Bacteria
-
 KAIST-UIUC researchers develop a treatment platform to disable the ‘biofilm’ shield of superbugs
< (From left) Ph.D. Candidate Joo Hun Lee (co-author), Professor Hyunjoon Kong (co-corresponding author) and Postdoctoral Researcher Yujin Ahn (co-first author) from the Department of Chemical and Biomolecular Engineering of the University of Illinois at Urbana-Champaign and Ju Yeon Chung (co-first author) from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung (co-corresponding author) from the Department of Biological Sciences of KAIST >
A major cause of hospital-acquired infections, the super bacteria Methicillin-resistant Staphylococcus aureus (MRSA), not only exhibits strong resistance to existing antibiotics but also forms a dense biofilm that blocks the effects of external treatments. To meet this challenge, KAIST researchers, in collaboration with an international team, successfully developed a platform that utilizes microbubbles to deliver gene-targeted nanoparticles capable of break ing down the biofilms, offering an innovative solution for treating infections resistant to conventional antibiotics.
KAIST (represented by President Kwang Hyung Lee) announced on May 29 that a research team led by Professor Hyun Jung Chung from the Department of Biological Sciences, in collaboration with Professor Hyunjoon Kong's team at the University of Illinois, has developed a microbubble-based nano-gene delivery platform (BTN MB) that precisely delivers gene suppressors into bacteria to effectively remove biofilms formed by MRSA.
The research team first designed short DNA oligonucleotides that simultaneously suppress three major MRSA genes, related to—biofilm formation (icaA), cell division (ftsZ), and antibiotic resistance (mecA)—and engineered nanoparticles (BTN) to effectively deliver them into the bacteria.
< Figure 1. Effective biofilm treatment using biofilm-targeting nanoparticles controlled by microbubbler system. Schematic illustration of BTN delivery with microbubbles (MB), enabling effective permeation of ASOs targeting bacterial genes within biofilms infecting skin wounds. Gene silencing of targets involved in biofilm formation, bacterial proliferation, and antibiotic resistance leads to effective biofilm removal and antibacterial efficacy in vivo. >
In addition, microbubbles (MB) were used to increase the permeability of the microbial membrane, specifically the biofilm formed by MRSA. By combining these two technologies, the team implemented a dual-strike strategy that fundamentally blocks bacterial growth and prevents resistance acquisition.
This treatment system operates in two stages. First, the MBs induce pressure changes within the bacterial biofilm, allowing the BTNs to penetrate. Then, the BTNs slip through the gaps in the biofilm and enter the bacteria, delivering the gene suppressors precisely. This leads to gene regulation within MRSA, simultaneously blocking biofilm regeneration, cell proliferation, and antibiotic resistance expression.
In experiments conducted in a porcine skin model and a mouse wound model infected with MRSA biofilm, the BTN MB treatment group showed a significant reduction in biofilm thickness, as well as remarkable decreases in bacterial count and inflammatory responses.
< Figure 2. (a) Schematic illustration on the evaluation of treatment efficacy of BTN-MB gene therapy. (b) Reduction in MRSA biofilm mass via simultaneous inhibition of multiple genes. (c, d) Antibacterial efficacy of BTN-MB over time in a porcine skin infection biofilm model. (e) Schematic of the experimental setup to verify antibacterial efficacy in a mouse skin wound infection model. (f) Wound healing effects in mice. (g) Antibacterial effects at the wound site. (h) Histological analysis results. >
These results are difficult to achieve with conventional antibiotic monotherapy and demonstrate the potential for treating a wide range of resistant bacterial infections.
Professor Hyun Jung Chung of KAIST, who led the research, stated, “This study presents a new therapeutic solution that combines nanotechnology, gene suppression, and physical delivery strategies to address superbug infections that existing antibiotics cannot resolve. We will continue our research with the aim of expanding its application to systemic infections and various other infectious diseases.”
< (From left) Ju Yeon Chung from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung from the Department of Biological Sciences >
The study was co-first authored by Ju Yeon Chung, a graduate student in the Department of Biological Sciences at KAIST, and Dr. Yujin Ahn from the University of Illinois. The study was published online on May 19 in the journal, Advanced Functional Materials.
