits 3D live imaging. Organoids provide effective alternatives to animal testing in the drug development processes, and can rapidly determine personalized medicine. On the other hand, active researches are ongoing to utilize organoids for organ replacement.
< Figure 1. Schematic illustration of holotomography compared to X-ray CT. Similar to CT, they share the commonality of measuring the optical properties of an unlabeled specimen in three dimensions. Instead of X-rays, holotomography irradiates light in the visible range, and provides refractive index measurements of transparent specimens rather than absorptivity. While CT obtains three-dimensional information only through mechanical rotation of the irradiating light, holotomography can replace this by applying wavefront control technology in the visible range. >
Organelle-level observation of 3D biological specimens such as organoids and stem cell colonies without staining or preprocessing holds significant implications for both innovating basic research and bioindustrial applications related to regenerative medicine and bioindustrial applications.
Holotomography (HT) is a 3D optical microscopy that implements 3D reconstruction analogous to that of X-ray computed tomography (CT). Although HT and CT share a similar theoretical background, HT facilitates high-resolution examination inside cells and tissues, instead of the human body. HT obtains 3D images of cells and tissues at the organelle level without chemical or genetic labeling, thus overcomes various challenges of existing methods in bio research and industry. Its potential is highlighted in research fields where sample physiology must not be disrupted, such as regenerative medicine, personalized medicine, and infertility treatment.
< Figure 2. Label-free 3D imaging of diverse live cells. Time-lapse image of Hep3B cells illustrating subcellular morphology changes upon H2O2 treatment, followed by cellular recovery after returning to the regular cell culture medium. >
This paper introduces the advantages and broad applicability of HT to biomedical researchers, while presenting an overview of principles and future technical challenges to optical researchers. It showcases various cases of applying HT in studies such as 3D biology, regenerative medicine, and cancer research, as well as suggesting future optical development. Also, it categorizes HT based on the light source, to describe the principles, limitations, and improvements of each category in detail. Particularly, the paper addresses strategies for deepening cell and organoid studies by introducing artificial intelligence (AI) to HT.
Due to its potential to drive advanced bioindustry, HT is attracting interest and investment from universities and corporates worldwide. The KAIST research team has been leading this international field by developing core technologies and carrying out key application researches throughout the last decade.
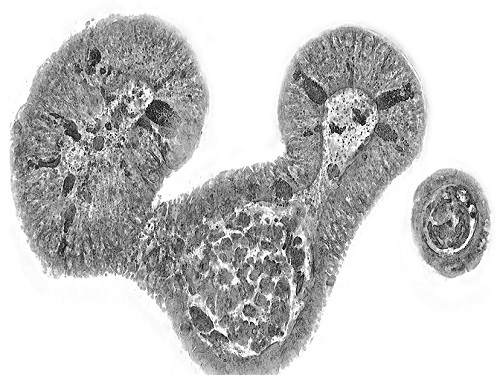
< Figure 3. Various types of cells and organelles that make up the imaging barrier of a living intestinal organoid can be observed using holotomography. >
This paper, co-authored by Dr. Geon Kim from KAIST Research Center for Natural Sciences, Professor Ki-Jun Yoon's team from the Department of Biological Sciences, Director Bon-Kyoung Koo's team from the Institute for Basic Science (IBS) Center for Genome Engineering, and Dr. Seongsoo Lee's team from the Korea Basic Science Institute (KBSI), was published in 'Nature Reviews Methods Primers' on the 25th of July. This research was supported by the Leader Grant and Basic Science Research Program of the National Research Foundation, the Hologram Core Technology Development Grant of the Ministry of Science and ICT, the Nano and Material Technology Development Project, and the Health and Medical R&D Project of the Ministry of Health and Welfare.
2024.07.30 View 6509
KAIST presents strategies for Holotomography in advanced bio research
Measuring and analyzing three-dimensional (3D) images of live cells and tissues is considered crucial in advanced fields of biology and medicine. Organoids, which are 3D structures that mimic organs, are particular examples that significantly benefits 3D live imaging. Organoids provide effective alternatives to animal testing in the drug development processes, and can rapidly determine personalized medicine. On the other hand, active researches are ongoing to utilize organoids for organ replacement.
< Figure 1. Schematic illustration of holotomography compared to X-ray CT. Similar to CT, they share the commonality of measuring the optical properties of an unlabeled specimen in three dimensions. Instead of X-rays, holotomography irradiates light in the visible range, and provides refractive index measurements of transparent specimens rather than absorptivity. While CT obtains three-dimensional information only through mechanical rotation of the irradiating light, holotomography can replace this by applying wavefront control technology in the visible range. >
Organelle-level observation of 3D biological specimens such as organoids and stem cell colonies without staining or preprocessing holds significant implications for both innovating basic research and bioindustrial applications related to regenerative medicine and bioindustrial applications.
Holotomography (HT) is a 3D optical microscopy that implements 3D reconstruction analogous to that of X-ray computed tomography (CT). Although HT and CT share a similar theoretical background, HT facilitates high-resolution examination inside cells and tissues, instead of the human body. HT obtains 3D images of cells and tissues at the organelle level without chemical or genetic labeling, thus overcomes various challenges of existing methods in bio research and industry. Its potential is highlighted in research fields where sample physiology must not be disrupted, such as regenerative medicine, personalized medicine, and infertility treatment.
