BIO
-
 KAIST develops technology for selective RNA modification in living cells and animals
· A team led by Professor Won Do Heo from the Department of Biological Sciences, KAIST, has developed a pioneering technology that selectively acetylates specific RNA molecules in living cells and tissues.
· The platform uses RNA-targeting CRISPR tools in combination with RNA-modifying enzymes to chemically modify only the intended RNA.
· The method opens new possibilities for gene therapy by enabling precise control of disease-related RNA without affecting the rest of the transcriptome.
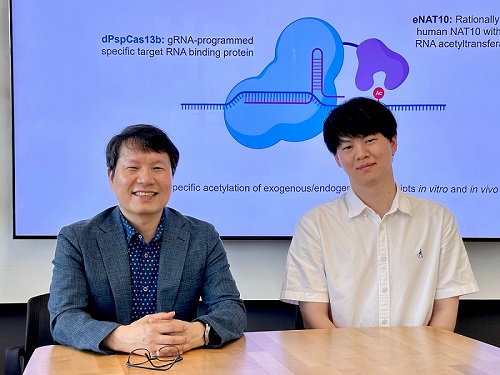
< Photo 1. (From left) Professor Won Do Heo and Jihwan Yu, a Ph.D. Candidate of the Department of Biological Sciences >
CRISPR-Cas13, a powerful RNA-targeting technology is gaining increasing attention as a next-generation gene therapy platform due to its precision and reduced side effects. Utilizing this system, researchers at KAIST have now developed the world’s first technology capable of selectively acetylating (chemically modifying) specific RNA molecules among countless transcripts within living cells. This breakthrough enables precise, programmable control of RNA function and is expected to open new avenues in RNA-based therapeutic development.
KAIST (President Kwang Hyung Lee) announced that a research team led by Professor Won Do Heo in the Department of Biological Sciences has recently developed a groundbreaking technology capable of selectively acetylating specific RNA molecules within the human body using the CRISPR-Cas13 system—an RNA-targeting platform gaining increasing attention in the fields of gene regulation and RNA-based therapeutics.
RNA molecules can undergo chemical modifications—the addition of specific chemical groups—which alter their function and behavior without changing the underlying nucleotide sequence. However, some of these modifications, a critical layer of post-transcriptional gene regulation, remain poorly understood. Among them, N4-acetylcytidine (ac4C) has been particularly enigmatic, with ongoing debate about its existence and function in human messenger RNA (mRNA), the RNA that encodes proteins.
To address this gap, the KAIST research team developed a targeted RNA acetylation system, named dCas13-eNAT10. This platform combines a catalytically inactive Cas13 enzyme (dCas13) that guides the system to specific RNA targets, with a hyperactive variant of the NAT10 enzyme (eNAT10), which performs RNA acetylation. This approach enables precise acetylation of only the desired RNA molecules among the vast pool of transcripts within the cell.
< Figure 1. Development of hyperactive variant eNAT10 through NAT10 protein engineering. By engineering the NAT10 protein, which performs RNA acetylation in human cells, based on its domain and structure, eNAT10 was developed, showing approximately a 3-fold increase in RNA acetylation activity compared to the wild-type enzyme. >
Using this system, the researchers demonstrated that guide RNAs could direct the dCas13-eNAT10 complex to acetylate specific RNA targets, and acetylation significantly increased protein expression from the modified mRNA. Moreover, the study revealed, for the first time, that RNA acetylation plays a role in intracellular RNA localization, facilitating the export of RNA from the nucleus to the cytoplasm—a critical step in gene expression regulation.
To validate its therapeutic potential, the team successfully delivered the targeted RNA acetylation system into the livers of live mice using adeno-associated virus (AAV), a commonly used gene therapy vector. This marks the first demonstration of in vivo RNA modification, extending the applicability of RNA chemical modification tools from cell culture models to living organisms.
< Figure 2. Acetylation of various RNA in cells using dCas13-eNAT10 fusion protein. Utilizing the CRISPR-Cas13 system, which can precisely target specific RNA through guide RNA, a dCas13-eNAT10 fusion protein was created, demonstrating its ability to specifically acetylate various endogenous RNA at different locations within cells. >
Professor Won Do Heo, who previously developed COVID-19 treatment technology using RNA gene scissors and technology to activate RNA gene scissors with light, stated, "Existing RNA chemical modification research faced difficulties in controlling specificity, temporality, and spatiality. However, this new technology allows selective acetylation of desired RNA, opening the door for accurate and detailed research into the functions of RNA acetylation." He added, "The RNA chemical modification technology developed in this study can be widely used as an RNA-based therapeutic agent and a tool for regulating RNA functions in living organisms in the future."
< Figure 3. In vivo delivery of targeted RNA acetylation system. The targeted RNA acetylation system was encoded in an AAV vector, commonly used in gene therapy, and delivered intravenously to adult mice, showing that target RNA in liver tissue was specifically acetylated according to the guide RNA. >
This research, with Ph.D. candidate Jihwan Yu from the Department of Biological Sciences at KAIST as the first author, was published in the journal Nature Chemical Biology on June 2, 2025. (Title: Programmable RNA acetylation with CRISPR-Cas13, Impact factor: 12.9, DOI: https://doi.org/10.1038/s41589-025-01922-3)
This research was supported by the Samsung Future Technology Foundation and the Bio & Medical Technology Development Program of the National Research Foundation of Korea.
2025.06.10 View 1068
KAIST develops technology for selective RNA modification in living cells and animals
· A team led by Professor Won Do Heo from the Department of Biological Sciences, KAIST, has developed a pioneering technology that selectively acetylates specific RNA molecules in living cells and tissues.
· The platform uses RNA-targeting CRISPR tools in combination with RNA-modifying enzymes to chemically modify only the intended RNA.
· The method opens new possibilities for gene therapy by enabling precise control of disease-related RNA without affecting the rest of the transcriptome.
< Photo 1. (From left) Professor Won Do Heo and Jihwan Yu, a Ph.D. Candidate of the Department of Biological Sciences >
CRISPR-Cas13, a powerful RNA-targeting technology is gaining increasing attention as a next-generation gene therapy platform due to its precision and reduced side effects. Utilizing this system, researchers at KAIST have now developed the world’s first technology capable of selectively acetylating (chemically modifying) specific RNA molecules among countless transcripts within living cells. This breakthrough enables precise, programmable control of RNA function and is expected to open new avenues in RNA-based therapeutic development.
KAIST (President Kwang Hyung Lee) announced that a research team led by Professor Won Do Heo in the Department of Biological Sciences has recently developed a groundbreaking technology capable of selectively acetylating specific RNA molecules within the human body using the CRISPR-Cas13 system—an RNA-targeting platform gaining increasing attention in the fields of gene regulation and RNA-based therapeutics.
RNA molecules can undergo chemical modifications—the addition of specific chemical groups—which alter their function and behavior without changing the underlying nucleotide sequence. However, some of these modifications, a critical layer of post-transcriptional gene regulation, remain poorly understood. Among them, N4-acetylcytidine (ac4C) has been particularly enigmatic, with ongoing debate about its existence and function in human messenger RNA (mRNA), the RNA that encodes proteins.
To address this gap, the KAIST research team developed a targeted RNA acetylation system, named dCas13-eNAT10. This platform combines a catalytically inactive Cas13 enzyme (dCas13) that guides the system to specific RNA targets, with a hyperactive variant of the NAT10 enzyme (eNAT10), which performs RNA acetylation. This approach enables precise acetylation of only the desired RNA molecules among the vast pool of transcripts within the cell.
< Figure 1. Development of hyperactive variant eNAT10 through NAT10 protein engineering. By engineering the NAT10 protein, which performs RNA acetylation in human cells, based on its domain and structure, eNAT10 was developed, showing approximately a 3-fold increase in RNA acetylation activity compared to the wild-type enzyme. >
Using this system, the researchers demonstrated that guide RNAs could direct the dCas13-eNAT10 complex to acetylate specific RNA targets, and acetylation significantly increased protein expression from the modified mRNA. Moreover, the study revealed, for the first time, that RNA acetylation plays a role in intracellular RNA localization, facilitating the export of RNA from the nucleus to the cytoplasm—a critical step in gene expression regulation.
To validate its therapeutic potential, the team successfully delivered the targeted RNA acetylation system into the livers of live mice using adeno-associated virus (AAV), a commonly used gene therapy vector. This marks the first demonstration of in vivo RNA modification, extending the applicability of RNA chemical modification tools from cell culture models to living organisms.
< Figure 2. Acetylation of various RNA in cells using dCas13-eNAT10 fusion protein. Utilizing the CRISPR-Cas13 system, which can precisely target specific RNA through guide RNA, a dCas13-eNAT10 fusion protein was created, demonstrating its ability to specifically acetylate various endogenous RNA at different locations within cells. >
Professor Won Do Heo, who previously developed COVID-19 treatment technology using RNA gene scissors and technology to activate RNA gene scissors with light, stated, "Existing RNA chemical modification research faced difficulties in controlling specificity, temporality, and spatiality. However, this new technology allows selective acetylation of desired RNA, opening the door for accurate and detailed research into the functions of RNA acetylation." He added, "The RNA chemical modification technology developed in this study can be widely used as an RNA-based therapeutic agent and a tool for regulating RNA functions in living organisms in the future."