※ Paper Title: Microbubble-Controlled Delivery of Biofilm-Targeting Nanoparticles to Treat MRSA Infection ※ DOI: https://doi.org/10.1002/adfm.202508291
This study was supported by the National Research Foundation and the Ministry of Health and Welfare, Republic of Korea; and the National Science Foundation and National Institutes of Health, USA.
2025.05.29 View 1884
KAIST-UIUC researchers develop a treatment platform to disable the ‘biofilm’ shield of superbugs
< (From left) Ph.D. Candidate Joo Hun Lee (co-author), Professor Hyunjoon Kong (co-corresponding author) and Postdoctoral Researcher Yujin Ahn (co-first author) from the Department of Chemical and Biomolecular Engineering of the University of Illinois at Urbana-Champaign and Ju Yeon Chung (co-first author) from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung (co-corresponding author) from the Department of Biological Sciences of KAIST >
A major cause of hospital-acquired infections, the super bacteria Methicillin-resistant Staphylococcus aureus (MRSA), not only exhibits strong resistance to existing antibiotics but also forms a dense biofilm that blocks the effects of external treatments. To meet this challenge, KAIST researchers, in collaboration with an international team, successfully developed a platform that utilizes microbubbles to deliver gene-targeted nanoparticles capable of break ing down the biofilms, offering an innovative solution for treating infections resistant to conventional antibiotics.
KAIST (represented by President Kwang Hyung Lee) announced on May 29 that a research team led by Professor Hyun Jung Chung from the Department of Biological Sciences, in collaboration with Professor Hyunjoon Kong's team at the University of Illinois, has developed a microbubble-based nano-gene delivery platform (BTN MB) that precisely delivers gene suppressors into bacteria to effectively remove biofilms formed by MRSA.
The research team first designed short DNA oligonucleotides that simultaneously suppress three major MRSA genes, related to—biofilm formation (icaA), cell division (ftsZ), and antibiotic resistance (mecA)—and engineered nanoparticles (BTN) to effectively deliver them into the bacteria.
< Figure 1. Effective biofilm treatment using biofilm-targeting nanoparticles controlled by microbubbler system. Schematic illustration of BTN delivery with microbubbles (MB), enabling effective permeation of ASOs targeting bacterial genes within biofilms infecting skin wounds. Gene silencing of targets involved in biofilm formation, bacterial proliferation, and antibiotic resistance leads to effective biofilm removal and antibacterial efficacy in vivo. >
In addition, microbubbles (MB) were used to increase the permeability of the microbial membrane, specifically the biofilm formed by MRSA. By combining these two technologies, the team implemented a dual-strike strategy that fundamentally blocks bacterial growth and prevents resistance acquisition.
This treatment system operates in two stages. First, the MBs induce pressure changes within the bacterial biofilm, allowing the BTNs to penetrate. Then, the BTNs slip through the gaps in the biofilm and enter the bacteria, delivering the gene suppressors precisely. This leads to gene regulation within MRSA, simultaneously blocking biofilm regeneration, cell proliferation, and antibiotic resistance expression.
In experiments conducted in a porcine skin model and a mouse wound model infected with MRSA biofilm, the BTN MB treatment group showed a significant reduction in biofilm thickness, as well as remarkable decreases in bacterial count and inflammatory responses.
< Figure 2. (a) Schematic illustration on the evaluation of treatment efficacy of BTN-MB gene therapy. (b) Reduction in MRSA biofilm mass via simultaneous inhibition of multiple genes. (c, d) Antibacterial efficacy of BTN-MB over time in a porcine skin infection biofilm model. (e) Schematic of the experimental setup to verify antibacterial efficacy in a mouse skin wound infection model. (f) Wound healing effects in mice. (g) Antibacterial effects at the wound site. (h) Histological analysis results. >
These results are difficult to achieve with conventional antibiotic monotherapy and demonstrate the potential for treating a wide range of resistant bacterial infections.
Professor Hyun Jung Chung of KAIST, who led the research, stated, “This study presents a new therapeutic solution that combines nanotechnology, gene suppression, and physical delivery strategies to address superbug infections that existing antibiotics cannot resolve. We will continue our research with the aim of expanding its application to systemic infections and various other infectious diseases.”
< (From left) Ju Yeon Chung from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung from the Department of Biological Sciences >
The study was co-first authored by Ju Yeon Chung, a graduate student in the Department of Biological Sciences at KAIST, and Dr. Yujin Ahn from the University of Illinois. The study was published online on May 19 in the journal, Advanced Functional Materials.