< Figure 2. Label-free 3D imaging of diverse live cells. Time-lapse image of Hep3B cells illustrating subcellular morphology changes upon H2O2 treatment, followed by cellular recovery after returning to the regular cell culture medium. >
This paper introduces the advantages and broad applicability of HT to biomedical researchers, while presenting an overview of principles and future technical challenges to optical researchers. It showcases various cases of applying HT in studies such as 3D biology, regenerative medicine, and cancer research, as well as suggesting future optical development. Also, it categorizes HT based on the light source, to describe the principles, limitations, and improvements of each category in detail. Particularly, the paper addresses strategies for deepening cell and organoid studies by introducing artificial intelligence (AI) to HT.
Due to its potential to drive advanced bioindustry, HT is attracting interest and investment from universities and corporates worldwide. The KAIST research team has been leading this international field by developing core technologies and carrying out key application researches throughout the last decade.
< Figure 3. Various types of cells and organelles that make up the imaging barrier of a living intestinal organoid can be observed using holotomography. >
This paper, co-authored by Dr. Geon Kim from KAIST Research Center for Natural Sciences, Professor Ki-Jun Yoon's team from the Department of Biological Sciences, Director Bon-Kyoung Koo's team from the Institute for Basic Science (IBS) Center for Genome Engineering, and Dr. Seongsoo Lee's team from the Korea Basic Science Institute (KBSI), was published in 'Nature Reviews Methods Primers' on the 25th of July. This research was supported by the Leader Grant and Basic Science Research Program of the National Research Foundation, the Hologram Core Technology Development Grant of the Ministry of Science and ICT, the Nano and Material Technology Development Project, and the Health and Medical R&D Project of the Ministry of Health and Welfare.
2024.07.30 View 6509 -
 KAIST Develops Microbial Liquid Egg Substitute
A team of researchers published a paper on developing a substitute for eggs using microorganisms, grabbing international attention. It is expected that the development of egg substitutes using non-animal raw materials will solve the problems of factory farming, which causes problems like increased emission of greenhouse gas and waste, and contribute to building a sustainable food system that allows easy protein intake.
KAIST (President Kwang-Hyung Lee) announced that Research Professor Kyeong Rok Choi from the Biological Process Research Center and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering have published a paper on the development of an "Eco-Friendly Liquid Egg Substitute Derived from Microorganisms."
Eggs play a crucial role in various culinary applications due to their unique physicochemical properties such as gelling, foaming, and emulsifying, while also providing essential nutrients. However, traditional egg production is not only unethical and resource-intensive but also has significant environmental impacts such as greenhouse gas emissions and waste issues. Additionally, factors such as wars and trade regulations have led to significant increases in egg prices, highlighting food security concerns. In response to these issues, there has been growing interest in egg substitutes made from non-animal sources to establish a sustainable food system.
Although there has been progress in developing non-animal protein-based egg substitutes, no substitute has been able to fully replicate the essential functional properties of liquid eggs, such as gelling and foaming, while also providing complete nutrition. In this context, the research team aimed to develop a liquid egg substitute using microbial biomass, which has a protein content comparable to that of meat per unit dry mass. Various microorganisms, such as yeast, Bacillus, lactic acid bacteria, and other probiotics, have been proven safe through long-term human consumption. Microbial biomass requires fewer resources like water and land during production, and possesses high-quality nutrients, making it a promising sustainable food resource.
< Figure 1. Comparison of heat treatment results of microbial pellets and microbial lysates >
However, the semi-solid microbial biomass recovered through microbial cultivation was observed to turn liquid upon heating, unlike liquid egg. To address this, the research team devised a microbial lysate by breaking down the cell walls and cell membranes of microorganisms, which correspond to the eggshell. They found that the microbial lysate's proteins coagulated when heated and formed a gel similar to that of liquid egg. The gel formed from the heated microbial lysate was found to have microscopic structures and physical properties similar to those of boiled eggs. The addition of microbial-derived edible enzymes or plant-based materials allowed for the adjustment of its properties, enabling the creation of various textures.
Furthermore, the researchers demonstrated that the microbial lysate could form stable foams widely used in baking, such as meringues (made from egg whites). They successfully baked meringue cookies using this lysate, showing its potential as a functional liquid egg substitute.
Distinguished Professor Sang Yup Lee stated, "This substitute has excellent nutritional components, making it suitable for regular food consumption. It is especially promising as emergency food for long-term space travel, wartime situations, and other emergencies. More importantly, it contributes to securing a sustainable food system."
< Figure 2. Example of foaming ability of microbial lysate and meringue cookie production >
< Figure 3. Example of foaming ability of microbial lysate and meringue cookie production >
The paper was published online in the journal npj Science of Food, issued by Nature.
- Paper Title: Microbial lysates repurposed as liquid egg substitutes
- Authors: Kyeong Rok Choi (first author), Da-Hee Ahn, Seok Yeong Jung, YuHyun Lee, and Sang Yup Lee (corresponding author)
This research was supported by the Ministry of Science and ICT's project for developing eco-friendly chemical technologies to replace petroleum (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and the Rural Development Administration's Agricultural Microorganisms Project Group (Director: Professor Pan-sik Jang, Seoul National University) for developing protein production technology from inorganic substances through microbial metabolic system control (Project Leader: Research Professor Kyeong Rok Choi, KAIST).