< Figure 3. In vivo delivery of targeted RNA acetylation system. The targeted RNA acetylation system was encoded in an AAV vector, commonly used in gene therapy, and delivered intravenously to adult mice, showing that target RNA in liver tissue was specifically acetylated according to the guide RNA. >
This research, with Ph.D. candidate Jihwan Yu from the Department of Biological Sciences at KAIST as the first author, was published in the journal Nature Chemical Biology on June 2, 2025. (Title: Programmable RNA acetylation with CRISPR-Cas13, Impact factor: 12.9, DOI: https://doi.org/10.1038/s41589-025-01922-3)
This research was supported by the Samsung Future Technology Foundation and the Bio & Medical Technology Development Program of the National Research Foundation of Korea.
2025.06.10 View 1068 -
 KAIST-UIUC researchers develop a treatment platform to disable the ‘biofilm’ shield of superbugs
< (From left) Ph.D. Candidate Joo Hun Lee (co-author), Professor Hyunjoon Kong (co-corresponding author) and Postdoctoral Researcher Yujin Ahn (co-first author) from the Department of Chemical and Biomolecular Engineering of the University of Illinois at Urbana-Champaign and Ju Yeon Chung (co-first author) from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung (co-corresponding author) from the Department of Biological Sciences of KAIST >
A major cause of hospital-acquired infections, the super bacteria Methicillin-resistant Staphylococcus aureus (MRSA), not only exhibits strong resistance to existing antibiotics but also forms a dense biofilm that blocks the effects of external treatments. To meet this challenge, KAIST researchers, in collaboration with an international team, successfully developed a platform that utilizes microbubbles to deliver gene-targeted nanoparticles capable of break ing down the biofilms, offering an innovative solution for treating infections resistant to conventional antibiotics.
KAIST (represented by President Kwang Hyung Lee) announced on May 29 that a research team led by Professor Hyun Jung Chung from the Department of Biological Sciences, in collaboration with Professor Hyunjoon Kong's team at the University of Illinois, has developed a microbubble-based nano-gene delivery platform (BTN MB) that precisely delivers gene suppressors into bacteria to effectively remove biofilms formed by MRSA.
The research team first designed short DNA oligonucleotides that simultaneously suppress three major MRSA genes, related to—biofilm formation (icaA), cell division (ftsZ), and antibiotic resistance (mecA)—and engineered nanoparticles (BTN) to effectively deliver them into the bacteria.
< Figure 1. Effective biofilm treatment using biofilm-targeting nanoparticles controlled by microbubbler system. Schematic illustration of BTN delivery with microbubbles (MB), enabling effective permeation of ASOs targeting bacterial genes within biofilms infecting skin wounds. Gene silencing of targets involved in biofilm formation, bacterial proliferation, and antibiotic resistance leads to effective biofilm removal and antibacterial efficacy in vivo. >
In addition, microbubbles (MB) were used to increase the permeability of the microbial membrane, specifically the biofilm formed by MRSA. By combining these two technologies, the team implemented a dual-strike strategy that fundamentally blocks bacterial growth and prevents resistance acquisition.
This treatment system operates in two stages. First, the MBs induce pressure changes within the bacterial biofilm, allowing the BTNs to penetrate. Then, the BTNs slip through the gaps in the biofilm and enter the bacteria, delivering the gene suppressors precisely. This leads to gene regulation within MRSA, simultaneously blocking biofilm regeneration, cell proliferation, and antibiotic resistance expression.
In experiments conducted in a porcine skin model and a mouse wound model infected with MRSA biofilm, the BTN MB treatment group showed a significant reduction in biofilm thickness, as well as remarkable decreases in bacterial count and inflammatory responses.
< Figure 2. (a) Schematic illustration on the evaluation of treatment efficacy of BTN-MB gene therapy. (b) Reduction in MRSA biofilm mass via simultaneous inhibition of multiple genes. (c, d) Antibacterial efficacy of BTN-MB over time in a porcine skin infection biofilm model. (e) Schematic of the experimental setup to verify antibacterial efficacy in a mouse skin wound infection model. (f) Wound healing effects in mice. (g) Antibacterial effects at the wound site. (h) Histological analysis results. >
These results are difficult to achieve with conventional antibiotic monotherapy and demonstrate the potential for treating a wide range of resistant bacterial infections.
Professor Hyun Jung Chung of KAIST, who led the research, stated, “This study presents a new therapeutic solution that combines nanotechnology, gene suppression, and physical delivery strategies to address superbug infections that existing antibiotics cannot resolve. We will continue our research with the aim of expanding its application to systemic infections and various other infectious diseases.”
< (From left) Ju Yeon Chung from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung from the Department of Biological Sciences >
The study was co-first authored by Ju Yeon Chung, a graduate student in the Department of Biological Sciences at KAIST, and Dr. Yujin Ahn from the University of Illinois. The study was published online on May 19 in the journal, Advanced Functional Materials.
※ Paper Title: Microbubble-Controlled Delivery of Biofilm-Targeting Nanoparticles to Treat MRSA Infection ※ DOI: https://doi.org/10.1002/adfm.202508291
This study was supported by the National Research Foundation and the Ministry of Health and Welfare, Republic of Korea; and the National Science Foundation and National Institutes of Health, USA.
2025.05.29 View 1644
KAIST-UIUC researchers develop a treatment platform to disable the ‘biofilm’ shield of superbugs
< (From left) Ph.D. Candidate Joo Hun Lee (co-author), Professor Hyunjoon Kong (co-corresponding author) and Postdoctoral Researcher Yujin Ahn (co-first author) from the Department of Chemical and Biomolecular Engineering of the University of Illinois at Urbana-Champaign and Ju Yeon Chung (co-first author) from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung (co-corresponding author) from the Department of Biological Sciences of KAIST >
A major cause of hospital-acquired infections, the super bacteria Methicillin-resistant Staphylococcus aureus (MRSA), not only exhibits strong resistance to existing antibiotics but also forms a dense biofilm that blocks the effects of external treatments. To meet this challenge, KAIST researchers, in collaboration with an international team, successfully developed a platform that utilizes microbubbles to deliver gene-targeted nanoparticles capable of break ing down the biofilms, offering an innovative solution for treating infections resistant to conventional antibiotics.
KAIST (represented by President Kwang Hyung Lee) announced on May 29 that a research team led by Professor Hyun Jung Chung from the Department of Biological Sciences, in collaboration with Professor Hyunjoon Kong's team at the University of Illinois, has developed a microbubble-based nano-gene delivery platform (BTN MB) that precisely delivers gene suppressors into bacteria to effectively remove biofilms formed by MRSA.
The research team first designed short DNA oligonucleotides that simultaneously suppress three major MRSA genes, related to—biofilm formation (icaA), cell division (ftsZ), and antibiotic resistance (mecA)—and engineered nanoparticles (BTN) to effectively deliver them into the bacteria.
< Figure 1. Effective biofilm treatment using biofilm-targeting nanoparticles controlled by microbubbler system. Schematic illustration of BTN delivery with microbubbles (MB), enabling effective permeation of ASOs targeting bacterial genes within biofilms infecting skin wounds. Gene silencing of targets involved in biofilm formation, bacterial proliferation, and antibiotic resistance leads to effective biofilm removal and antibacterial efficacy in vivo. >
In addition, microbubbles (MB) were used to increase the permeability of the microbial membrane, specifically the biofilm formed by MRSA. By combining these two technologies, the team implemented a dual-strike strategy that fundamentally blocks bacterial growth and prevents resistance acquisition.
This treatment system operates in two stages. First, the MBs induce pressure changes within the bacterial biofilm, allowing the BTNs to penetrate. Then, the BTNs slip through the gaps in the biofilm and enter the bacteria, delivering the gene suppressors precisely. This leads to gene regulation within MRSA, simultaneously blocking biofilm regeneration, cell proliferation, and antibiotic resistance expression.
In experiments conducted in a porcine skin model and a mouse wound model infected with MRSA biofilm, the BTN MB treatment group showed a significant reduction in biofilm thickness, as well as remarkable decreases in bacterial count and inflammatory responses.
< Figure 2. (a) Schematic illustration on the evaluation of treatment efficacy of BTN-MB gene therapy. (b) Reduction in MRSA biofilm mass via simultaneous inhibition of multiple genes. (c, d) Antibacterial efficacy of BTN-MB over time in a porcine skin infection biofilm model. (e) Schematic of the experimental setup to verify antibacterial efficacy in a mouse skin wound infection model. (f) Wound healing effects in mice. (g) Antibacterial effects at the wound site. (h) Histological analysis results. >
These results are difficult to achieve with conventional antibiotic monotherapy and demonstrate the potential for treating a wide range of resistant bacterial infections.