※ Paper Title: Microbubble-Controlled Delivery of Biofilm-Targeting Nanoparticles to Treat MRSA Infection ※ DOI: https://doi.org/10.1002/adfm.202508291
This study was supported by the National Research Foundation and the Ministry of Health and Welfare, Republic of Korea; and the National Science Foundation and National Institutes of Health, USA.
2025.05.29 View 1884 -
 KAIST Develops Healthcare Device Tracking Chronic Diabetic Wounds
A KAIST research team has developed an effective wireless system that monitors the wound healing process by tracking the spatiotemporal temperature changes and heat transfer characteristics of damaged areas such as diabetic wounds.
On the 5th of March, KAIST (represented by President Kwang Hyung Lee) announced that the research team led by Professor Kyeongha Kwon from KAIST’s School of Electrical Engineering, in association with Chung-Ang University professor Hanjun Ryu, developed digital healthcare technology that tracks the wound healing process in real time, which allows appropriate treatments to be administered.
< Figure 1. Schematic illustrations and diagrams of real-time wound monitoring systems. >
The skin serves as a barrier protecting the body from harmful substances, therefore damage to the skin may cause severe health risks to patients in need of intensive care. Especially in the case of diabetic patients, chronic wounds are easily formed due to complications in normal blood circulation and the wound healing process. In the United States alone, hundreds of billions of dollars of medical costs stem from regenerating the skin from such wounds. While various methods exist to promote wound healing, personalized management is essential depending on the condition of each patient's wounds.
Accordingly, the research team tracked the heating response within the wound by utilizing the differences in temperature between the damaged area and the surrounding healthy skin. They then measured heat transfer characteristics to observe moisture changes near the skin surface, ultimately establishing a basis for understanding the formation process of scar tissue. The team conducted experiments using diabetic mice models regarding the delay in wound healing under pathological conditions, and it was demonstrated that the collected data accurately tracks the wound healing process and the formation of scar tissue.
To minimize the tissue damage that may occur in the process of removing the tracking device after healing, the system integrates biodegradable sensor modules capable of natural decomposition within the body. These biodegradable modules disintegrate within the body after use, thus reducing the risk of additional discomfort or tissue damage upon device removal. Furthermore, the device could one day be used for monitoring inside the wound area as there is no need for removal.
Professor Kyeongha Kwon, who led the research, anticipates that continuous monitoring of wound temperature and heat transfer characteristics will enable medical professionals to more accurately assess the status of diabetic patients' wounds and provide appropriate treatment. He further predicted that the implementation of biodegradable sensors allows for the safe decomposition of the device after wound healing without the need for removal, making live monitoring possible not only in hospitals but also at home.
The research team plans to integrate antimicrobial materials into this device, aiming to expand its technological capabilities to enable the observation and prevention of inflammatory responses, bacterial infections, and other complications. The goal is to provide a multi-purpose wound monitoring platform capable of real-time antimicrobial monitoring in hospitals or homes by detecting changes in temperature and heat transfer characteristics indicative of infection levels.
< Image 1. Image of the bioresorbable temperature sensor >
The results of this study were published on February 19th in the international journal Advanced Healthcare Materials and selected as the inside back cover article, titled "Materials and Device Designs for Wireless Monitoring of Temperature and Thermal Transport Properties of Wound Beds during Healing."
This research was conducted with support from the Basic Research Program, the Regional Innovation Center Program, and the BK21 Program.
2024.03.11 View 7962
KAIST Develops Healthcare Device Tracking Chronic Diabetic Wounds
A KAIST research team has developed an effective wireless system that monitors the wound healing process by tracking the spatiotemporal temperature changes and heat transfer characteristics of damaged areas such as diabetic wounds.
On the 5th of March, KAIST (represented by President Kwang Hyung Lee) announced that the research team led by Professor Kyeongha Kwon from KAIST’s School of Electrical Engineering, in association with Chung-Ang University professor Hanjun Ryu, developed digital healthcare technology that tracks the wound healing process in real time, which allows appropriate treatments to be administered.
