Biorefinery
-
 KAIST presents strategies for environmentally friendly and sustainable polyamides production
- Provides current research trends in bio-based polyamide production
- Research on bio-based polyamides production gains importance for achieving a carbon-neutral society
Global industries focused on carbon neutrality, under the slogan "Net-Zero," are gaining increasing attention. In particular, research on microbial production of polymers, replacing traditional chemical methods with biological approaches, is actively progressing.
Polyamides, represented by nylon, are linear polymers widely used in various industries such as automotive, electronics, textiles, and medical fields. They possess beneficial properties such as high tensile strength, electrical insulation, heat resistance, wear resistance, and biocompatibility. Since the commercialization of nylon in 1938, approximately 7 million tons of polyamides are produced worldwide annually. Considering their broad applications and significance, producing polyamides through bio-based methods holds considerable environmental and industrial importance.
KAIST (President Kwang-Hyung Lee) announced that a research team led by Distinguished Professor Sang Yup Lee, including Dr. Jong An Lee and doctoral candidate Ji Yeon Kim from the Department of Chemical and Biomolecular Engineering, published a paper titled "Current Advancements in Bio-Based Production of Polyamides”. The paper was featured on the cover of the monthly issue of "Trends in Chemistry” by Cell Press.
As part of climate change response technologies, bio-refineries involve using biotechnological and chemical methods to produce industrially important chemicals and biofuels from renewable biomass without relying on fossil resources. Notably, systems metabolic engineering, pioneered by KAIST's Distinguished Professor Sang Yup Lee, is a research field that effectively manipulates microbial metabolic pathways to produce valuable chemicals, forming the core technology for bio-refineries. The research team has successfully developed high-performance strains producing a variety of compounds, including succinic acid, biodegradable plastics, biofuels, and natural products, using systems metabolic engineering tools and strategies.
The research team predicted that if bio-based polyamide production technology, which is widely used in the production of clothing and textiles, becomes widespread, it will attract attention as a future technology that can respond to the climate crisis due to its environment-friendly production technology. In this study, the research team comprehensively reviewed the bio-based polyamide production strategies. They provided insights into the advancements in polyamide monomer production using metabolically engineered microorganisms and highlighted the recent trends in bio-based polyamide advancements utilizing these monomers. Additionally, they reviewed the strategies for synthesizing bio-based polyamides through chemical conversion of natural oils and discussed the biodegradability and recycling of the polyamides. Furthermore, the paper presented the future direction in which metabolic engineering can be applied for the bio-based polyamide production, contributing to environmentally friendly and sustainable society.
Ji Yeon Kim, the co-first author of this paper from KAIST, stated "The importance of utilizing systems metabolic engineering tools and strategies for bio-based polyamides production is becoming increasingly prominent in achieving carbon neutrality."
Professor Sang Yup Lee emphasized, "Amid growing concerns about climate change, the significance of environmentally friendly and sustainable industrial development is greater than ever. Systems metabolic engineering is expected to have a significant impact not only on the chemical industry but also in various fields."
< [Figure 1] A schematic overview of the overall process for polyamides production >
This paper by Dr. Jong An Lee, PhD student Ji Yeon Kim, Dr. Jung Ho Ahn, and Master Yeah-Ji Ahn from the Department of Chemical and Biomolecular Engineering at KAIST was published in the December issue of 'Trends in Chemistry', an authoritative review journal in the field of chemistry published by Cell. It was published on December 7 as the cover paper and featured review.
※ Paper title: Current advancements in the bio-based production of polyamides
※ Author information: Jong An Lee, Ji Yeon Kim, Jung Ho Ahn, Yeah-Ji Ahn, and Sang Yup Lee
This research was conducted with the support from the development of platform technologies of microbial cell factories for the next-generation biorefineries project and C1 gas refinery program by Korean Ministry of Science and ICT.