2024.07.05 View 7780
KAIST Develops Microbial Liquid Egg Substitute
A team of researchers published a paper on developing a substitute for eggs using microorganisms, grabbing international attention. It is expected that the development of egg substitutes using non-animal raw materials will solve the problems of factory farming, which causes problems like increased emission of greenhouse gas and waste, and contribute to building a sustainable food system that allows easy protein intake.
KAIST (President Kwang-Hyung Lee) announced that Research Professor Kyeong Rok Choi from the Biological Process Research Center and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering have published a paper on the development of an "Eco-Friendly Liquid Egg Substitute Derived from Microorganisms."
Eggs play a crucial role in various culinary applications due to their unique physicochemical properties such as gelling, foaming, and emulsifying, while also providing essential nutrients. However, traditional egg production is not only unethical and resource-intensive but also has significant environmental impacts such as greenhouse gas emissions and waste issues. Additionally, factors such as wars and trade regulations have led to significant increases in egg prices, highlighting food security concerns. In response to these issues, there has been growing interest in egg substitutes made from non-animal sources to establish a sustainable food system.
Although there has been progress in developing non-animal protein-based egg substitutes, no substitute has been able to fully replicate the essential functional properties of liquid eggs, such as gelling and foaming, while also providing complete nutrition. In this context, the research team aimed to develop a liquid egg substitute using microbial biomass, which has a protein content comparable to that of meat per unit dry mass. Various microorganisms, such as yeast, Bacillus, lactic acid bacteria, and other probiotics, have been proven safe through long-term human consumption. Microbial biomass requires fewer resources like water and land during production, and possesses high-quality nutrients, making it a promising sustainable food resource.
< Figure 1. Comparison of heat treatment results of microbial pellets and microbial lysates >
However, the semi-solid microbial biomass recovered through microbial cultivation was observed to turn liquid upon heating, unlike liquid egg. To address this, the research team devised a microbial lysate by breaking down the cell walls and cell membranes of microorganisms, which correspond to the eggshell. They found that the microbial lysate's proteins coagulated when heated and formed a gel similar to that of liquid egg. The gel formed from the heated microbial lysate was found to have microscopic structures and physical properties similar to those of boiled eggs. The addition of microbial-derived edible enzymes or plant-based materials allowed for the adjustment of its properties, enabling the creation of various textures.
Furthermore, the researchers demonstrated that the microbial lysate could form stable foams widely used in baking, such as meringues (made from egg whites). They successfully baked meringue cookies using this lysate, showing its potential as a functional liquid egg substitute.
Distinguished Professor Sang Yup Lee stated, "This substitute has excellent nutritional components, making it suitable for regular food consumption. It is especially promising as emergency food for long-term space travel, wartime situations, and other emergencies. More importantly, it contributes to securing a sustainable food system."
< Figure 2. Example of foaming ability of microbial lysate and meringue cookie production >
< Figure 3. Example of foaming ability of microbial lysate and meringue cookie production >
The paper was published online in the journal npj Science of Food, issued by Nature.
- Paper Title: Microbial lysates repurposed as liquid egg substitutes
- Authors: Kyeong Rok Choi (first author), Da-Hee Ahn, Seok Yeong Jung, YuHyun Lee, and Sang Yup Lee (corresponding author)
This research was supported by the Ministry of Science and ICT's project for developing eco-friendly chemical technologies to replace petroleum (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and the Rural Development Administration's Agricultural Microorganisms Project Group (Director: Professor Pan-sik Jang, Seoul National University) for developing protein production technology from inorganic substances through microbial metabolic system control (Project Leader: Research Professor Kyeong Rok Choi, KAIST).
2024.07.05 View 7780 -
 Revolutionary 'scLENS' Unveiled to Decode Complex Single-Cell Genomic Data
Unlocking biological information from complex single-cell genomic data has just become easier and more precise, thanks to the innovative 'scLENS' tool developed by the Biomedical Mathematics Group within the IBS Center for Mathematical and Computational Sciences led by Chief Investigator Jae Kyoung Kim, who is also a professor at KAIST. This new finding represents a significant leap forward in the field of single-cell transcriptomics.
Single-cell genomic analysis is an advanced technique that measures gene expression at the individual cell level, revealing cellular changes and interactions that are not observable with traditional genomic analysis methods. When applied to cancer tissues, this analysis can delineate the composition of diverse cell types within a tumor, providing insights into how cancer progresses and identifying key genes involved during each stage of progression.
Despite the immense potential of single-cell genomic analysis, handling the vast amount of data that it generates has always been challenging. The amount of data covers the expression of tens of thousands of genes across hundreds to thousands of individual cells. This not only results in large datasets but also introduces noise-related distortions, which arise in part due to current measurement limitations.
< Figure 1. Overview of scLENS (single-cell Low-dimensional embedding using the effective Noise Subtract) >
(Left) Current dimensionality reduction methods for scRNA-seq data involve conventional data preprocessing steps, such as log normalization, followed by manual selection of signals from the scaled data. However, this study reveals that the high levels of sparsity and variability in scRNA-seq data can lead to signal distortion during the data preprocessing, compromising the accuracy of downstream analyses.