< Figure 1. Schematic illustrations and diagrams of real-time wound monitoring systems. >
The skin serves as a barrier protecting the body from harmful substances, therefore damage to the skin may cause severe health risks to patients in need of intensive care. Especially in the case of diabetic patients, chronic wounds are easily formed due to complications in normal blood circulation and the wound healing process. In the United States alone, hundreds of billions of dollars of medical costs stem from regenerating the skin from such wounds. While various methods exist to promote wound healing, personalized management is essential depending on the condition of each patient's wounds.
Accordingly, the research team tracked the heating response within the wound by utilizing the differences in temperature between the damaged area and the surrounding healthy skin. They then measured heat transfer characteristics to observe moisture changes near the skin surface, ultimately establishing a basis for understanding the formation process of scar tissue. The team conducted experiments using diabetic mice models regarding the delay in wound healing under pathological conditions, and it was demonstrated that the collected data accurately tracks the wound healing process and the formation of scar tissue.
To minimize the tissue damage that may occur in the process of removing the tracking device after healing, the system integrates biodegradable sensor modules capable of natural decomposition within the body. These biodegradable modules disintegrate within the body after use, thus reducing the risk of additional discomfort or tissue damage upon device removal. Furthermore, the device could one day be used for monitoring inside the wound area as there is no need for removal.
Professor Kyeongha Kwon, who led the research, anticipates that continuous monitoring of wound temperature and heat transfer characteristics will enable medical professionals to more accurately assess the status of diabetic patients' wounds and provide appropriate treatment. He further predicted that the implementation of biodegradable sensors allows for the safe decomposition of the device after wound healing without the need for removal, making live monitoring possible not only in hospitals but also at home.
The research team plans to integrate antimicrobial materials into this device, aiming to expand its technological capabilities to enable the observation and prevention of inflammatory responses, bacterial infections, and other complications. The goal is to provide a multi-purpose wound monitoring platform capable of real-time antimicrobial monitoring in hospitals or homes by detecting changes in temperature and heat transfer characteristics indicative of infection levels.
< Image 1. Image of the bioresorbable temperature sensor >
The results of this study were published on February 19th in the international journal Advanced Healthcare Materials and selected as the inside back cover article, titled "Materials and Device Designs for Wireless Monitoring of Temperature and Thermal Transport Properties of Wound Beds during Healing."
This research was conducted with support from the Basic Research Program, the Regional Innovation Center Program, and the BK21 Program.
2024.03.11 View 7962 -
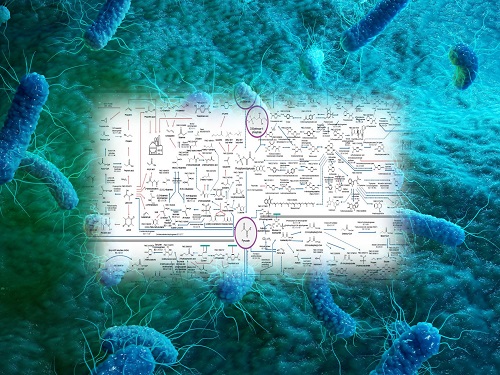 Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 16637
Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 16637 -
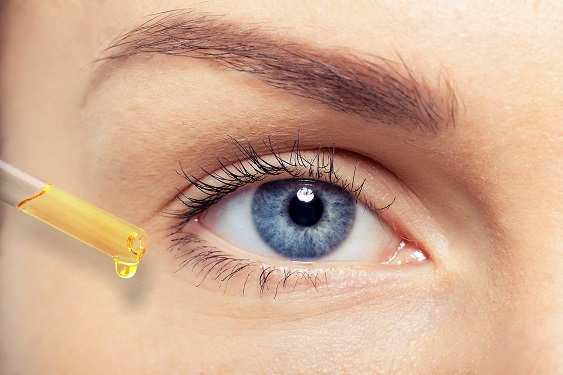 Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 11262
Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 11262 -
 'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media
Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
Their results were made available online on Jan. 18 in Biosensors and Bioelectronics, ahead of publication in the journal’s April issue.
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. “By using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model, we demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures,” said Professor Sungho Jo from the School of Computing.
Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample — the spectral fingerprint — allowing researchers to identify its molecules. The surface-enhanced version places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. “Moreover, strong signals of surrounding media are also enhanced to overwhelm target signals, requiring time-consuming and tedious bacterial separation steps,” said Professor Yeon Sik Jung from the Department of Materials Science and Engineering.