< [Figure 2] Cover paper of the December issue of Trends in Chemistry >
2023.12.21 View 8186
KAIST presents strategies for environmentally friendly and sustainable polyamides production
- Provides current research trends in bio-based polyamide production
- Research on bio-based polyamides production gains importance for achieving a carbon-neutral society
Global industries focused on carbon neutrality, under the slogan "Net-Zero," are gaining increasing attention. In particular, research on microbial production of polymers, replacing traditional chemical methods with biological approaches, is actively progressing.
Polyamides, represented by nylon, are linear polymers widely used in various industries such as automotive, electronics, textiles, and medical fields. They possess beneficial properties such as high tensile strength, electrical insulation, heat resistance, wear resistance, and biocompatibility. Since the commercialization of nylon in 1938, approximately 7 million tons of polyamides are produced worldwide annually. Considering their broad applications and significance, producing polyamides through bio-based methods holds considerable environmental and industrial importance.
KAIST (President Kwang-Hyung Lee) announced that a research team led by Distinguished Professor Sang Yup Lee, including Dr. Jong An Lee and doctoral candidate Ji Yeon Kim from the Department of Chemical and Biomolecular Engineering, published a paper titled "Current Advancements in Bio-Based Production of Polyamides”. The paper was featured on the cover of the monthly issue of "Trends in Chemistry” by Cell Press.
As part of climate change response technologies, bio-refineries involve using biotechnological and chemical methods to produce industrially important chemicals and biofuels from renewable biomass without relying on fossil resources. Notably, systems metabolic engineering, pioneered by KAIST's Distinguished Professor Sang Yup Lee, is a research field that effectively manipulates microbial metabolic pathways to produce valuable chemicals, forming the core technology for bio-refineries. The research team has successfully developed high-performance strains producing a variety of compounds, including succinic acid, biodegradable plastics, biofuels, and natural products, using systems metabolic engineering tools and strategies.
The research team predicted that if bio-based polyamide production technology, which is widely used in the production of clothing and textiles, becomes widespread, it will attract attention as a future technology that can respond to the climate crisis due to its environment-friendly production technology. In this study, the research team comprehensively reviewed the bio-based polyamide production strategies. They provided insights into the advancements in polyamide monomer production using metabolically engineered microorganisms and highlighted the recent trends in bio-based polyamide advancements utilizing these monomers. Additionally, they reviewed the strategies for synthesizing bio-based polyamides through chemical conversion of natural oils and discussed the biodegradability and recycling of the polyamides. Furthermore, the paper presented the future direction in which metabolic engineering can be applied for the bio-based polyamide production, contributing to environmentally friendly and sustainable society.
Ji Yeon Kim, the co-first author of this paper from KAIST, stated "The importance of utilizing systems metabolic engineering tools and strategies for bio-based polyamides production is becoming increasingly prominent in achieving carbon neutrality."
Professor Sang Yup Lee emphasized, "Amid growing concerns about climate change, the significance of environmentally friendly and sustainable industrial development is greater than ever. Systems metabolic engineering is expected to have a significant impact not only on the chemical industry but also in various fields."
< [Figure 1] A schematic overview of the overall process for polyamides production >
This paper by Dr. Jong An Lee, PhD student Ji Yeon Kim, Dr. Jung Ho Ahn, and Master Yeah-Ji Ahn from the Department of Chemical and Biomolecular Engineering at KAIST was published in the December issue of 'Trends in Chemistry', an authoritative review journal in the field of chemistry published by Cell. It was published on December 7 as the cover paper and featured review.
※ Paper title: Current advancements in the bio-based production of polyamides
※ Author information: Jong An Lee, Ji Yeon Kim, Jung Ho Ahn, Yeah-Ji Ahn, and Sang Yup Lee
This research was conducted with the support from the development of platform technologies of microbial cell factories for the next-generation biorefineries project and C1 gas refinery program by Korean Ministry of Science and ICT.