(Right) To address this issue, the researchers integrated L2 normalization into the conventional preprocessing pipeline, effectively mitigating signal distortion. Moreover, they developed a novel signal detection algorithm that eliminates the need for user intervention by leveraging random matrix theory-based noise filtering and signal robustness testing. By incorporating these techniques, scLENS enables accurate and automated analysis of scRNA-seq data, overcoming the limitations of existing dimensionality reduction methods.
Corresponding author Jae Kyoung Kim highlighted, “There has been a remarkable advancement in experimental technologies for analyzing single-cell transcriptomes over the past decade. However, due to limitations in data analysis methods, there has been a struggle to fully utilize valuable data obtained through extensive cost and time."
Researchers have developed numerous analysis methods over the years to discern biological signals from this noise. However, the accuracy of these methods has been less than satisfactory. A critical issue is that determining signal and noise thresholds often depends on subjective decisions from the users.
The newly developed scLENS tool harnesses Random Matrix Theory and Signal robustness test to automatically differentiate signals from noise without relying on subjective user input.
First author Hyun Kim stated, "Previously, users had to arbitrarily decide the threshold for signal and noise, which compromised the reproducibility of analysis results and introduced subjectivity. scLENS eliminates this problem by automatically detecting signals using only the inherent structure of the data."
During the development of scLENS, researchers identified the fundamental reasons for inaccuracies in existing analysis methods. They found that commonly used data preprocessing methods distort both biological signals and noise. The new preprocessing approach that scLENS offers is free from such distortions.
By resolving issues related to noise threshold determined by subjective user choice and signal distortion in conventional data preprocessing, scLENS significantly outperforms existing methods in accuracy. Additionally, scLENS automates the laborious process of signal dimension selection, allowing researchers to extract biological signals conveniently and automatically.
CI Kim added, "scLENS solves major issues in single-cell transcriptome data analysis, substantially improving the accuracy and efficiency throughout the analysis process. This is a prime example of how fundamental mathematical theories can drive innovation in life sciences research, allowing researchers to more quickly and accurately answer biological questions and uncover secrets of life that were previously hidden."
This research was published in the international journal 'Nature Communications' on April 27.
Terminology
* Single-cell RNA sequencing (scRNA-seq): A technique used to measure gene expression levels in individual cells, providing insights into cell heterogeneity and rare cell types.
* Dimensionality reduction: A method to reduce the number of features or variables in a dataset while preserving the most important information, making data analysis more manageable and interpretable.
* Random matrix theory: A mathematical framework used to model and analyze the properties of large, random matrices, which can be applied to filter out noise in high-dimensional data.
* Signal robustness test: Among the signals, this test selects signals that are robust to the slight perturbation in data because real biological signals should be invariant for such slight modification in the data.
2024.05.09 View 6808
Revolutionary 'scLENS' Unveiled to Decode Complex Single-Cell Genomic Data
Unlocking biological information from complex single-cell genomic data has just become easier and more precise, thanks to the innovative 'scLENS' tool developed by the Biomedical Mathematics Group within the IBS Center for Mathematical and Computational Sciences led by Chief Investigator Jae Kyoung Kim, who is also a professor at KAIST. This new finding represents a significant leap forward in the field of single-cell transcriptomics.
Single-cell genomic analysis is an advanced technique that measures gene expression at the individual cell level, revealing cellular changes and interactions that are not observable with traditional genomic analysis methods. When applied to cancer tissues, this analysis can delineate the composition of diverse cell types within a tumor, providing insights into how cancer progresses and identifying key genes involved during each stage of progression.
Despite the immense potential of single-cell genomic analysis, handling the vast amount of data that it generates has always been challenging. The amount of data covers the expression of tens of thousands of genes across hundreds to thousands of individual cells. This not only results in large datasets but also introduces noise-related distortions, which arise in part due to current measurement limitations.
< Figure 1. Overview of scLENS (single-cell Low-dimensional embedding using the effective Noise Subtract) >
(Left) Current dimensionality reduction methods for scRNA-seq data involve conventional data preprocessing steps, such as log normalization, followed by manual selection of signals from the scaled data. However, this study reveals that the high levels of sparsity and variability in scRNA-seq data can lead to signal distortion during the data preprocessing, compromising the accuracy of downstream analyses.
(Right) To address this issue, the researchers integrated L2 normalization into the conventional preprocessing pipeline, effectively mitigating signal distortion. Moreover, they developed a novel signal detection algorithm that eliminates the need for user intervention by leveraging random matrix theory-based noise filtering and signal robustness testing. By incorporating these techniques, scLENS enables accurate and automated analysis of scRNA-seq data, overcoming the limitations of existing dimensionality reduction methods.
Corresponding author Jae Kyoung Kim highlighted, “There has been a remarkable advancement in experimental technologies for analyzing single-cell transcriptomes over the past decade. However, due to limitations in data analysis methods, there has been a struggle to fully utilize valuable data obtained through extensive cost and time."
Researchers have developed numerous analysis methods over the years to discern biological signals from this noise. However, the accuracy of these methods has been less than satisfactory. A critical issue is that determining signal and noise thresholds often depends on subjective decisions from the users.
The newly developed scLENS tool harnesses Random Matrix Theory and Signal robustness test to automatically differentiate signals from noise without relying on subjective user input.
First author Hyun Kim stated, "Previously, users had to arbitrarily decide the threshold for signal and noise, which compromised the reproducibility of analysis results and introduced subjectivity. scLENS eliminates this problem by automatically detecting signals using only the inherent structure of the data."