To parse through the noisy signals, the researchers implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such an ability is critical for analyzing one-dimensional spectral data, according to Professor Jo.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that were almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” Professor Jo said. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
The National R&D Program, through a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, supported this research.
-PublicationEojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deepneural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
-ProfileProfessor Yeon Sik JungDepartment of Materials Science and EngineeringKAIST
Professor Sungho JoSchool of ComputingKAIST
2022.03.04 View 23115
'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media
Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
Their results were made available online on Jan. 18 in Biosensors and Bioelectronics, ahead of publication in the journal’s April issue.
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. “By using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model, we demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures,” said Professor Sungho Jo from the School of Computing.
Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample — the spectral fingerprint — allowing researchers to identify its molecules. The surface-enhanced version places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. “Moreover, strong signals of surrounding media are also enhanced to overwhelm target signals, requiring time-consuming and tedious bacterial separation steps,” said Professor Yeon Sik Jung from the Department of Materials Science and Engineering.
To parse through the noisy signals, the researchers implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such an ability is critical for analyzing one-dimensional spectral data, according to Professor Jo.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that were almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” Professor Jo said. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
The National R&D Program, through a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, supported this research.
-PublicationEojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deepneural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
-ProfileProfessor Yeon Sik JungDepartment of Materials Science and EngineeringKAIST
Professor Sungho JoSchool of ComputingKAIST
2022.03.04 View 23115 -
 Natural Rainbow Colorants Microbially Produced
Integrated strategies of systems metabolic engineering and membrane engineering led to the production of natural rainbow colorants comprising seven natural colorants from bacteria for the first time
A research group at KAIST has engineered bacterial strains capable of producing three carotenoids and four violacein derivatives, completing the seven colors in the rainbow spectrum. The research team integrated systems metabolic engineering and membrane engineering strategies for the production of seven natural rainbow colorants in engineered Escherichia coli strains. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Colorants are widely used in our lives and are directly related to human health when we eat food additives and wear cosmetics. However, most of these colorants are made from petroleum, causing unexpected side effects and health problems. Furthermore, they raise environmental concerns such as water pollution from dyeing fabric in the textiles industry. For these reasons, the demand for the production of natural colorants using microorganisms has increased, but could not be readily realized due to the high cost and low yield of the bioprocesses.
These challenges inspired the metabolic engineers at KAIST including researchers Dr. Dongsoo Yang and Dr. Seon Young Park, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team reported the study entitled “Production of rainbow colorants by metabolically engineered Escherichia coli” in Advanced Science online on May 5. It was selected as the journal cover of the July 7 issue.
This research reports for the first time the production of rainbow colorants comprising three carotenoids and four violacein derivatives from glucose or glycerol via systems metabolic engineering and membrane engineering. The research group focused on the production of hydrophobic natural colorants useful for lipophilic food and dyeing garments. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, three carotenoids comprising astaxanthin (red), -carotene (orange), and zeaxanthin (yellow), and four violacein derivatives comprising proviolacein (green), prodeoxyviolacein (blue), violacein (navy), and deoxyviolacein (purple) could be produced. Thus, the production of natural colorants covering the complete rainbow spectrum was achieved.
When hydrophobic colorants are produced from microorganisms, the colorants are accumulated inside the cell. As the accumulation capacity is limited, the hydrophobic colorants could not be produced with concentrations higher than the limit. In this regard, the researchers engineered the cell morphology and generated inner-membrane vesicles (spherical membranous structures) to increase the intracellular capacity for accumulating the natural colorants. To further promote production, the researchers generated outer-membrane vesicles to secrete the natural colorants, thus succeeding in efficiently producing all of seven rainbow colorants. It was even more impressive that the production of natural green and navy colorants was achieved for the first time.
“The production of the seven natural rainbow colorants that can replace the current petroleum-based synthetic colorants was achieved for the first time,” said Dr. Dongsoo Yang. He explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals. “As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Lee.
This work was supported by the "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01550602)" Rural Development Administration, Republic of Korea.
-Publication:Dongsoo Yang, Seon Young Park, and Sang Yup Lee. Production of rainbow colorants by metabolically engineered Escherichia coli. Advanced Science, 2100743.