< [Figure 2] Cover paper of the December issue of Trends in Chemistry >
2023.12.21 View 8186 -
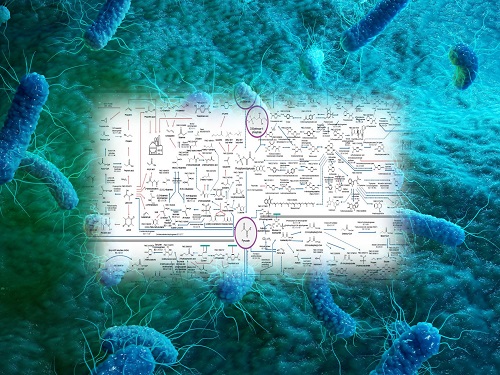 Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 18098
Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 18098 -
 E. coli Engineered to Grow on CO₂ and Formic Acid as Sole Carbon Sources
- An E. coli strain that can grow to a relatively high cell density solely on CO₂ and formic acid was developed by employing metabolic engineering. -
Most biorefinery processes have relied on the use of biomass as a raw material for the production of chemicals and materials. Even though the use of CO₂ as a carbon source in biorefineries is desirable, it has not been possible to make common microbial strains such as E. coli grow on CO₂.
Now, a metabolic engineering research group at KAIST has developed a strategy to grow an E. coli strain to higher cell density solely on CO₂ and formic acid. Formic acid is a one carbon carboxylic acid, and can be easily produced from CO₂ using a variety of methods. Since it is easier to store and transport than CO₂, formic acid can be considered a good liquid-form alternative of CO₂.
With support from the C1 Gas Refinery R&D Center and the Ministry of Science and ICT, a research team led by Distinguished Professor Sang Yup Lee stepped up their work to develop an engineered E. coli strain capable of growing up to 11-fold higher cell density than those previously reported, using CO₂ and formic acid as sole carbon sources. This work was published in Nature Microbiology on September 28.
Despite the recent reports by several research groups on the development of E. coli strains capable of growing on CO₂ and formic acid, the maximum cell growth remained too low (optical density of around 1) and thus the production of chemicals from CO₂ and formic acid has been far from realized.
The team previously reported the reconstruction of the tetrahydrofolate cycle and reverse glycine cleavage pathway to construct an engineered E. coli strain that can sustain growth on CO₂ and formic acid. To further enhance the growth, the research team introduced the previously designed synthetic CO₂ and formic acid assimilation pathway, and two formate dehydrogenases.
Metabolic fluxes were also fine-tuned, the gluconeogenic flux enhanced, and the levels of cytochrome bo3 and bd-I ubiquinol oxidase for ATP generation were optimized. This engineered E. coli strain was able to grow to a relatively high OD600 of 7~11, showing promise as a platform strain growing solely on CO₂ and formic acid.
Professor Lee said, “We engineered E. coli that can grow to a higher cell density only using CO₂ and formic acid. We think that this is an important step forward, but this is not the end. The engineered strain we developed still needs further engineering so that it can grow faster to a much higher density.”
Professor Lee’s team is continuing to develop such a strain. “In the future, we would be delighted to see the production of chemicals from an engineered E. coli strain using CO₂ and formic acid as sole carbon sources,” he added.
-Profile:Distinguished Professor Sang Yup Leehttp://mbel.kaist.ac.krDepartment of Chemical and Biomolecular EngineeringKAIST
2020.09.29 View 13076
E. coli Engineered to Grow on CO₂ and Formic Acid as Sole Carbon Sources
- An E. coli strain that can grow to a relatively high cell density solely on CO₂ and formic acid was developed by employing metabolic engineering. -
Most biorefinery processes have relied on the use of biomass as a raw material for the production of chemicals and materials. Even though the use of CO₂ as a carbon source in biorefineries is desirable, it has not been possible to make common microbial strains such as E. coli grow on CO₂.