Professor Hyun Jung Chung of KAIST, who led the research, stated, “This study presents a new therapeutic solution that combines nanotechnology, gene suppression, and physical delivery strategies to address superbug infections that existing antibiotics cannot resolve. We will continue our research with the aim of expanding its application to systemic infections and various other infectious diseases.”
< (From left) Ju Yeon Chung from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung from the Department of Biological Sciences >
The study was co-first authored by Ju Yeon Chung, a graduate student in the Department of Biological Sciences at KAIST, and Dr. Yujin Ahn from the University of Illinois. The study was published online on May 19 in the journal, Advanced Functional Materials.
※ Paper Title: Microbubble-Controlled Delivery of Biofilm-Targeting Nanoparticles to Treat MRSA Infection ※ DOI: https://doi.org/10.1002/adfm.202508291
This study was supported by the National Research Foundation and the Ministry of Health and Welfare, Republic of Korea; and the National Science Foundation and National Institutes of Health, USA.
2025.05.29 View 1644 -
 KAIST to Develop a Korean-style ChatGPT Platform Specifically Geared Toward Medical Diagnosis and Drug Discovery
On May 23rd, KAIST (President Kwang-Hyung Lee) announced that its Digital Bio-Health AI Research Center (Director: Professor JongChul Ye of KAIST Kim Jaechul Graduate School of AI) has been selected for the Ministry of Science and ICT's 'AI Top-Tier Young Researcher Support Program (AI Star Fellowship Project).' With a total investment of ₩11.5 billion from May 2025 to December 2030, the center will embark on the full-scale development of AI technology and a platform capable of independently inferring and determining the kinds of diseases, and discovering new drugs.
< Photo. On May 20th, a kick-off meeting for the AI Star Fellowship Project was held at KAIST Kim Jaechul Graduate School of AI’s Yangjae Research Center with the KAIST research team and participating organizations of Samsung Medical Center, NAVER Cloud, and HITS. [From left to right in the front row] Professor Jaegul Joo (KAIST), Professor Yoonjae Choi (KAIST), Professor Woo Youn Kim (KAIST/HITS), Professor JongChul Ye (KAIST), Professor Sungsoo Ahn (KAIST), Dr. Haanju Yoo (NAVER Cloud), Yoonho Lee (KAIST), HyeYoon Moon (Samsung Medical Center), Dr. Su Min Kim (Samsung Medical Center) >
This project aims to foster an innovative AI research ecosystem centered on young researchers and develop an inferential AI agent that can utilize and automatically expand specialized knowledge systems in the bio and medical fields.
Professor JongChul Ye of the Kim Jaechul Graduate School of AI will serve as the lead researcher, with young researchers from KAIST including Professors Yoonjae Choi, Kimin Lee, Sungsoo Ahn, and Chanyoung Park, along with mid-career researchers like Professors Jaegul Joo and Woo Youn Kim, jointly undertaking the project. They will collaborate with various laboratories within KAIST to conduct comprehensive research covering the entire cycle from the theoretical foundations of AI inference to its practical application.
Specifically, the main goals include: - Building high-performance inference models that integrate diverse medical knowledge systems to enhance the precision and reliability of diagnosis and treatment. - Developing a convergence inference platform that efficiently combines symbol-based inference with neural network models. - Securing AI technology for new drug development and biomarker discovery based on 'cell ontology.'
Furthermore, through close collaboration with industry and medical institutions such as Samsung Medical Center, NAVER Cloud, and HITS Co., Ltd., the project aims to achieve: - Clinical diagnostic AI utilizing medical knowledge systems. - AI-based molecular target exploration for new drug development. - Commercialization of an extendible AI inference platform.
Professor JongChul Ye, Director of KAIST's Digital Bio-Health AI Research Center, stated, "At a time when competition in AI inference model development is intensifying, it is a great honor for KAIST to lead the development of AI technology specialized in the bio and medical fields with world-class young researchers." He added, "We will do our best to ensure that the participating young researchers reach a world-leading level in terms of research achievements after the completion of this seven-year project starting in 2025."
The AI Star Fellowship is a newly established program where post-doctoral researchers and faculty members within seven years of appointment participate as project leaders (PLs) to independently lead research. Multiple laboratories within a university and demand-side companies form a consortium to operate the program.
Through this initiative, KAIST plans to nurture bio-medical convergence AI talent and simultaneously promote the commercialization of core technologies in collaboration with Samsung Medical Center, NAVER Cloud, and HITS.
2025.05.26 View 3053
KAIST to Develop a Korean-style ChatGPT Platform Specifically Geared Toward Medical Diagnosis and Drug Discovery
On May 23rd, KAIST (President Kwang-Hyung Lee) announced that its Digital Bio-Health AI Research Center (Director: Professor JongChul Ye of KAIST Kim Jaechul Graduate School of AI) has been selected for the Ministry of Science and ICT's 'AI Top-Tier Young Researcher Support Program (AI Star Fellowship Project).' With a total investment of ₩11.5 billion from May 2025 to December 2030, the center will embark on the full-scale development of AI technology and a platform capable of independently inferring and determining the kinds of diseases, and discovering new drugs.
< Photo. On May 20th, a kick-off meeting for the AI Star Fellowship Project was held at KAIST Kim Jaechul Graduate School of AI’s Yangjae Research Center with the KAIST research team and participating organizations of Samsung Medical Center, NAVER Cloud, and HITS. [From left to right in the front row] Professor Jaegul Joo (KAIST), Professor Yoonjae Choi (KAIST), Professor Woo Youn Kim (KAIST/HITS), Professor JongChul Ye (KAIST), Professor Sungsoo Ahn (KAIST), Dr. Haanju Yoo (NAVER Cloud), Yoonho Lee (KAIST), HyeYoon Moon (Samsung Medical Center), Dr. Su Min Kim (Samsung Medical Center) >
This project aims to foster an innovative AI research ecosystem centered on young researchers and develop an inferential AI agent that can utilize and automatically expand specialized knowledge systems in the bio and medical fields.
Professor JongChul Ye of the Kim Jaechul Graduate School of AI will serve as the lead researcher, with young researchers from KAIST including Professors Yoonjae Choi, Kimin Lee, Sungsoo Ahn, and Chanyoung Park, along with mid-career researchers like Professors Jaegul Joo and Woo Youn Kim, jointly undertaking the project. They will collaborate with various laboratories within KAIST to conduct comprehensive research covering the entire cycle from the theoretical foundations of AI inference to its practical application.
Specifically, the main goals include: - Building high-performance inference models that integrate diverse medical knowledge systems to enhance the precision and reliability of diagnosis and treatment. - Developing a convergence inference platform that efficiently combines symbol-based inference with neural network models. - Securing AI technology for new drug development and biomarker discovery based on 'cell ontology.'
Furthermore, through close collaboration with industry and medical institutions such as Samsung Medical Center, NAVER Cloud, and HITS Co., Ltd., the project aims to achieve: - Clinical diagnostic AI utilizing medical knowledge systems. - AI-based molecular target exploration for new drug development. - Commercialization of an extendible AI inference platform.
Professor JongChul Ye, Director of KAIST's Digital Bio-Health AI Research Center, stated, "At a time when competition in AI inference model development is intensifying, it is a great honor for KAIST to lead the development of AI technology specialized in the bio and medical fields with world-class young researchers." He added, "We will do our best to ensure that the participating young researchers reach a world-leading level in terms of research achievements after the completion of this seven-year project starting in 2025."
The AI Star Fellowship is a newly established program where post-doctoral researchers and faculty members within seven years of appointment participate as project leaders (PLs) to independently lead research. Multiple laboratories within a university and demand-side companies form a consortium to operate the program.
Through this initiative, KAIST plans to nurture bio-medical convergence AI talent and simultaneously promote the commercialization of core technologies in collaboration with Samsung Medical Center, NAVER Cloud, and HITS.