-Profile Distinguished Professor Sang Yup LeeMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2021.06.09 View 12486
Natural Rainbow Colorants Microbially Produced
Integrated strategies of systems metabolic engineering and membrane engineering led to the production of natural rainbow colorants comprising seven natural colorants from bacteria for the first time
A research group at KAIST has engineered bacterial strains capable of producing three carotenoids and four violacein derivatives, completing the seven colors in the rainbow spectrum. The research team integrated systems metabolic engineering and membrane engineering strategies for the production of seven natural rainbow colorants in engineered Escherichia coli strains. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Colorants are widely used in our lives and are directly related to human health when we eat food additives and wear cosmetics. However, most of these colorants are made from petroleum, causing unexpected side effects and health problems. Furthermore, they raise environmental concerns such as water pollution from dyeing fabric in the textiles industry. For these reasons, the demand for the production of natural colorants using microorganisms has increased, but could not be readily realized due to the high cost and low yield of the bioprocesses.
These challenges inspired the metabolic engineers at KAIST including researchers Dr. Dongsoo Yang and Dr. Seon Young Park, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team reported the study entitled “Production of rainbow colorants by metabolically engineered Escherichia coli” in Advanced Science online on May 5. It was selected as the journal cover of the July 7 issue.
This research reports for the first time the production of rainbow colorants comprising three carotenoids and four violacein derivatives from glucose or glycerol via systems metabolic engineering and membrane engineering. The research group focused on the production of hydrophobic natural colorants useful for lipophilic food and dyeing garments. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, three carotenoids comprising astaxanthin (red), -carotene (orange), and zeaxanthin (yellow), and four violacein derivatives comprising proviolacein (green), prodeoxyviolacein (blue), violacein (navy), and deoxyviolacein (purple) could be produced. Thus, the production of natural colorants covering the complete rainbow spectrum was achieved.
When hydrophobic colorants are produced from microorganisms, the colorants are accumulated inside the cell. As the accumulation capacity is limited, the hydrophobic colorants could not be produced with concentrations higher than the limit. In this regard, the researchers engineered the cell morphology and generated inner-membrane vesicles (spherical membranous structures) to increase the intracellular capacity for accumulating the natural colorants. To further promote production, the researchers generated outer-membrane vesicles to secrete the natural colorants, thus succeeding in efficiently producing all of seven rainbow colorants. It was even more impressive that the production of natural green and navy colorants was achieved for the first time.
“The production of the seven natural rainbow colorants that can replace the current petroleum-based synthetic colorants was achieved for the first time,” said Dr. Dongsoo Yang. He explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals. “As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Lee.
This work was supported by the "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01550602)" Rural Development Administration, Republic of Korea.
-Publication:Dongsoo Yang, Seon Young Park, and Sang Yup Lee. Production of rainbow colorants by metabolically engineered Escherichia coli. Advanced Science, 2100743.
-Profile Distinguished Professor Sang Yup LeeMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2021.06.09 View 12486 -
 New drug targeting method for microbial pathogens developed using in silico cell
A ripple effect is expected on the new antibacterial discovery using “in silico” cells
Featured as a journal cover paper of Molecular BioSystems
A research team of Distinguished Professor Sang Yup Lee at KAIST recently constructed an in silico cell of a microbial pathogen that is resistant to antibiotics and developed a new drug targeting method that could effectively disrupt the pathogen"s growth using the in silico cell.
Hyun Uk Kim, a graduate research assistant at the Department of Chemical and Biomolecular Engineering, KAIST, conducted this study as a part of his thesis research, and the study was featured as a journal cover paper in the February issue of Molecular BioSystems this year, published by The Royal Society of Chemistry based in Europe.
It was relatively easy to treat infectious microbes using antibiotics in the past. However, the overdose of antibiotics has caused pathogens to increase their resistance to various antibiotics, and it has become more difficult to cure infectious diseases these days.
A representative microbial pathogen is Acinetobacter baumannaii. Originally isolated from soils and water, this microorganism did not have resistance to antibiotics, and hence it was easy to eradicate them if infected. However, within a decade, this miroorganism has transformed into a dreadful super-bacterium resistant to antibiotics and caused many casualties among the U.S. and French soldiers who were injured from the recent Iraqi war and infected with Acinetobacter baumannaii.
Professor Lee’s group constructed an in silico cell of this A. baumannii by computationally collecting, integrating, and analyzing the biological information of the bacterium, scattered over various databases and literatures, in order to study this organism"s genomic features and system-wide metabolic characteristics. Furthermore, they employed this in silico cell for integrative approaches, including several network analysis and analysis of essential reactions and metabolites, to predict drug targets that effectively disrupt the pathogen"s growth. Final drug targets are the ones that selectively kill pathogens without harming human body.