Now, a metabolic engineering research group at KAIST has developed a strategy to grow an E. coli strain to higher cell density solely on CO₂ and formic acid. Formic acid is a one carbon carboxylic acid, and can be easily produced from CO₂ using a variety of methods. Since it is easier to store and transport than CO₂, formic acid can be considered a good liquid-form alternative of CO₂.
With support from the C1 Gas Refinery R&D Center and the Ministry of Science and ICT, a research team led by Distinguished Professor Sang Yup Lee stepped up their work to develop an engineered E. coli strain capable of growing up to 11-fold higher cell density than those previously reported, using CO₂ and formic acid as sole carbon sources. This work was published in Nature Microbiology on September 28.
Despite the recent reports by several research groups on the development of E. coli strains capable of growing on CO₂ and formic acid, the maximum cell growth remained too low (optical density of around 1) and thus the production of chemicals from CO₂ and formic acid has been far from realized.
The team previously reported the reconstruction of the tetrahydrofolate cycle and reverse glycine cleavage pathway to construct an engineered E. coli strain that can sustain growth on CO₂ and formic acid. To further enhance the growth, the research team introduced the previously designed synthetic CO₂ and formic acid assimilation pathway, and two formate dehydrogenases.
Metabolic fluxes were also fine-tuned, the gluconeogenic flux enhanced, and the levels of cytochrome bo3 and bd-I ubiquinol oxidase for ATP generation were optimized. This engineered E. coli strain was able to grow to a relatively high OD600 of 7~11, showing promise as a platform strain growing solely on CO₂ and formic acid.
Professor Lee said, “We engineered E. coli that can grow to a higher cell density only using CO₂ and formic acid. We think that this is an important step forward, but this is not the end. The engineered strain we developed still needs further engineering so that it can grow faster to a much higher density.”
Professor Lee’s team is continuing to develop such a strain. “In the future, we would be delighted to see the production of chemicals from an engineered E. coli strain using CO₂ and formic acid as sole carbon sources,” he added.
-Profile:Distinguished Professor Sang Yup Leehttp://mbel.kaist.ac.krDepartment of Chemical and Biomolecular EngineeringKAIST
2020.09.29 View 13076 -
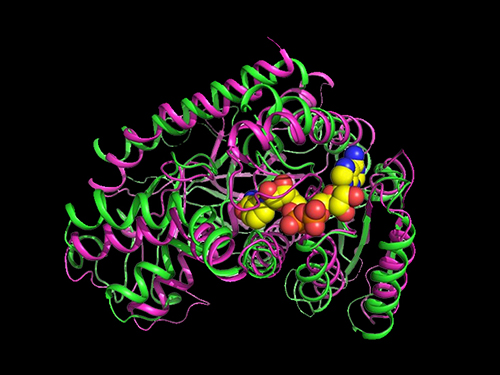 Researchers Present a Microbial Strain Capable of Massive Succinic Acid Production
A research team led by Distinguished Professor Sang Yup Lee reported the production of a microbial strain capable of the massive production of succinic acid with the highest production efficiency to date. This strategy of integrating systems metabolic engineering with enzyme engineering will be useful for the production of industrially competitive bio-based chemicals. Their strategy was described in Nature Communications on April 23.
The bio-based production of industrial chemicals from renewable non-food biomass has become increasingly important as a sustainable substitute for conventional petroleum-based production processes relying on fossil resources. Here, systems metabolic engineering, which is the key component for biorefinery technology, is utilized to effectively engineer the complex metabolic pathways of microorganisms to enable the efficient production of industrial chemicals.
Succinic acid, a four-carbon dicarboxylic acid, is one of the most promising platform chemicals serving as a precursor for industrially important chemicals. Among microorganisms producing succinic acid, Mannheimia succiniciproducens has been proven to be one of the best strains for succinic acid production.
The research team has developed a bio-based succinic acid production technology using the M. succiniciproducens strain isolated from the rumen of Korean cow for over 20 years and succeeded in developing a strain capable of producing succinic acid with the highest production efficiency.