During the development of scLENS, researchers identified the fundamental reasons for inaccuracies in existing analysis methods. They found that commonly used data preprocessing methods distort both biological signals and noise. The new preprocessing approach that scLENS offers is free from such distortions.
By resolving issues related to noise threshold determined by subjective user choice and signal distortion in conventional data preprocessing, scLENS significantly outperforms existing methods in accuracy. Additionally, scLENS automates the laborious process of signal dimension selection, allowing researchers to extract biological signals conveniently and automatically.
CI Kim added, "scLENS solves major issues in single-cell transcriptome data analysis, substantially improving the accuracy and efficiency throughout the analysis process. This is a prime example of how fundamental mathematical theories can drive innovation in life sciences research, allowing researchers to more quickly and accurately answer biological questions and uncover secrets of life that were previously hidden."
This research was published in the international journal 'Nature Communications' on April 27.
Terminology
* Single-cell RNA sequencing (scRNA-seq): A technique used to measure gene expression levels in individual cells, providing insights into cell heterogeneity and rare cell types.
* Dimensionality reduction: A method to reduce the number of features or variables in a dataset while preserving the most important information, making data analysis more manageable and interpretable.
* Random matrix theory: A mathematical framework used to model and analyze the properties of large, random matrices, which can be applied to filter out noise in high-dimensional data.
* Signal robustness test: Among the signals, this test selects signals that are robust to the slight perturbation in data because real biological signals should be invariant for such slight modification in the data.
2024.05.09 View 6808 -
 KAIST Research team develops anti-icing film that only requires sunlight
A KAIST research team has developed an anti-icing and de-icing film coating technology that can apply the photothermal effect of gold nanoparticles to industrial sites without the need for heating wires, periodic spray or oil coating of anti-freeze substances, and substrate design alterations.
The group led by Professor Hyoungsoo Kim from the Department of Mechanical Engineering (Fluid & Interface Laboratory) and Professor Dong Ki Yoon from the Department of Chemistry (Soft Material Assembly Group) revealed on January 3 to have together developed an original technique that can uniformly pattern gold nanorod (GNR) particles in quadrants through simple evaporation, and have used this to develop an anti-icing and de-icing surface.
Many scientists in recent years have tried to control substrate surfaces through various coating techniques, and those involving the patterning of functional nanomaterials have gained special attention. In particular, GNR is considered a promising candidate nanomaterial for its biocompatibility, chemical stability, relatively simple synthesis, and its stable and unique property of surface plasmon resonance. To maximize the performance of GNR, it is important to achieve a high uniformity during film deposition, and a high level of rod alignment. However, achieving both criteria has thus far been a difficult challenge.
< Figure 1. Conceptual image to display Hydrodynamic mechanisms for the formation of a homogeneous quadrant cellulose nanocrystal(CNC) matrix. >
To solve this, the joint research team utilized cellulose nanocrystal (CNC), a next-generation functional nanomaterial that can easily be extracted from nature. By co-assembling GNR on CNC quadrant templates, the team could uniformly dry the film and successfully obtain a GNR film with a uniform alignment in a ring-shape. Compared to existing coffee-ring films, the highly uniform and aligned GNR film developed through this research showed enhanced plasmonic photothermal properties, and the team showed that it could carry out anti-icing and de-icing functions by simply irradiating light in the visible wavelength range.
< Figure 2. Optical and thermal performance evaluation results of gold nanorod film and demonstration of plasmonic heater for anti-icing and de-icing. >
Professor Hyoungsoo Kim said, “This technique can be applied to plastic, as well as flexible surfaces. By using it on exterior materials and films, it can generate its own heat energy, which would greatly save energy through voluntary thermal energy harvesting across various applications including cars, aircrafts, and windows in residential or commercial spaces, where frosting becomes a serious issue in the winter.” Professor Dong Ki Yoon added, “This research is significant in that we can now freely pattern the CNC-GNR composite, which was previously difficult to create into films, over a large area. We can utilize this as an anti-icing material, and if we were to take advantage of the plasmonic properties of gold, we can also use it like stained-glass to decorate glass surfaces.”
This research was conducted by Ph.D. candidate Jeongsu Pyeon from the Department of Mechanical Engineering, and his co-first author Dr. Soon Mo Park (a KAIST graduate, currently a post-doctoral associate at Cornell University), and was pushed in the online volume of Nature Communication on December 8, 2023 under the title “Plasmonic Metasurfaces of Cellulose Nanocrystal Matrices with Quadrants of Aligned Gold Nanorods for Photothermal Anti-Icing." Recognized for its achievement, the research was also selected as an editor’s highlight for the journals Materials Science and Chemistry, and Inorganic and Physical Chemistry.
This research was supported by the Individual Basic Mid-Sized Research Fund from the National Research Foundation of Korea and the Center for Multiscale Chiral Architectures.
2024.01.16 View 12258
KAIST Research team develops anti-icing film that only requires sunlight
A KAIST research team has developed an anti-icing and de-icing film coating technology that can apply the photothermal effect of gold nanoparticles to industrial sites without the need for heating wires, periodic spray or oil coating of anti-freeze substances, and substrate design alterations.