2025.05.26 View 3053 -
 KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 4964
KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 4964 -
 KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
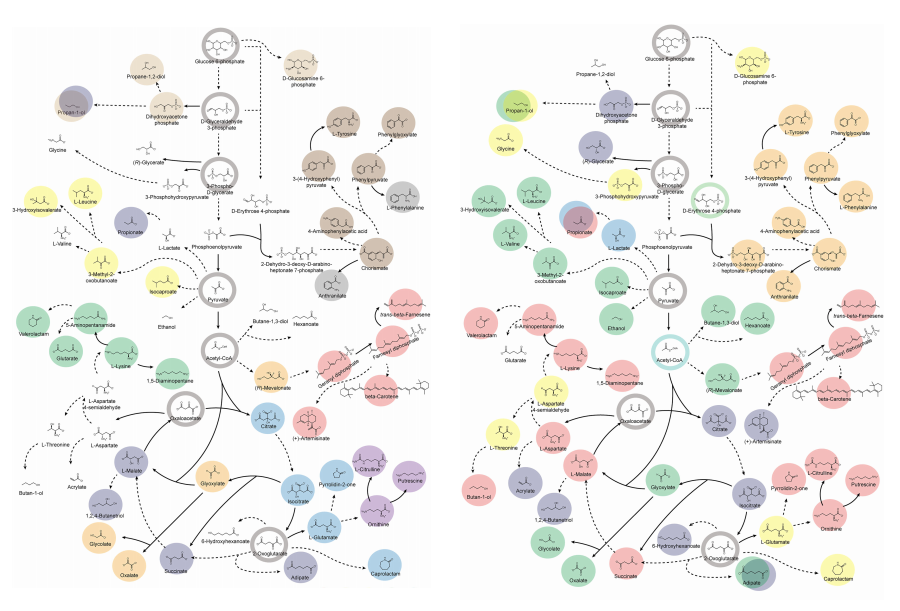
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3617
KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3617 -
 KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
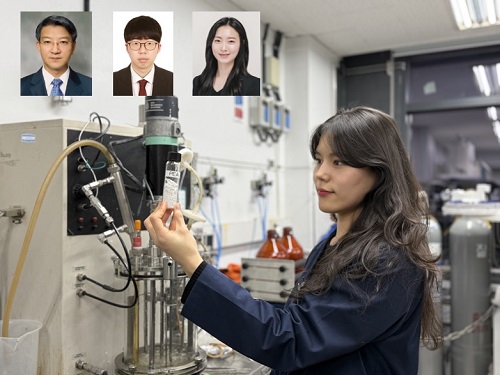
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 4781
KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 4781 -
 KAIST develops a new, bone-like material that strengthens with use in collaboration with GIT
Materials used in apartment buildings, vehicles, and other structures deteriorate over time under repeated loads, leading to failure and breakage. A joint research team from Korea and the United States has successfully developed a bioinspired material that becomes stronger with use, taking inspiration from the way bones synthesize minerals from bodily fluids under stress, increasing bone density.
< (From left) Professor Sung Hoon Kang of the Department of Materials Science and Engineering, Johns Hopkins University Ph.D. candidates Bohan Sun and Grant Kitchen, Professor Yuhang Hu and Ph.D. candidate Dongjung He of Georgia Institute of Technology >
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of February that a research team led by Professor Sung Hoon Kang from the Department of Materials Science and Engineering, in collaboration with Johns Hopkins University and the Georgia Institute of Technology, had developed a new material that strengthens with repeated use, similar to how bones become stronger with exercise.
Professor Kang’s team sought to address the issue of conventional materials degrading with repeated use. Inspired by the biological process where stress triggers cells to form minerals that strengthen bones, the team developed a material that synthesizes minerals under stress without relying on cellular activity. This innovation is expected to enable applications in a variety of fields.
To replace the function of cells, the research team created a porous piezoelectric substrate that converts mechanical force into electricity and actually generates more charge under greater force. They then synthesized a composite material by infusing it with an electrolyte containing mineral components similar to those in blood.
< Figure 1. Schematic diagram of the biomimetic concept based on bone and pitcher plants, the reversible strengthening mechanism, the process of fabricating porous composites, the mechanical property changes with increasing stiffness and energy dissipation after cyclic loading, and the reprogrammable self-folding mechanism and applications >
After subjecting the material to periodic forces and measuring changes in its properties, they observed that its stiffness increased proportionally with the frequency and magnitude of stress and that its energy dissipation capability improved.
The reason for such properties was found to be due to minerals forming inside the porous material under repeated stress, as observed through micro-CT imaging of its internal structure. When subjected to large forces, these minerals fractured and dissipated energy, only to reform under further cyclic stress.
Unlike conventional materials that weaken with repeated use, this new material simultaneously enhances stiffness and impact absorption over time.
< Figure 2. Comparison of the changes in properties of the newly developed new material (LIPPS) with other materials under cyclic loading. (A) Graph showing the relative change rate of energy dissipation after cyclic loading and the relative change rate of elastic modulus upon unloading. LIPPS is in a new area that existing materials have not reached, and shows the characteristics of simultaneous increases in elastic modulus and energy dissipation. (B) Graph comparing the performance of LIPPS with current state-of-the-art mechanically adaptive materials. (Left) The maximum property change rate compared to the baseline after cyclic loading, LIPPS shows much higher changes in elastic modulus, dissipated energy density and ratio, toughness (impact resistance), and stored energy density than the existing adaptive materials. (Right) The absolute value range of the reported properties before and after cyclic loading shows that LIPPS has higher elastic modulus and toughness than the existing adaptive materials. >
Moreover, because its properties improve in proportion to the magnitude and frequency of applied stress, it can self-adjust to achieve mechanical property distributions suitable for different structural applications. It also possesses self-healing capabilities.
Professor Kang stated, "This newly developed material, which strengthens and absorbs impact better with repeated use compared to conventional materials, holds great potential for applications in artificial joints, as well as in aircraft, ships, automobiles, and structural engineering."
This study, with Professor Sung Hoon Kang as the corresponding author, was published in Science Advances (Vol. 11, Issue 6, February).
(Paper title: “A material dynamically enhancing both load-bearing and energy-dissipation capability under cyclic loading”) DOI: 10.1126/sciadv.adt3979
This research was conducted as a joint effort with Johns Hopkins University's Extreme Materials Institute and the Georgia Institute of Technology, supported by the National Research Foundation of Korea’s Brain Pool Plus program.
2025.02.22 View 3201
KAIST develops a new, bone-like material that strengthens with use in collaboration with GIT
Materials used in apartment buildings, vehicles, and other structures deteriorate over time under repeated loads, leading to failure and breakage. A joint research team from Korea and the United States has successfully developed a bioinspired material that becomes stronger with use, taking inspiration from the way bones synthesize minerals from bodily fluids under stress, increasing bone density.
< (From left) Professor Sung Hoon Kang of the Department of Materials Science and Engineering, Johns Hopkins University Ph.D. candidates Bohan Sun and Grant Kitchen, Professor Yuhang Hu and Ph.D. candidate Dongjung He of Georgia Institute of Technology >
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of February that a research team led by Professor Sung Hoon Kang from the Department of Materials Science and Engineering, in collaboration with Johns Hopkins University and the Georgia Institute of Technology, had developed a new material that strengthens with repeated use, similar to how bones become stronger with exercise.
Professor Kang’s team sought to address the issue of conventional materials degrading with repeated use. Inspired by the biological process where stress triggers cells to form minerals that strengthen bones, the team developed a material that synthesizes minerals under stress without relying on cellular activity. This innovation is expected to enable applications in a variety of fields.
To replace the function of cells, the research team created a porous piezoelectric substrate that converts mechanical force into electricity and actually generates more charge under greater force. They then synthesized a composite material by infusing it with an electrolyte containing mineral components similar to those in blood.
< Figure 1. Schematic diagram of the biomimetic concept based on bone and pitcher plants, the reversible strengthening mechanism, the process of fabricating porous composites, the mechanical property changes with increasing stiffness and energy dissipation after cyclic loading, and the reprogrammable self-folding mechanism and applications >
After subjecting the material to periodic forces and measuring changes in its properties, they observed that its stiffness increased proportionally with the frequency and magnitude of stress and that its energy dissipation capability improved.
The reason for such properties was found to be due to minerals forming inside the porous material under repeated stress, as observed through micro-CT imaging of its internal structure. When subjected to large forces, these minerals fractured and dissipated energy, only to reform under further cyclic stress.
Unlike conventional materials that weaken with repeated use, this new material simultaneously enhances stiffness and impact absorption over time.
< Figure 2. Comparison of the changes in properties of the newly developed new material (LIPPS) with other materials under cyclic loading. (A) Graph showing the relative change rate of energy dissipation after cyclic loading and the relative change rate of elastic modulus upon unloading. LIPPS is in a new area that existing materials have not reached, and shows the characteristics of simultaneous increases in elastic modulus and energy dissipation. (B) Graph comparing the performance of LIPPS with current state-of-the-art mechanically adaptive materials. (Left) The maximum property change rate compared to the baseline after cyclic loading, LIPPS shows much higher changes in elastic modulus, dissipated energy density and ratio, toughness (impact resistance), and stored energy density than the existing adaptive materials. (Right) The absolute value range of the reported properties before and after cyclic loading shows that LIPPS has higher elastic modulus and toughness than the existing adaptive materials. >
Moreover, because its properties improve in proportion to the magnitude and frequency of applied stress, it can self-adjust to achieve mechanical property distributions suitable for different structural applications. It also possesses self-healing capabilities.
Professor Kang stated, "This newly developed material, which strengthens and absorbs impact better with repeated use compared to conventional materials, holds great potential for applications in artificial joints, as well as in aircraft, ships, automobiles, and structural engineering."
This study, with Professor Sung Hoon Kang as the corresponding author, was published in Science Advances (Vol. 11, Issue 6, February).