They carried out systems metabolic engineering to optimize the succinic acid production pathway of the M. succiniciproducens strain by determining the crystal structure of key enzymes important for succinic acid production and performing protein engineering to develop enzymes with better catalytic performance.
As a result, 134 g per liter of succinic acid was produced from the fermentation of an engineered strain using glucose, glycerol, and carbon dioxide. They were able to achieve 21 g per liter per hour of succinic acid production, which is one of the key factors determining the economic feasibility of the overall production process. This is the world’s best succinic acid production efficiency reported to date. Previous production methods averaged 1~3 g per liter per hour.
Distinguished professor Sang Yup Lee explained that his team’s work will significantly contribute to transforming the current petrochemical-based industry into an eco-friendly bio-based one.
“Our research on the highly efficient bio-based production of succinic acid from renewable non-food resources and carbon dioxide has provided a basis for reducing our strong dependence on fossil resources, which is the main cause of the environmental crisis,” Professor Lee said.
This work was supported by the Technology Development Program to Solve Climate Changes via Systems Metabolic Engineering for Biorefineries and the C1 Gas Refinery Program from the Ministry of Science and ICT through the National Research Foundation of Korea.
2020.05.06 View 11374
Researchers Present a Microbial Strain Capable of Massive Succinic Acid Production
A research team led by Distinguished Professor Sang Yup Lee reported the production of a microbial strain capable of the massive production of succinic acid with the highest production efficiency to date. This strategy of integrating systems metabolic engineering with enzyme engineering will be useful for the production of industrially competitive bio-based chemicals. Their strategy was described in Nature Communications on April 23.
The bio-based production of industrial chemicals from renewable non-food biomass has become increasingly important as a sustainable substitute for conventional petroleum-based production processes relying on fossil resources. Here, systems metabolic engineering, which is the key component for biorefinery technology, is utilized to effectively engineer the complex metabolic pathways of microorganisms to enable the efficient production of industrial chemicals.
Succinic acid, a four-carbon dicarboxylic acid, is one of the most promising platform chemicals serving as a precursor for industrially important chemicals. Among microorganisms producing succinic acid, Mannheimia succiniciproducens has been proven to be one of the best strains for succinic acid production.
The research team has developed a bio-based succinic acid production technology using the M. succiniciproducens strain isolated from the rumen of Korean cow for over 20 years and succeeded in developing a strain capable of producing succinic acid with the highest production efficiency.
They carried out systems metabolic engineering to optimize the succinic acid production pathway of the M. succiniciproducens strain by determining the crystal structure of key enzymes important for succinic acid production and performing protein engineering to develop enzymes with better catalytic performance.
As a result, 134 g per liter of succinic acid was produced from the fermentation of an engineered strain using glucose, glycerol, and carbon dioxide. They were able to achieve 21 g per liter per hour of succinic acid production, which is one of the key factors determining the economic feasibility of the overall production process. This is the world’s best succinic acid production efficiency reported to date. Previous production methods averaged 1~3 g per liter per hour.
Distinguished professor Sang Yup Lee explained that his team’s work will significantly contribute to transforming the current petrochemical-based industry into an eco-friendly bio-based one.
“Our research on the highly efficient bio-based production of succinic acid from renewable non-food resources and carbon dioxide has provided a basis for reducing our strong dependence on fossil resources, which is the main cause of the environmental crisis,” Professor Lee said.
This work was supported by the Technology Development Program to Solve Climate Changes via Systems Metabolic Engineering for Biorefineries and the C1 Gas Refinery Program from the Ministry of Science and ICT through the National Research Foundation of Korea.