Here, essential reactions refer to enzymatic reactions required for normal metabolic functioning in organisms, while essential metabolites indicate chemical compounds required in the metabolism for proper functioning, and their removal brings about the effect of simultaneously disrupting their associated enzymes that interact with them.
This study attempted to predict highly reliable drug targets by systematically scanning biological components, including metabolic genes, enzymatic reactions, that constitute an in silico cell in a short period of time.
This research achievement is highly regarded as it, for the first time, systematically scanned essential metabolites for the effective drug targets using the concept of systems biology, and paved the way for a new antibacterial discovery. This study is also expected to contribute to elucidating the infectious mechanism caused by pathogens.
"Although tons of genomic information is poured in at this moment, application research that efficiently converts this preliminary information into actually useful information is still lagged behind. In this regard, this study is meaningful in that medically useful information is generated from the genomic information of Acinetobacter baumannii," says Professor Lee. "In particular, development of this organism"s in silico cell allows generation of new knowledge regarding essential genes and enzymatic reactions under specific conditions," he added.
This study was supported by the Korean Systems Biology Project of the Ministry of Education, Science and Technology, and the patent for the development of in silico cells of microbial pathogens and drug targeting methods has been filed.
[Picture 1 Cells in silico]
[Picture 2 A process of generating drug targets without harming human body while effectively disrupting the growth of a pathogen, after predicting metabolites from in silico cells]
2010.04.05 View 19470
New drug targeting method for microbial pathogens developed using in silico cell
A ripple effect is expected on the new antibacterial discovery using “in silico” cells
Featured as a journal cover paper of Molecular BioSystems
A research team of Distinguished Professor Sang Yup Lee at KAIST recently constructed an in silico cell of a microbial pathogen that is resistant to antibiotics and developed a new drug targeting method that could effectively disrupt the pathogen"s growth using the in silico cell.
Hyun Uk Kim, a graduate research assistant at the Department of Chemical and Biomolecular Engineering, KAIST, conducted this study as a part of his thesis research, and the study was featured as a journal cover paper in the February issue of Molecular BioSystems this year, published by The Royal Society of Chemistry based in Europe.
It was relatively easy to treat infectious microbes using antibiotics in the past. However, the overdose of antibiotics has caused pathogens to increase their resistance to various antibiotics, and it has become more difficult to cure infectious diseases these days.
A representative microbial pathogen is Acinetobacter baumannaii. Originally isolated from soils and water, this microorganism did not have resistance to antibiotics, and hence it was easy to eradicate them if infected. However, within a decade, this miroorganism has transformed into a dreadful super-bacterium resistant to antibiotics and caused many casualties among the U.S. and French soldiers who were injured from the recent Iraqi war and infected with Acinetobacter baumannaii.
Professor Lee’s group constructed an in silico cell of this A. baumannii by computationally collecting, integrating, and analyzing the biological information of the bacterium, scattered over various databases and literatures, in order to study this organism"s genomic features and system-wide metabolic characteristics. Furthermore, they employed this in silico cell for integrative approaches, including several network analysis and analysis of essential reactions and metabolites, to predict drug targets that effectively disrupt the pathogen"s growth. Final drug targets are the ones that selectively kill pathogens without harming human body.
Here, essential reactions refer to enzymatic reactions required for normal metabolic functioning in organisms, while essential metabolites indicate chemical compounds required in the metabolism for proper functioning, and their removal brings about the effect of simultaneously disrupting their associated enzymes that interact with them.
This study attempted to predict highly reliable drug targets by systematically scanning biological components, including metabolic genes, enzymatic reactions, that constitute an in silico cell in a short period of time.
This research achievement is highly regarded as it, for the first time, systematically scanned essential metabolites for the effective drug targets using the concept of systems biology, and paved the way for a new antibacterial discovery. This study is also expected to contribute to elucidating the infectious mechanism caused by pathogens.
"Although tons of genomic information is poured in at this moment, application research that efficiently converts this preliminary information into actually useful information is still lagged behind. In this regard, this study is meaningful in that medically useful information is generated from the genomic information of Acinetobacter baumannii," says Professor Lee. "In particular, development of this organism"s in silico cell allows generation of new knowledge regarding essential genes and enzymatic reactions under specific conditions," he added.