The group led by Professor Hyoungsoo Kim from the Department of Mechanical Engineering (Fluid & Interface Laboratory) and Professor Dong Ki Yoon from the Department of Chemistry (Soft Material Assembly Group) revealed on January 3 to have together developed an original technique that can uniformly pattern gold nanorod (GNR) particles in quadrants through simple evaporation, and have used this to develop an anti-icing and de-icing surface.
Many scientists in recent years have tried to control substrate surfaces through various coating techniques, and those involving the patterning of functional nanomaterials have gained special attention. In particular, GNR is considered a promising candidate nanomaterial for its biocompatibility, chemical stability, relatively simple synthesis, and its stable and unique property of surface plasmon resonance. To maximize the performance of GNR, it is important to achieve a high uniformity during film deposition, and a high level of rod alignment. However, achieving both criteria has thus far been a difficult challenge.
< Figure 1. Conceptual image to display Hydrodynamic mechanisms for the formation of a homogeneous quadrant cellulose nanocrystal(CNC) matrix. >
To solve this, the joint research team utilized cellulose nanocrystal (CNC), a next-generation functional nanomaterial that can easily be extracted from nature. By co-assembling GNR on CNC quadrant templates, the team could uniformly dry the film and successfully obtain a GNR film with a uniform alignment in a ring-shape. Compared to existing coffee-ring films, the highly uniform and aligned GNR film developed through this research showed enhanced plasmonic photothermal properties, and the team showed that it could carry out anti-icing and de-icing functions by simply irradiating light in the visible wavelength range.
< Figure 2. Optical and thermal performance evaluation results of gold nanorod film and demonstration of plasmonic heater for anti-icing and de-icing. >
Professor Hyoungsoo Kim said, “This technique can be applied to plastic, as well as flexible surfaces. By using it on exterior materials and films, it can generate its own heat energy, which would greatly save energy through voluntary thermal energy harvesting across various applications including cars, aircrafts, and windows in residential or commercial spaces, where frosting becomes a serious issue in the winter.” Professor Dong Ki Yoon added, “This research is significant in that we can now freely pattern the CNC-GNR composite, which was previously difficult to create into films, over a large area. We can utilize this as an anti-icing material, and if we were to take advantage of the plasmonic properties of gold, we can also use it like stained-glass to decorate glass surfaces.”
This research was conducted by Ph.D. candidate Jeongsu Pyeon from the Department of Mechanical Engineering, and his co-first author Dr. Soon Mo Park (a KAIST graduate, currently a post-doctoral associate at Cornell University), and was pushed in the online volume of Nature Communication on December 8, 2023 under the title “Plasmonic Metasurfaces of Cellulose Nanocrystal Matrices with Quadrants of Aligned Gold Nanorods for Photothermal Anti-Icing." Recognized for its achievement, the research was also selected as an editor’s highlight for the journals Materials Science and Chemistry, and Inorganic and Physical Chemistry.
This research was supported by the Individual Basic Mid-Sized Research Fund from the National Research Foundation of Korea and the Center for Multiscale Chiral Architectures.
2024.01.16 View 12258 -
 A KAIST Research Team Develops High-Performance Stretchable Solar Cells
With the market for wearable electric devices growing rapidly, stretchable solar cells that can function under strain have received considerable attention as an energy source. To build such solar cells, it is necessary that their photoactive layer, which converts light into electricity, shows high electrical performance while possessing mechanical elasticity. However, satisfying both of these two requirements is challenging, making stretchable solar cells difficult to develop.
On December 26, a KAIST research team from the Department of Chemical and Biomolecular Engineering (CBE) led by Professor Bumjoon Kim announced the development of a new conductive polymer material that achieved both high electrical performance and elasticity while introducing the world’s highest-performing stretchable organic solar cell.
Organic solar cells are devices whose photoactive layer, which is responsible for the conversion of light into electricity, is composed of organic materials. Compared to existing non-organic material-based solar cells, they are lighter and flexible, making them highly applicable for wearable electrical devices. Solar cells as an energy source are particularly important for building electrical devices, but high-efficiency solar cells often lack flexibility, and their application in wearable devices have therefore been limited to this point.
The team led by Professor Kim conjugated a highly stretchable polymer to an electrically conductive polymer with excellent electrical properties through chemical bonding, and developed a new conductive polymer with both electrical conductivity and mechanical stretchability. This polymer meets the highest reported level of photovoltaic conversion efficiency (19%) using organic solar cells, while also showing 10 times the stretchability of existing devices. The team thereby built the world’s highest performing stretchable solar cell that can be stretched up to 40% during operation, and demonstrated its applicability for wearable devices.
< Figure 1. Chemical structure of the newly developed conductive polymer and performance of stretchable organic solar cells using the material. >
Professor Kim said, “Through this research, we not only developed the world’s best performing stretchable organic solar cell, but it is also significant that we developed a new polymer that can be applicable as a base material for various electronic devices that needs to be malleable and/or elastic.”
< Figure 2. Photovoltaic efficiency and mechanical stretchability of newly developed polymers compared to existing polymers. >
This research, conducted by KAIST researchers Jin-Woo Lee and Heung-Goo Lee as first co-authors in cooperation with teams led by Professor Taek-Soo Kim from the Department of Mechanical Engineering and Professor Sheng Li from the Department of CBE, was published in Joule on December 1 (Paper Title: Rigid and Soft Block-Copolymerized Conjugated Polymers Enable High-Performance Intrinsically-Stretchable Organic Solar Cells).