(Paper title: “A material dynamically enhancing both load-bearing and energy-dissipation capability under cyclic loading”) DOI: 10.1126/sciadv.adt3979
This research was conducted as a joint effort with Johns Hopkins University's Extreme Materials Institute and the Georgia Institute of Technology, supported by the National Research Foundation of Korea’s Brain Pool Plus program.
2025.02.22 View 3201 -
 Formosa Group of Taiwan to Establish Bio R&D Center at KAIST Investing 12.5 M USD
KAIST (President Kwang-Hyung Lee) announced on February 17th that it signed an agreement for cooperation in the bio-medical field with Formosa Group, one of the three largest companies in Taiwan.
< Formosa Group Chairman Sandy Wang and KAIST President Kwang-Hyung Lee at the signing ceremony >
Formosa Group Executive Committee member and Chairman Sandy Wang, who leads the group's bio and eco-friendly energy sectors, decided to establish a bio-medical research center within KAIST and invest approximately KRW 18 billion or more over 5 years. In addition, to commercialize the research results, KAIST and Formosa Group will establish a joint venture in Korea with KAIST Holdings, a KAIST-funded company.
The cooperation between the two organizations began in early 2023 when KAIST signed a comprehensive exchange and cooperation agreement (MOU) with Ming Chi University of Science and Technology (明志科技大學), Chang Gung University (長庚大學), and Chang Gung Memorial Hospital (長庚記念醫院), which are established and supported by Formosa Group. Afterwards, Chairman Sandy Wang visited KAIST in May 2024 and signed a more specific business agreement (MOA).
KAIST Holdings is a holding company established by KAIST, a government-funded organization, to attract investment and conduct business, and will pursue the establishment of a joint venture with a 50:50 equity structure in cooperation with Formosa Group. KAIST Holdings will invest KAIST’s intellectual property rights, and Formosa Group will invest a corresponding amount of funds.
The KAIST-Formosa joint venture will provide research funds to the KAIST-Formosa Bio-Medical Research Center to be established in the future, secure the right to implement the intellectual property rights generated, and promote full-scale business.
The KAIST-Formosa Bio-Medical Research Center will establish a ‘brain organoid bank’ created by obtaining tissues from hundreds of patients with degenerative brain diseases, thereby securing high-dimensional data that will reveal the fundamental causes of aging and disease. It is expected that KAIST’s world-class artificial intelligence technology will analyze large-scale patient data to find the causes of aging and disease.
Through this business, it is expected that by 2030, five years from now, it will discover more than 10 types of intractable brain disease treatments and expand to more than 20 businesses, including human cell-centered diagnostics and preclinical businesses, and secure infrastructure and intellectual property rights that can create value worth approximately KRW 250 billion.
The Chang Gung Memorial Hospital in Taiwan has 10,000 beds and handles 35,000 patients per day, and systematically accumulates patient tissue and clinical data. Chang Gung Memorial Hospital will differentiate the tissues of patients with degenerative brain diseases and send them to the KAIST-Formosa Bio-Medical Research Center, which will then produce brain organoids to be used for disease research and new drug development. This will allow the world’s largest patient tissue data bank to be established.
Dean Daesoo Kim of the College of Life Science and Bioengineering at KAIST said, “This collaboration between KAIST and Formosa Group is a new research collaboration model that goes beyond joint research to establish a joint venture and global commercialization of developed technologies, and it is significant in that it can serve as an opportunity to promote biomedical research and development.”
With this agreement, KAIST, which has been promoting the KAIST Advanced Regenerative Medicine Engineering Center in Osong K-Bio Square, has secured a practical global partner.
< Representatives of the Formosa Group and KAIST >
KAIST’s Senior Vice President for Planning and Budget, Professor Kyung-Soo Kim emphasized, “KAIST has made great efforts to secure an edge in state-of-the-art biomedical fields such as stem cells and gene editing technology, by attracting the world’s best experts and discovering global cooperation partners, and these results can ultimately be linked to the Osong K-Bio Square project.”
SVP Kim then predicted, “In particular, the practical cooperation with Taiwan’s best Formosa Chang Gung Memorial Hospital, which has abundant clinical experience in stem cell treatment, will be an important axis of KAIST’s bio innovation strategy.”
Formosa Chairman Sandy Wang emphasized that this investment and cooperation is built on trust in KAIST’s R&D capabilities and the passion of its researchers. And added that through this, the Formosa Group will practice corporate social responsibility and take an important first step together with KAIST to protect the welfare and health of humanity. She also went on the say that she expects to see the cooperation expanded to various fields such as mobility and semiconductors based on the successes begotten from the cooperation in the bio field.
KAIST President Kwang-Hyung Lee said, “I evaluate this agreement as one of the most important events that will spearhead KAIST into overseas biotechnology stages,” and added, “I expect that this cooperation will be an opportunity for Taiwan and Korea, both of which have IT industry-centered structures, to create new growth engines in the bio industry.” Meanwhile, Formosa Group is a company founded by Chairman Sandy Wang’s father, Chairman Yung-Ching Wang. It is the world’s No. 1 plastic PVC producer and is leading core industries of the Taiwanese economy, including semiconductors, steel, heavy industry, bio, and batteries. Chairman Yung-Ching Wang was respected by the Taiwanese people for his exemplary return of wealth to society under the belief that the companies and assets he founded “belong to the people.”
2025.02.17 View 3863
Formosa Group of Taiwan to Establish Bio R&D Center at KAIST Investing 12.5 M USD
KAIST (President Kwang-Hyung Lee) announced on February 17th that it signed an agreement for cooperation in the bio-medical field with Formosa Group, one of the three largest companies in Taiwan.
< Formosa Group Chairman Sandy Wang and KAIST President Kwang-Hyung Lee at the signing ceremony >
Formosa Group Executive Committee member and Chairman Sandy Wang, who leads the group's bio and eco-friendly energy sectors, decided to establish a bio-medical research center within KAIST and invest approximately KRW 18 billion or more over 5 years. In addition, to commercialize the research results, KAIST and Formosa Group will establish a joint venture in Korea with KAIST Holdings, a KAIST-funded company.
The cooperation between the two organizations began in early 2023 when KAIST signed a comprehensive exchange and cooperation agreement (MOU) with Ming Chi University of Science and Technology (明志科技大學), Chang Gung University (長庚大學), and Chang Gung Memorial Hospital (長庚記念醫院), which are established and supported by Formosa Group. Afterwards, Chairman Sandy Wang visited KAIST in May 2024 and signed a more specific business agreement (MOA).
KAIST Holdings is a holding company established by KAIST, a government-funded organization, to attract investment and conduct business, and will pursue the establishment of a joint venture with a 50:50 equity structure in cooperation with Formosa Group. KAIST Holdings will invest KAIST’s intellectual property rights, and Formosa Group will invest a corresponding amount of funds.
The KAIST-Formosa joint venture will provide research funds to the KAIST-Formosa Bio-Medical Research Center to be established in the future, secure the right to implement the intellectual property rights generated, and promote full-scale business.
The KAIST-Formosa Bio-Medical Research Center will establish a ‘brain organoid bank’ created by obtaining tissues from hundreds of patients with degenerative brain diseases, thereby securing high-dimensional data that will reveal the fundamental causes of aging and disease. It is expected that KAIST’s world-class artificial intelligence technology will analyze large-scale patient data to find the causes of aging and disease.
Through this business, it is expected that by 2030, five years from now, it will discover more than 10 types of intractable brain disease treatments and expand to more than 20 businesses, including human cell-centered diagnostics and preclinical businesses, and secure infrastructure and intellectual property rights that can create value worth approximately KRW 250 billion.
The Chang Gung Memorial Hospital in Taiwan has 10,000 beds and handles 35,000 patients per day, and systematically accumulates patient tissue and clinical data. Chang Gung Memorial Hospital will differentiate the tissues of patients with degenerative brain diseases and send them to the KAIST-Formosa Bio-Medical Research Center, which will then produce brain organoids to be used for disease research and new drug development. This will allow the world’s largest patient tissue data bank to be established.
Dean Daesoo Kim of the College of Life Science and Bioengineering at KAIST said, “This collaboration between KAIST and Formosa Group is a new research collaboration model that goes beyond joint research to establish a joint venture and global commercialization of developed technologies, and it is significant in that it can serve as an opportunity to promote biomedical research and development.”
With this agreement, KAIST, which has been promoting the KAIST Advanced Regenerative Medicine Engineering Center in Osong K-Bio Square, has secured a practical global partner.
< Representatives of the Formosa Group and KAIST >
KAIST’s Senior Vice President for Planning and Budget, Professor Kyung-Soo Kim emphasized, “KAIST has made great efforts to secure an edge in state-of-the-art biomedical fields such as stem cells and gene editing technology, by attracting the world’s best experts and discovering global cooperation partners, and these results can ultimately be linked to the Osong K-Bio Square project.”