2020.05.06 View 11374 -
 A powerful strategy for developing microbial cell factories by employing synthetic small RNAs
The current systems for the production of chemicals, fuels and materials heavily rely on the use of fossil resources. Due to the increasing concerns on climate change and other environmental problems, however, there has been much interest in developing biorefineries for the production of such chemicals, fuels and materials from renewable resources. For the biorefineries to be competitive with the traditional fossil resource-based refineries, development of high performance microorganisms is the most important as it will affect the overall economics of the process most significantly. Metabolic engineering, which can be defined as purposeful modification of cellular metabolic and regulatory networks with an aim to improve the production of a desired product, has been successfully employed to improve the performance of the cell. However, it is not trivial to engineer the cellular metabolism and regulatory circuits in the cell due to their high complexity.
In metabolic engineering, it is important to find the genes that need to be amplified and attenuated in order to increase the product formation rate while minimizing the production of undesirable byproducts. Gene knock-out experiments are often performed to delete those metabolic fluxes that will consequently result in the increase of the desired product formation. However, gene knock-out experiments require much effort and time to perform, and are difficult to do for a large number of genes. Furthermore, the gene knock-out experiments performed in one strain cannot be transferred to another organism and thus the whole experimental process has to be repeated. This is a big problem in developing a high performance microbial cell factory because it is required to find the best platform strain among many different strains. Therefore, researchers have been eager to develop a strategy that allows rapid identification of multiple genes to be attenuated in multiple strains at the same time.
A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the development of a strategy for efficiently developing microbial cell factories by employing synthetic small RNAs (sRNAs). They first reported the development of such system in Nature Biotechnology last February. This strategy of employing synthetic sRNAs in metabolic engineering has been receiving great interest worldwide as it allows easy, rapid, high-throughput, tunable, and un-doable knock-down of multiple genes in multiple strains at the same time. The research team published a paper online on August 8 as a cover page (September issue) in Nature Protocols, describing the detailed strategy and protocol to employ synthetic sRNAs for metabolic engineering.
In this paper, researchers described the detailed step-by-step protocol for synthetic sRNA-based gene expression control, including the sRNA design principles. Tailor-made synthetic sRNAs can be easily manipulated by using conventional gene cloning method. The use of synthetic sRNAs for gene expression regulation provides several advantages such as portability, conditionality, and tunability in high-throughput experiments. Plasmid-based synthetic sRNA expression system does not leave any scar on the chromosome, and can be easily transferred to many other host strains to be examined. Thus, the construction of libraries and examination of different host strains are much easier than the conventional hard-coded gene manipulation systems. Also, the expression of genes can be conditionally repressed by controlling the production of synthetic sRNAs. Synthetic sRNAs possessing different repression efficiencies make it possible to finely tune the gene expression levels as well. Furthermore, synthetic sRNAs allow knock-down of the expression of essential genes, which was not possible by conventional gene knock-out experiments.
Synthetic sRNAs can be utilized for diverse experiments where gene expression regulation is needed. One of promising applications is high-throughput screening of the target genes to be manipulated and multiple strains simultaneously to enhance the production of chemicals and materials of interest. Such simultaneous optimization of gene targets and strains has been one of the big challenges in metabolic engineering. Another application is to fine tune the expression of the screened genes for flux optimization, which would enhance chemical production further by balancing the flux between biomass formation and target chemical production. Synthetic sRNAs can also be applied to finely regulating genetic interactions in a circuit or network, which is essential in synthetic biology. Once a sRNA scaffold-harboring plasmid is constructed, tailor-made, synthetic sRNAs can be made within 3-4 days, followed by the desired application experiments.
Dr. Eytan Zlotorynski, an editor at Nature Protocols, said "This paper describes the detailed protocol for the design and applications of synthetic sRNA. The method, which has many advantages, is likely to become common practice, and prove useful for metabolic engineering and synthetic biology studies."
This paper published in Nature Protocols will be useful for all researchers in academia and industry who are interested in the use of synthetic sRNAs for fundamental and applied biological and biotechnological studies.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001-2012M1A2A2026556) and the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea.