This research was supported by the National Research Foundation of Korea.
2024.01.04 View 10163
A KAIST Research Team Develops High-Performance Stretchable Solar Cells
With the market for wearable electric devices growing rapidly, stretchable solar cells that can function under strain have received considerable attention as an energy source. To build such solar cells, it is necessary that their photoactive layer, which converts light into electricity, shows high electrical performance while possessing mechanical elasticity. However, satisfying both of these two requirements is challenging, making stretchable solar cells difficult to develop.
On December 26, a KAIST research team from the Department of Chemical and Biomolecular Engineering (CBE) led by Professor Bumjoon Kim announced the development of a new conductive polymer material that achieved both high electrical performance and elasticity while introducing the world’s highest-performing stretchable organic solar cell.
Organic solar cells are devices whose photoactive layer, which is responsible for the conversion of light into electricity, is composed of organic materials. Compared to existing non-organic material-based solar cells, they are lighter and flexible, making them highly applicable for wearable electrical devices. Solar cells as an energy source are particularly important for building electrical devices, but high-efficiency solar cells often lack flexibility, and their application in wearable devices have therefore been limited to this point.
The team led by Professor Kim conjugated a highly stretchable polymer to an electrically conductive polymer with excellent electrical properties through chemical bonding, and developed a new conductive polymer with both electrical conductivity and mechanical stretchability. This polymer meets the highest reported level of photovoltaic conversion efficiency (19%) using organic solar cells, while also showing 10 times the stretchability of existing devices. The team thereby built the world’s highest performing stretchable solar cell that can be stretched up to 40% during operation, and demonstrated its applicability for wearable devices.
< Figure 1. Chemical structure of the newly developed conductive polymer and performance of stretchable organic solar cells using the material. >
Professor Kim said, “Through this research, we not only developed the world’s best performing stretchable organic solar cell, but it is also significant that we developed a new polymer that can be applicable as a base material for various electronic devices that needs to be malleable and/or elastic.”
< Figure 2. Photovoltaic efficiency and mechanical stretchability of newly developed polymers compared to existing polymers. >
This research, conducted by KAIST researchers Jin-Woo Lee and Heung-Goo Lee as first co-authors in cooperation with teams led by Professor Taek-Soo Kim from the Department of Mechanical Engineering and Professor Sheng Li from the Department of CBE, was published in Joule on December 1 (Paper Title: Rigid and Soft Block-Copolymerized Conjugated Polymers Enable High-Performance Intrinsically-Stretchable Organic Solar Cells).
This research was supported by the National Research Foundation of Korea.
2024.01.04 View 10163 -
 KAIST proposes alternatives to chemical factories through “iBridge”
- A computer simulation program “iBridge” was developed at KAIST that can put together microbial cell factories quickly and efficiently to produce cosmetics and food additives, and raw materials for nylons
- Eco-friendly and sustainable fermentation process to establish an alternative to chemical plants
As climate change and environmental concerns intensify, sustainable microbial cell factories garner significant attention as candidates to replace chemical plants. To develop microorganisms to be used in the microbial cell factories, it is crucial to modify their metabolic processes to induce efficient target chemical production by modulating its gene expressions. Yet, the challenge persists in determining which gene expressions to amplify and suppress, and the experimental verification of these modification targets is a time- and resource-intensive process even for experts. The challenges were addressed by a team of researchers at KAIST (President Kwang-Hyung Lee) led by Distinguished Professor Sang Yup Lee.
It was announced on the 9th by the school that a method for building a microbial factory at low cost, quickly and efficiently, was presented by a novel computer simulation program developed by the team under Professor Lee’s guidance, which is named “iBridge”. This innovative system is designed to predict gene targets to either overexpress or downregulate in the goal of producing a desired compound to enable the cost-effective and efficient construction of microbial cell factories specifically tailored for producing the chemical compound in demand from renewable biomass.
Systems metabolic engineering is a field of research and engineering pioneered by KAIST’s Distinguished Professor Sang Yup Lee that seeks to produce valuable compounds in industrial demands using microorganisms that are re-configured by a combination of methods including, but not limited to, metabolic engineering, synthetic biology, systems biology, and fermentation engineering.
In order to improve microorganisms’ capability to produce useful compounds, it is essential to delete, suppress, or overexpress microbial genes. However, it is difficult even for the experts to identify the gene targets to modify without experimental confirmations for each of them, which can take up immeasurable amount of time and resources.
The newly developed iBridge identifies positive and negative metabolites within cells, which exert positive and/or negative impact on formation of the products, by calculating the sum of covariances of their outgoing (consuming) reaction fluxes for a target chemical. Subsequently, it pinpoints "bridge" reactions responsible for converting negative metabolites into positive ones as candidates for overexpression, while identifying the opposites as targets for downregulation.
The research team successfully utilized the iBridge simulation to establish E. coli microbial cell factories each capable of producing three of the compounds that are in high demands at a production capacity that has not been reported around the world. They developed E. coli strains that can each produce panthenol, a moisturizing agent found in many cosmetics, putrescine, which is one of the key components in nylon production, and 4-hydroxyphenyllactic acid, an anti-bacterial food additive. In addition to these three compounds, the study presents predictions for overexpression and suppression genes to construct microbial factories for 298 other industrially valuable compounds.