SVP Kim then predicted, “In particular, the practical cooperation with Taiwan’s best Formosa Chang Gung Memorial Hospital, which has abundant clinical experience in stem cell treatment, will be an important axis of KAIST’s bio innovation strategy.”
Formosa Chairman Sandy Wang emphasized that this investment and cooperation is built on trust in KAIST’s R&D capabilities and the passion of its researchers. And added that through this, the Formosa Group will practice corporate social responsibility and take an important first step together with KAIST to protect the welfare and health of humanity. She also went on the say that she expects to see the cooperation expanded to various fields such as mobility and semiconductors based on the successes begotten from the cooperation in the bio field.
KAIST President Kwang-Hyung Lee said, “I evaluate this agreement as one of the most important events that will spearhead KAIST into overseas biotechnology stages,” and added, “I expect that this cooperation will be an opportunity for Taiwan and Korea, both of which have IT industry-centered structures, to create new growth engines in the bio industry.” Meanwhile, Formosa Group is a company founded by Chairman Sandy Wang’s father, Chairman Yung-Ching Wang. It is the world’s No. 1 plastic PVC producer and is leading core industries of the Taiwanese economy, including semiconductors, steel, heavy industry, bio, and batteries. Chairman Yung-Ching Wang was respected by the Taiwanese people for his exemplary return of wealth to society under the belief that the companies and assets he founded “belong to the people.”
2025.02.17 View 3863 -
 KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 26441
KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 26441 -
 KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences>
A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for developing innovative therapeutic technologies.
On the 22nd of January, KAIST (represented by President Kwang Hyung Lee) announced that the joint research team led by Professors Seyun Kim, Gwangrog Lee, and Won-Ki Cho from the Department of Biological Sciences had uncovered essential mechanisms controlling gene expression in animal cells.
Inositol phosphate metabolites produced by inositol metabolism enzymes serve as vital secondary messengers in eukaryotic cell signaling systems and are broadly implicated in cancer, obesity, diabetes, and neurological disorders.
The research team demonstrated that the inositol polyphosphate multikinase (IPMK) enzyme, a key player in the inositol metabolism system, acts as a critical transcriptional activator within the core gene expression networks of animal cells. Notably, although IPMK was previously reported to play an important role in the transcription process governed by serum response factor (SRF), a representative transcription factor in animal cells, the precise mechanism of its action was unclear.
SRF is a transcription factor directly controlling the expression of at least 200–300 genes, regulating cell growth, proliferation, apoptosis, and motility, and is indispensable for organ development, such as in the heart.
The team discovered that IPMK binds directly to SRF, altering the three-dimensional structure of the SRF protein. This interaction facilitates the transcriptional activity of various genes through the SRF activated by IPMK, demonstrating that IPMK acts as a critical regulatory switch to enhance SRF's protein activity.
< Figure 1. The serum response factor (SRF) protein, a key transcription factor in animal cells, directly binds to inositol polyphosphate multikinase (IPMK) enzyme and undergoes structural change to acquire DNA binding ability, and precisely regulates growth and differentiation of animal cells through transcriptional activation. >
The team further verified that disruptions in the direct interaction between IPMK and SRF lead to the reduced functionality and activity of SRF, causing severe impairments in gene expression.
By highlighting the significance of the intrinsically disordered region (IDR) in SRF, the researchers underscored the biological importance of intrinsically disordered proteins (IDPs). Unlike most proteins that adopt distinct structures through folding, IDPs, including those with IDRs, do not exhibit specific structures but play crucial biological roles, attracting significant attention in the scientific community.
Professor Seyun Kim commented, "This study provides a vital mechanism proving that IPMK, a key enzyme in the inositol metabolism system, is a major transcriptional activator in the core gene expression network of animal cells. By understanding fundamental processes such as cancer development and metastasis, tissue differentiation from stem cells, and neural activation through SRF, we hope this discovery will lead to the broad application of innovative therapeutic technologies."
The findings were published on January 7th in the international journal Nucleic Acids Research (IF=16.7, top 1.8% in Biochemistry and Molecular Biology), under the title “Single-molecule analysis reveals that IPMK enhances the DNA-binding activity of the transcription factor SRF" (DOI: 10.1093/nar/gkae1281).
This research was supported by the National Research Foundation of Korea's Mid-career Research Program, Leading Research Center Program, and Global Research Laboratory Program, as well as by the Suh Kyungbae Science Foundation and the Samsung Future Technology Development Program.
2025.01.24 View 9897
KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences>
A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for developing innovative therapeutic technologies.
On the 22nd of January, KAIST (represented by President Kwang Hyung Lee) announced that the joint research team led by Professors Seyun Kim, Gwangrog Lee, and Won-Ki Cho from the Department of Biological Sciences had uncovered essential mechanisms controlling gene expression in animal cells.
Inositol phosphate metabolites produced by inositol metabolism enzymes serve as vital secondary messengers in eukaryotic cell signaling systems and are broadly implicated in cancer, obesity, diabetes, and neurological disorders.
The research team demonstrated that the inositol polyphosphate multikinase (IPMK) enzyme, a key player in the inositol metabolism system, acts as a critical transcriptional activator within the core gene expression networks of animal cells. Notably, although IPMK was previously reported to play an important role in the transcription process governed by serum response factor (SRF), a representative transcription factor in animal cells, the precise mechanism of its action was unclear.
SRF is a transcription factor directly controlling the expression of at least 200–300 genes, regulating cell growth, proliferation, apoptosis, and motility, and is indispensable for organ development, such as in the heart.
The team discovered that IPMK binds directly to SRF, altering the three-dimensional structure of the SRF protein. This interaction facilitates the transcriptional activity of various genes through the SRF activated by IPMK, demonstrating that IPMK acts as a critical regulatory switch to enhance SRF's protein activity.
< Figure 1. The serum response factor (SRF) protein, a key transcription factor in animal cells, directly binds to inositol polyphosphate multikinase (IPMK) enzyme and undergoes structural change to acquire DNA binding ability, and precisely regulates growth and differentiation of animal cells through transcriptional activation. >
The team further verified that disruptions in the direct interaction between IPMK and SRF lead to the reduced functionality and activity of SRF, causing severe impairments in gene expression.
By highlighting the significance of the intrinsically disordered region (IDR) in SRF, the researchers underscored the biological importance of intrinsically disordered proteins (IDPs). Unlike most proteins that adopt distinct structures through folding, IDPs, including those with IDRs, do not exhibit specific structures but play crucial biological roles, attracting significant attention in the scientific community.
Professor Seyun Kim commented, "This study provides a vital mechanism proving that IPMK, a key enzyme in the inositol metabolism system, is a major transcriptional activator in the core gene expression network of animal cells. By understanding fundamental processes such as cancer development and metastasis, tissue differentiation from stem cells, and neural activation through SRF, we hope this discovery will lead to the broad application of innovative therapeutic technologies."
The findings were published on January 7th in the international journal Nucleic Acids Research (IF=16.7, top 1.8% in Biochemistry and Molecular Biology), under the title “Single-molecule analysis reveals that IPMK enhances the DNA-binding activity of the transcription factor SRF" (DOI: 10.1093/nar/gkae1281).
This research was supported by the National Research Foundation of Korea's Mid-career Research Program, Leading Research Center Program, and Global Research Laboratory Program, as well as by the Suh Kyungbae Science Foundation and the Samsung Future Technology Development Program.
2025.01.24 View 9897 -
 A Way for Smartwatches to Detect Depression Risks Devised by KAIST and U of Michigan Researchers
- A international joint research team of KAIST and the University of Michigan developed a digital biomarker for predicting symptoms of depression based on data collected by smartwatches
- It has the potential to be used as a medical technology to replace the economically burdensome fMRI measurement test
- It is expected to expand the scope of digital health data analysis
The CORONA virus pandemic also brought about a pandemic of mental illness. Approximately one billion people worldwide suffer from various psychiatric conditions. Korea is one of more serious cases, with approximately 1.8 million patients exhibiting depression and anxiety disorders, and the total number of patients with clinical mental diseases has increased by 37% in five years to approximately 4.65 million. A joint research team from Korea and the US has developed a technology that uses biometric data collected through wearable devices to predict tomorrow's mood and, further, to predict the possibility of developing symptoms of depression.
< Figure 1. Schematic diagram of the research results. Based on the biometric data collected by a smartwatch, a mathematical algorithm that solves the inverse problem to estimate the brain's circadian phase and sleep stages has been developed. This algorithm can estimate the degrees of circadian disruption, and these estimates can be used as the digital biomarkers to predict depression risks. >
KAIST (President Kwang Hyung Lee) announced on the 15th of January that the research team under Professor Dae Wook Kim from the Department of Brain and Cognitive Sciences and the team under Professor Daniel B. Forger from the Department of Mathematics at the University of Michigan in the United States have developed a technology to predict symptoms of depression such as sleep disorders, depression, loss of appetite, overeating, and decreased concentration in shift workers from the activity and heart rate data collected from smartwatches.