This study was supported by the Korean Systems Biology Project of the Ministry of Education, Science and Technology, and the patent for the development of in silico cells of microbial pathogens and drug targeting methods has been filed.
[Picture 1 Cells in silico]
[Picture 2 A process of generating drug targets without harming human body while effectively disrupting the growth of a pathogen, after predicting metabolites from in silico cells]
2010.04.05 View 19470 -
 KAIST, GS Caltex Jointly Develop New Bacteria to Produce Biobutanol
KAIST and GS Caltex, Korea"s second-largest refiner, have jointly developed a new strain of bacteria to produce biobutanol, which is regarded as a promising next-generation biofuel, KAIST authorities said on Monday (June 2).
A research team led by Prof. Sang-Yup Lee of the Chemical and Biomolecular Engineering Department and researchers of GS Caltex succeeded in developing an improved strain of bacteria which enables to produce a large amount of biobutanol in the process of fermenting biomass. The research team has applied for international patent for the new technology.
Biomass refers to living and recently dead biological material that can be used as fuel or for industrial production. It usually refers to plant matter grown for use as biofuel, but it also includes plant or animal matter used for production of fibers, chemicals or heat.
In the 1970s and 1980s when scientists began researching the possibilities of alternative fuels, bacteria were used in the process of fermenting biomass. This ABE (acetone, butanol, ethanol) fermentation process yields butanol, acetone, and ethanol in a ratio of 6:3:1, respectively. Acetone produced in this process is not usable.
The newly developed technology to produce biobutanol has an advantage of lowering production cost by eliminating the process to separate acetone from butanol. This has been made possible by improving the bacteria used for the fermentation in metabolic engineering terms, and producing butanol and ethanol only in a ratio of 6:1, while curbing the generation of acetone.
In comparison with bioethanol, also a biofuel mixture which is currently under widespread use in some countries, butanol is more easily transported with gasoline and diesel through pipelines because of its lower tendency to separate from the fuel when contaminated with water. Butanol is also less corrosive than ethanol, another reason its transport through pipeline is preferable.
Global interest in full utilization of biomass and development of other alternative energy including biobutanol has deepened in recent years, as crude oil prices have skyrocketed to record levels and climate changes resulting from the excessive use of fossil fuel have been causing various problems around the world.
2008.06.04 View 14473
KAIST, GS Caltex Jointly Develop New Bacteria to Produce Biobutanol
KAIST and GS Caltex, Korea"s second-largest refiner, have jointly developed a new strain of bacteria to produce biobutanol, which is regarded as a promising next-generation biofuel, KAIST authorities said on Monday (June 2).
A research team led by Prof. Sang-Yup Lee of the Chemical and Biomolecular Engineering Department and researchers of GS Caltex succeeded in developing an improved strain of bacteria which enables to produce a large amount of biobutanol in the process of fermenting biomass. The research team has applied for international patent for the new technology.
Biomass refers to living and recently dead biological material that can be used as fuel or for industrial production. It usually refers to plant matter grown for use as biofuel, but it also includes plant or animal matter used for production of fibers, chemicals or heat.
In the 1970s and 1980s when scientists began researching the possibilities of alternative fuels, bacteria were used in the process of fermenting biomass. This ABE (acetone, butanol, ethanol) fermentation process yields butanol, acetone, and ethanol in a ratio of 6:3:1, respectively. Acetone produced in this process is not usable.
The newly developed technology to produce biobutanol has an advantage of lowering production cost by eliminating the process to separate acetone from butanol. This has been made possible by improving the bacteria used for the fermentation in metabolic engineering terms, and producing butanol and ethanol only in a ratio of 6:1, while curbing the generation of acetone.
In comparison with bioethanol, also a biofuel mixture which is currently under widespread use in some countries, butanol is more easily transported with gasoline and diesel through pipelines because of its lower tendency to separate from the fuel when contaminated with water. Butanol is also less corrosive than ethanol, another reason its transport through pipeline is preferable.
Global interest in full utilization of biomass and development of other alternative energy including biobutanol has deepened in recent years, as crude oil prices have skyrocketed to record levels and climate changes resulting from the excessive use of fossil fuel have been causing various problems around the world.
2008.06.04 View 14473