Dr. Youngjoon Lee, the co-first author of this paper from KAIST, emphasized the accelerated construction of various microbial factories the newly developed simulation enabled. He stated, "With the use of this simulation, multiple microbial cell factories have been established significantly faster than it would have been using the conventional methods. Microbial cell factories producing a wider range of valuable compounds can now be constructed quickly using this technology."
Professor Sang Yup Lee said, "Systems metabolic engineering is a crucial technology for addressing the current climate change issues." He added, "This simulation could significantly expedite the transition from resorting to conventional chemical factories to utilizing environmentally friendly microbial factories."
< Figure. Conceptual diagram of the flow of iBridge simulation >
The team’s work on iBridge is described in a paper titled "Genome-Wide Identification of Overexpression and Downregulation Gene Targets Based on the Sum of Covariances of the Outgoing Reaction Fluxes" written by Dr. Won Jun Kim, and Dr. Youngjoon Lee of the Bioprocess Research Center and Professors Hyun Uk Kim and Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST. The paper was published via peer-review on the 6th of November on “Cell Systems” by Cell Press.
This research was conducted with the support from the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and Development of Platform Technology for the Production of Novel Aromatic Bioplastic using Microbial Cell Factories Project (Project Leader: Research Professor So Young Choi, KAIST) of the Korean Ministry of Science and ICT.
2023.11.09 View 9774
KAIST proposes alternatives to chemical factories through “iBridge”
- A computer simulation program “iBridge” was developed at KAIST that can put together microbial cell factories quickly and efficiently to produce cosmetics and food additives, and raw materials for nylons
- Eco-friendly and sustainable fermentation process to establish an alternative to chemical plants
As climate change and environmental concerns intensify, sustainable microbial cell factories garner significant attention as candidates to replace chemical plants. To develop microorganisms to be used in the microbial cell factories, it is crucial to modify their metabolic processes to induce efficient target chemical production by modulating its gene expressions. Yet, the challenge persists in determining which gene expressions to amplify and suppress, and the experimental verification of these modification targets is a time- and resource-intensive process even for experts. The challenges were addressed by a team of researchers at KAIST (President Kwang-Hyung Lee) led by Distinguished Professor Sang Yup Lee.
It was announced on the 9th by the school that a method for building a microbial factory at low cost, quickly and efficiently, was presented by a novel computer simulation program developed by the team under Professor Lee’s guidance, which is named “iBridge”. This innovative system is designed to predict gene targets to either overexpress or downregulate in the goal of producing a desired compound to enable the cost-effective and efficient construction of microbial cell factories specifically tailored for producing the chemical compound in demand from renewable biomass.
Systems metabolic engineering is a field of research and engineering pioneered by KAIST’s Distinguished Professor Sang Yup Lee that seeks to produce valuable compounds in industrial demands using microorganisms that are re-configured by a combination of methods including, but not limited to, metabolic engineering, synthetic biology, systems biology, and fermentation engineering.
In order to improve microorganisms’ capability to produce useful compounds, it is essential to delete, suppress, or overexpress microbial genes. However, it is difficult even for the experts to identify the gene targets to modify without experimental confirmations for each of them, which can take up immeasurable amount of time and resources.
The newly developed iBridge identifies positive and negative metabolites within cells, which exert positive and/or negative impact on formation of the products, by calculating the sum of covariances of their outgoing (consuming) reaction fluxes for a target chemical. Subsequently, it pinpoints "bridge" reactions responsible for converting negative metabolites into positive ones as candidates for overexpression, while identifying the opposites as targets for downregulation.
The research team successfully utilized the iBridge simulation to establish E. coli microbial cell factories each capable of producing three of the compounds that are in high demands at a production capacity that has not been reported around the world. They developed E. coli strains that can each produce panthenol, a moisturizing agent found in many cosmetics, putrescine, which is one of the key components in nylon production, and 4-hydroxyphenyllactic acid, an anti-bacterial food additive. In addition to these three compounds, the study presents predictions for overexpression and suppression genes to construct microbial factories for 298 other industrially valuable compounds.
Dr. Youngjoon Lee, the co-first author of this paper from KAIST, emphasized the accelerated construction of various microbial factories the newly developed simulation enabled. He stated, "With the use of this simulation, multiple microbial cell factories have been established significantly faster than it would have been using the conventional methods. Microbial cell factories producing a wider range of valuable compounds can now be constructed quickly using this technology."
Professor Sang Yup Lee said, "Systems metabolic engineering is a crucial technology for addressing the current climate change issues." He added, "This simulation could significantly expedite the transition from resorting to conventional chemical factories to utilizing environmentally friendly microbial factories."
< Figure. Conceptual diagram of the flow of iBridge simulation >
The team’s work on iBridge is described in a paper titled "Genome-Wide Identification of Overexpression and Downregulation Gene Targets Based on the Sum of Covariances of the Outgoing Reaction Fluxes" written by Dr. Won Jun Kim, and Dr. Youngjoon Lee of the Bioprocess Research Center and Professors Hyun Uk Kim and Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST. The paper was published via peer-review on the 6th of November on “Cell Systems” by Cell Press.
This research was conducted with the support from the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and Development of Platform Technology for the Production of Novel Aromatic Bioplastic using Microbial Cell Factories Project (Project Leader: Research Professor So Young Choi, KAIST) of the Korean Ministry of Science and ICT.
2023.11.09 View 9774