According to WHO, a promising new treatment direction for mental illness focuses on the sleep and circadian timekeeping system located in the hypothalamus of the brain, which directly affect impulsivity, emotional responses, decision-making, and overall mood.
However, in order to measure endogenous circadian rhythms and sleep states, blood or saliva must be drawn every 30 minutes throughout the night to measure changes in the concentration of the melatonin hormone in our bodies and polysomnography (PSG) must be performed. As such treatments requires hospitalization and most psychiatric patients only visit for outpatient treatment, there has been no significant progress in developing treatment methods that take these two factors into account. In addition, the cost of the PSG test, which is approximately $1000, leaves mental health treatment considering sleep and circadian rhythms out of reach for the socially disadvantaged.
The solution to overcome these problems is to employ wearable devices for the easier collection of biometric data such as heart rate, body temperature, and activity level in real time without spatial constraints. However, current wearable devices have the limitation of providing only indirect information on biomarkers required by medical staff, such as the phase of the circadian clock.
The joint research team developed a filtering technology that accurately estimates the phase of the circadian clock, which changes daily, such as heart rate and activity time series data collected from a smartwatch. This is an implementation of a digital twin that precisely describes the circadian rhythm in the brain, and it can be used to estimate circadian rhythm disruption.
< Figure 2. The suprachiasmatic nucleus located in the hypothalamus of the brain is the central biological clock that regulates the 24-hour physiological rhythm and plays a key role in maintaining the body’s circadian rhythm. If the phase of this biological clock is disrupted, it affects various parts of the brain, which can cause psychiatric conditions such as depression. >
The possibility of using the digital twin of this circadian clock to predict the symptoms of depression was verified through collaboration with the research team of Professor Srijan Sen of the Michigan Neuroscience Institute and Professor Amy Bohnert of the Department of Psychiatry of the University of Michigan.
The collaborative research team conducted a large-scale prospective cohort study involving approximately 800 shift workers and showed that the circadian rhythm disruption digital biomarker estimated through the technology can predict tomorrow's mood as well as six symptoms, including sleep problems, appetite changes, decreased concentration, and suicidal thoughts, which are representative symptoms of depression.
< Figure 3. The circadian rhythm of hormones such as melatonin regulates various physiological functions and behaviors such as heart rate and activity level. These physiological and behavioral signals can be measured in daily life through wearable devices. In order to estimate the body’s circadian rhythm inversely based on the measured biometric signals, a mathematical algorithm is needed. This algorithm plays a key role in accurately identifying the characteristics of circadian rhythms by extracting hidden physiological patterns from biosignals. >
Professor Dae Wook Kim said, "It is very meaningful to be able to conduct research that provides a clue for ways to apply wearable biometric data using mathematics that have not previously been utilized for actual disease management." He added, "We expect that this research will be able to present continuous and non-invasive mental health monitoring technology. This is expected to present a new paradigm for mental health care. By resolving some of the major problems socially disadvantaged people may face in current treatment practices, they may be able to take more active steps when experiencing symptoms of depression, such as seeking counsel before things get out of hand."
< Figure 4. A mathematical algorithm was devised to circumvent the problems of estimating the phase of the brain's biological clock and sleep stages inversely from the biodata collected by a smartwatch. This algorithm can estimate the degree of daily circadian rhythm disruption, and this estimate can be used as a digital biomarker to predict depression symptoms. >
The results of this study, in which Professor Dae Wook Kim of the Department of Brain and Cognitive Sciences at KAIST participated as the joint first author and corresponding author, were published in the online version of the international academic journal npj Digital Medicine on December 5, 2024. (Paper title: The real-world association between digital markers of circadian disruption and mental health risks) DOI: 10.1038/s41746-024-01348-6
This study was conducted with the support of the KAIST's Research Support Program for New Faculty Members, the US National Science Foundation, the US National Institutes of Health, and the US Army Research Institute MURI Program.
2025.01.20 View 7231
A Way for Smartwatches to Detect Depression Risks Devised by KAIST and U of Michigan Researchers
- A international joint research team of KAIST and the University of Michigan developed a digital biomarker for predicting symptoms of depression based on data collected by smartwatches
- It has the potential to be used as a medical technology to replace the economically burdensome fMRI measurement test
- It is expected to expand the scope of digital health data analysis
The CORONA virus pandemic also brought about a pandemic of mental illness. Approximately one billion people worldwide suffer from various psychiatric conditions. Korea is one of more serious cases, with approximately 1.8 million patients exhibiting depression and anxiety disorders, and the total number of patients with clinical mental diseases has increased by 37% in five years to approximately 4.65 million. A joint research team from Korea and the US has developed a technology that uses biometric data collected through wearable devices to predict tomorrow's mood and, further, to predict the possibility of developing symptoms of depression.
< Figure 1. Schematic diagram of the research results. Based on the biometric data collected by a smartwatch, a mathematical algorithm that solves the inverse problem to estimate the brain's circadian phase and sleep stages has been developed. This algorithm can estimate the degrees of circadian disruption, and these estimates can be used as the digital biomarkers to predict depression risks. >
KAIST (President Kwang Hyung Lee) announced on the 15th of January that the research team under Professor Dae Wook Kim from the Department of Brain and Cognitive Sciences and the team under Professor Daniel B. Forger from the Department of Mathematics at the University of Michigan in the United States have developed a technology to predict symptoms of depression such as sleep disorders, depression, loss of appetite, overeating, and decreased concentration in shift workers from the activity and heart rate data collected from smartwatches.
According to WHO, a promising new treatment direction for mental illness focuses on the sleep and circadian timekeeping system located in the hypothalamus of the brain, which directly affect impulsivity, emotional responses, decision-making, and overall mood.
However, in order to measure endogenous circadian rhythms and sleep states, blood or saliva must be drawn every 30 minutes throughout the night to measure changes in the concentration of the melatonin hormone in our bodies and polysomnography (PSG) must be performed. As such treatments requires hospitalization and most psychiatric patients only visit for outpatient treatment, there has been no significant progress in developing treatment methods that take these two factors into account. In addition, the cost of the PSG test, which is approximately $1000, leaves mental health treatment considering sleep and circadian rhythms out of reach for the socially disadvantaged.
The solution to overcome these problems is to employ wearable devices for the easier collection of biometric data such as heart rate, body temperature, and activity level in real time without spatial constraints. However, current wearable devices have the limitation of providing only indirect information on biomarkers required by medical staff, such as the phase of the circadian clock.
The joint research team developed a filtering technology that accurately estimates the phase of the circadian clock, which changes daily, such as heart rate and activity time series data collected from a smartwatch. This is an implementation of a digital twin that precisely describes the circadian rhythm in the brain, and it can be used to estimate circadian rhythm disruption.
< Figure 2. The suprachiasmatic nucleus located in the hypothalamus of the brain is the central biological clock that regulates the 24-hour physiological rhythm and plays a key role in maintaining the body’s circadian rhythm. If the phase of this biological clock is disrupted, it affects various parts of the brain, which can cause psychiatric conditions such as depression. >
The possibility of using the digital twin of this circadian clock to predict the symptoms of depression was verified through collaboration with the research team of Professor Srijan Sen of the Michigan Neuroscience Institute and Professor Amy Bohnert of the Department of Psychiatry of the University of Michigan.
The collaborative research team conducted a large-scale prospective cohort study involving approximately 800 shift workers and showed that the circadian rhythm disruption digital biomarker estimated through the technology can predict tomorrow's mood as well as six symptoms, including sleep problems, appetite changes, decreased concentration, and suicidal thoughts, which are representative symptoms of depression.
< Figure 3. The circadian rhythm of hormones such as melatonin regulates various physiological functions and behaviors such as heart rate and activity level. These physiological and behavioral signals can be measured in daily life through wearable devices. In order to estimate the body’s circadian rhythm inversely based on the measured biometric signals, a mathematical algorithm is needed. This algorithm plays a key role in accurately identifying the characteristics of circadian rhythms by extracting hidden physiological patterns from biosignals. >
Professor Dae Wook Kim said, "It is very meaningful to be able to conduct research that provides a clue for ways to apply wearable biometric data using mathematics that have not previously been utilized for actual disease management." He added, "We expect that this research will be able to present continuous and non-invasive mental health monitoring technology. This is expected to present a new paradigm for mental health care. By resolving some of the major problems socially disadvantaged people may face in current treatment practices, they may be able to take more active steps when experiencing symptoms of depression, such as seeking counsel before things get out of hand."
< Figure 4. A mathematical algorithm was devised to circumvent the problems of estimating the phase of the brain's biological clock and sleep stages inversely from the biodata collected by a smartwatch. This algorithm can estimate the degree of daily circadian rhythm disruption, and this estimate can be used as a digital biomarker to predict depression symptoms. >
The results of this study, in which Professor Dae Wook Kim of the Department of Brain and Cognitive Sciences at KAIST participated as the joint first author and corresponding author, were published in the online version of the international academic journal npj Digital Medicine on December 5, 2024. (Paper title: The real-world association between digital markers of circadian disruption and mental health risks) DOI: 10.1038/s41746-024-01348-6
This study was conducted with the support of the KAIST's Research Support Program for New Faculty Members, the US National Science Foundation, the US National Institutes of Health, and the US Army Research Institute MURI Program.