2013.10.31 View 11410
A powerful strategy for developing microbial cell factories by employing synthetic small RNAs
The current systems for the production of chemicals, fuels and materials heavily rely on the use of fossil resources. Due to the increasing concerns on climate change and other environmental problems, however, there has been much interest in developing biorefineries for the production of such chemicals, fuels and materials from renewable resources. For the biorefineries to be competitive with the traditional fossil resource-based refineries, development of high performance microorganisms is the most important as it will affect the overall economics of the process most significantly. Metabolic engineering, which can be defined as purposeful modification of cellular metabolic and regulatory networks with an aim to improve the production of a desired product, has been successfully employed to improve the performance of the cell. However, it is not trivial to engineer the cellular metabolism and regulatory circuits in the cell due to their high complexity.
In metabolic engineering, it is important to find the genes that need to be amplified and attenuated in order to increase the product formation rate while minimizing the production of undesirable byproducts. Gene knock-out experiments are often performed to delete those metabolic fluxes that will consequently result in the increase of the desired product formation. However, gene knock-out experiments require much effort and time to perform, and are difficult to do for a large number of genes. Furthermore, the gene knock-out experiments performed in one strain cannot be transferred to another organism and thus the whole experimental process has to be repeated. This is a big problem in developing a high performance microbial cell factory because it is required to find the best platform strain among many different strains. Therefore, researchers have been eager to develop a strategy that allows rapid identification of multiple genes to be attenuated in multiple strains at the same time.
A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the development of a strategy for efficiently developing microbial cell factories by employing synthetic small RNAs (sRNAs). They first reported the development of such system in Nature Biotechnology last February. This strategy of employing synthetic sRNAs in metabolic engineering has been receiving great interest worldwide as it allows easy, rapid, high-throughput, tunable, and un-doable knock-down of multiple genes in multiple strains at the same time. The research team published a paper online on August 8 as a cover page (September issue) in Nature Protocols, describing the detailed strategy and protocol to employ synthetic sRNAs for metabolic engineering.
In this paper, researchers described the detailed step-by-step protocol for synthetic sRNA-based gene expression control, including the sRNA design principles. Tailor-made synthetic sRNAs can be easily manipulated by using conventional gene cloning method. The use of synthetic sRNAs for gene expression regulation provides several advantages such as portability, conditionality, and tunability in high-throughput experiments. Plasmid-based synthetic sRNA expression system does not leave any scar on the chromosome, and can be easily transferred to many other host strains to be examined. Thus, the construction of libraries and examination of different host strains are much easier than the conventional hard-coded gene manipulation systems. Also, the expression of genes can be conditionally repressed by controlling the production of synthetic sRNAs. Synthetic sRNAs possessing different repression efficiencies make it possible to finely tune the gene expression levels as well. Furthermore, synthetic sRNAs allow knock-down of the expression of essential genes, which was not possible by conventional gene knock-out experiments.
Synthetic sRNAs can be utilized for diverse experiments where gene expression regulation is needed. One of promising applications is high-throughput screening of the target genes to be manipulated and multiple strains simultaneously to enhance the production of chemicals and materials of interest. Such simultaneous optimization of gene targets and strains has been one of the big challenges in metabolic engineering. Another application is to fine tune the expression of the screened genes for flux optimization, which would enhance chemical production further by balancing the flux between biomass formation and target chemical production. Synthetic sRNAs can also be applied to finely regulating genetic interactions in a circuit or network, which is essential in synthetic biology. Once a sRNA scaffold-harboring plasmid is constructed, tailor-made, synthetic sRNAs can be made within 3-4 days, followed by the desired application experiments.
Dr. Eytan Zlotorynski, an editor at Nature Protocols, said "This paper describes the detailed protocol for the design and applications of synthetic sRNA. The method, which has many advantages, is likely to become common practice, and prove useful for metabolic engineering and synthetic biology studies."
This paper published in Nature Protocols will be useful for all researchers in academia and industry who are interested in the use of synthetic sRNAs for fundamental and applied biological and biotechnological studies.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001-2012M1A2A2026556) and the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea.
2013.10.31 View 11410