2025.01.20 View 7231 -
 KAIST Develops Insect-Eye-Inspired Camera Capturing 9,120 Frames Per Second
< (From left) Bio and Brain Engineering PhD Student Jae-Myeong Kwon, Professor Ki-Hun Jeong, PhD Student Hyun-Kyung Kim, PhD Student Young-Gil Cha, and Professor Min H. Kim of the School of Computing >
The compound eyes of insects can detect fast-moving objects in parallel and, in low-light conditions, enhance sensitivity by integrating signals over time to determine motion. Inspired by these biological mechanisms, KAIST researchers have successfully developed a low-cost, high-speed camera that overcomes the limitations of frame rate and sensitivity faced by conventional high-speed cameras.
KAIST (represented by President Kwang Hyung Lee) announced on the 16th of January that a research team led by Professors Ki-Hun Jeong (Department of Bio and Brain Engineering) and Min H. Kim (School of Computing) has developed a novel bio-inspired camera capable of ultra-high-speed imaging with high sensitivity by mimicking the visual structure of insect eyes.
High-quality imaging under high-speed and low-light conditions is a critical challenge in many applications. While conventional high-speed cameras excel in capturing fast motion, their sensitivity decreases as frame rates increase because the time available to collect light is reduced.
To address this issue, the research team adopted an approach similar to insect vision, utilizing multiple optical channels and temporal summation. Unlike traditional monocular camera systems, the bio-inspired camera employs a compound-eye-like structure that allows for the parallel acquisition of frames from different time intervals.
< Figure 1. (A) Vision in a fast-eyed insect. Reflected light from swiftly moving objects sequentially stimulates the photoreceptors along the individual optical channels called ommatidia, of which the visual signals are separately and parallelly processed via the lamina and medulla. Each neural response is temporally summed to enhance the visual signals. The parallel processing and temporal summation allow fast and low-light imaging in dim light. (B) High-speed and high-sensitivity microlens array camera (HS-MAC). A rolling shutter image sensor is utilized to simultaneously acquire multiple frames by channel division, and temporal summation is performed in parallel to realize high speed and sensitivity even in a low-light environment. In addition, the frame components of a single fragmented array image are stitched into a single blurred frame, which is subsequently deblurred by compressive image reconstruction. >
During this process, light is accumulated over overlapping time periods for each frame, increasing the signal-to-noise ratio. The researchers demonstrated that their bio-inspired camera could capture objects up to 40 times dimmer than those detectable by conventional high-speed cameras.
The team also introduced a "channel-splitting" technique to significantly enhance the camera's speed, achieving frame rates thousands of times faster than those supported by the image sensors used in packaging. Additionally, a "compressed image restoration" algorithm was employed to eliminate blur caused by frame integration and reconstruct sharp images.
The resulting bio-inspired camera is less than one millimeter thick and extremely compact, capable of capturing 9,120 frames per second while providing clear images in low-light conditions.
< Figure 2. A high-speed, high-sensitivity biomimetic camera packaged in an image sensor. It is made small enough to fit on a finger, with a thickness of less than 1 mm. >
The research team plans to extend this technology to develop advanced image processing algorithms for 3D imaging and super-resolution imaging, aiming for applications in biomedical imaging, mobile devices, and various other camera technologies.
Hyun-Kyung Kim, a doctoral student in the Department of Bio and Brain Engineering at KAIST and the study's first author, stated, “We have experimentally validated that the insect-eye-inspired camera delivers outstanding performance in high-speed and low-light imaging despite its small size. This camera opens up possibilities for diverse applications in portable camera systems, security surveillance, and medical imaging.”
< Figure 3. Rotating plate and flame captured using the high-speed, high-sensitivity biomimetic camera. The rotating plate at 1,950 rpm was accurately captured at 9,120 fps. In addition, the pinch-off of the flame with a faint intensity of 880 µlux was accurately captured at 1,020 fps. >
This research was published in the international journal Science Advances in January 2025 (Paper Title: “Biologically-inspired microlens array camera for high-speed and high-sensitivity imaging”).
DOI: https://doi.org/10.1126/sciadv.ads3389
This study was supported by the Korea Research Institute for Defense Technology Planning and Advancement (KRIT) of the Defense Acquisition Program Administration (DAPA), the Ministry of Science and ICT, and the Ministry of Trade, Industry and Energy (MOTIE).
2025.01.16 View 7288
KAIST Develops Insect-Eye-Inspired Camera Capturing 9,120 Frames Per Second
< (From left) Bio and Brain Engineering PhD Student Jae-Myeong Kwon, Professor Ki-Hun Jeong, PhD Student Hyun-Kyung Kim, PhD Student Young-Gil Cha, and Professor Min H. Kim of the School of Computing >
The compound eyes of insects can detect fast-moving objects in parallel and, in low-light conditions, enhance sensitivity by integrating signals over time to determine motion. Inspired by these biological mechanisms, KAIST researchers have successfully developed a low-cost, high-speed camera that overcomes the limitations of frame rate and sensitivity faced by conventional high-speed cameras.
KAIST (represented by President Kwang Hyung Lee) announced on the 16th of January that a research team led by Professors Ki-Hun Jeong (Department of Bio and Brain Engineering) and Min H. Kim (School of Computing) has developed a novel bio-inspired camera capable of ultra-high-speed imaging with high sensitivity by mimicking the visual structure of insect eyes.
High-quality imaging under high-speed and low-light conditions is a critical challenge in many applications. While conventional high-speed cameras excel in capturing fast motion, their sensitivity decreases as frame rates increase because the time available to collect light is reduced.
To address this issue, the research team adopted an approach similar to insect vision, utilizing multiple optical channels and temporal summation. Unlike traditional monocular camera systems, the bio-inspired camera employs a compound-eye-like structure that allows for the parallel acquisition of frames from different time intervals.
< Figure 1. (A) Vision in a fast-eyed insect. Reflected light from swiftly moving objects sequentially stimulates the photoreceptors along the individual optical channels called ommatidia, of which the visual signals are separately and parallelly processed via the lamina and medulla. Each neural response is temporally summed to enhance the visual signals. The parallel processing and temporal summation allow fast and low-light imaging in dim light. (B) High-speed and high-sensitivity microlens array camera (HS-MAC). A rolling shutter image sensor is utilized to simultaneously acquire multiple frames by channel division, and temporal summation is performed in parallel to realize high speed and sensitivity even in a low-light environment. In addition, the frame components of a single fragmented array image are stitched into a single blurred frame, which is subsequently deblurred by compressive image reconstruction. >
During this process, light is accumulated over overlapping time periods for each frame, increasing the signal-to-noise ratio. The researchers demonstrated that their bio-inspired camera could capture objects up to 40 times dimmer than those detectable by conventional high-speed cameras.
The team also introduced a "channel-splitting" technique to significantly enhance the camera's speed, achieving frame rates thousands of times faster than those supported by the image sensors used in packaging. Additionally, a "compressed image restoration" algorithm was employed to eliminate blur caused by frame integration and reconstruct sharp images.
The resulting bio-inspired camera is less than one millimeter thick and extremely compact, capable of capturing 9,120 frames per second while providing clear images in low-light conditions.
< Figure 2. A high-speed, high-sensitivity biomimetic camera packaged in an image sensor. It is made small enough to fit on a finger, with a thickness of less than 1 mm. >
The research team plans to extend this technology to develop advanced image processing algorithms for 3D imaging and super-resolution imaging, aiming for applications in biomedical imaging, mobile devices, and various other camera technologies.
Hyun-Kyung Kim, a doctoral student in the Department of Bio and Brain Engineering at KAIST and the study's first author, stated, “We have experimentally validated that the insect-eye-inspired camera delivers outstanding performance in high-speed and low-light imaging despite its small size. This camera opens up possibilities for diverse applications in portable camera systems, security surveillance, and medical imaging.”
< Figure 3. Rotating plate and flame captured using the high-speed, high-sensitivity biomimetic camera. The rotating plate at 1,950 rpm was accurately captured at 9,120 fps. In addition, the pinch-off of the flame with a faint intensity of 880 µlux was accurately captured at 1,020 fps. >
This research was published in the international journal Science Advances in January 2025 (Paper Title: “Biologically-inspired microlens array camera for high-speed and high-sensitivity imaging”).
DOI: https://doi.org/10.1126/sciadv.ads3389
This study was supported by the Korea Research Institute for Defense Technology Planning and Advancement (KRIT) of the Defense Acquisition Program Administration (DAPA), the Ministry of Science and ICT, and the Ministry of Trade, Industry and Energy (MOTIE).
2025.01.16 View 7288