research
Researchers have found a potential link between non-inherited somatic mutations in the brain and the progression of Alzheimer’s disease
Researchers have identified somatic mutations in the brain that could contribute to the development of Alzheimer’s disease (AD). Their findings were published in the journal Nature Communications last week.
Decades worth of research has identified inherited mutations that lead to early-onset familial AD. Inherited mutations, however, are behind at most half the cases of late onset sporadic AD, in which there is no family history of the disease. But the genetic factors causing the other half of these sporadic cases have been unclear.
Professor Jeong Ho Lee at the Graduate School of Medical Science and Engineering and colleagues analysed the DNA present in post-mortem hippocampal formations and in blood samples from people aged 70 to 96 with AD and age-matched controls. They specifically looked for non-inherited somatic mutations in their brains using high-depth whole exome sequencing.
The team developed a bioinformatics pipeline that enabled them to detect low-level brain somatic single nucleotide variations (SNVs) – mutations that involve the substitution of a single nucleotide with another nucleotide. Brain somatic SNVs have been reported on and accumulate throughout our lives and can sometimes be associated with a range of neurological diseases.
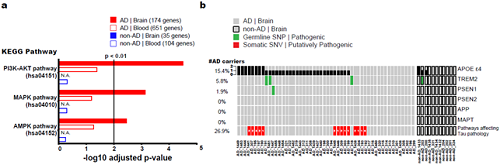
The number of somatic SNVs did not differ between individuals with AD and non-demented controls. Interestingly, somatic SNVs in AD brains arise about 4.8 times more slowly than in blood. When the team performed gene-set enrichment tests, 26.9 percent of the AD brain samples had pathogenic brain somatic SNVs known to be linked to hyperphosphorylation of tau proteins, which is one of major hallmarks of AD.
Then, they pinpointed a pathogenic SNV in the PIN1 gene, a cis/trans isomerase that balances phosphorylation in tau proteins, found in one AD patient’s brain. They found the mutation was 4.9 time more abundant in AT8-positive – a marker for hyper-phosphorylated tau proteins– neurons in the entorhinal cortex than the bulk hippocampal tissue. Furthermore, in a series of functional assays, they observed the mutation causing a loss of function in PIN1 and such haploinsufficiency increased the phosphorylation and aggregation of tau proteins.
“Our study provides new insights into the molecular genetic factors behind Alzheimer’s disease and other neurodegenerative diseases potentially linked to somatic mutations in the brain,” said Professor Lee.
The team is planning to expand their study to a larger cohort in order to establish stronger links between these brain somatic mutations and the pathogenesis of Alzheimer’s disease.

< 20190719023938_78765_1841401221.jpg >
< 20190719023954_91863_87911845.png >
(Figure 2. Pathogenic brain somatic mutations associated with tau phosphorylation are significantly enriched in AD brains.)

< 20190719024006_74119_1572542400.jpg >
(Figure 3. A pathogenic brain somatic mutation in PIN1 (c. 477 C>T) is a loss-of-function and related functional assays show its haploinsufficiency increases phosphorylation and aggregation of tau.)
-
research “One Experiment Is All It Takes”: KAIST Team Revolutionizes Drug Interaction Testing, Replacing 60,000 Studies
A groundbreaking new method developed by researchers at KAIST and Chungnam National University could drastically streamline drug interaction testing — replacing dozens of traditional experiments with just one. The research, led by Professor Jae Kyoung Kim of KAIST Department of Mathematical Sciences & IBS Biomedical Mathematics Group and Professor Sang Kyum Kim of Chungnam National University's College of Pharmacy, introduces a novel analysis technique called 50-BOA, published in Natu
2025-06-16 -
research KAIST Develops Virtual Staining Technology for 3D Histopathology
Moving beyond traditional methods of observing thinly sliced and stained cancer tissues, a collaborative international research team led by KAIST has successfully developed a groundbreaking technology. This innovation uses advanced optical techniques combined with an artificial intelligence-based deep learning algorithm to create realistic, virtually stained 3D images of cancer tissue without the need for serial sectioning nor staining. This breakthrough is anticipated to pave the way for next-g
2025-05-26 -
research No More Touch Issues on Rainy Days! KAIST Develops Human-Like Tactile Sensor
Recent advancements in robotics have enabled machines to handle delicate objects like eggs with precision, thanks to highly integrated pressure sensors that provide detailed tactile feedback. However, even the most advanced robots struggle to accurately detect pressure in complex environments involving water, bending, or electromagnetic interference. A research team at KAIST has successfully developed a pressure sensor that operates stably without external interference, even on wet surfaces li
2025-03-14 -
research KAIST Research Team Develops an AI Framework Capable of Overcoming the Strength-Ductility Dilemma in Additive-manufactured Titanium Alloys
<(From Left) Ph.D. Student Jaejung Park and Professor Seungchul Lee of KAIST Department of Mechanical Engineering and , Professor Hyoung Seop Kim of POSTECH, and M.S.–Ph.D. Integrated Program Student Jeong Ah Lee of POSTECH. > The KAIST research team led by Professor Seungchul Lee from Department of Mechanical Engineering, in collaboration with Professor Hyoung Seop Kim’s team at POSTECH, successfully overcame the strength–ductility dilemma of Ti 6Al 4V alloy using a
2025-02-21 -
research KAIST Develops Stretchable Displays Featuring 25% Expansion Without Image Distortion
Stretchable displays, praised for their spatial efficiency, design flexibility, and human-like flexibility, are seen as the next generation of display technology. A team of Korean researchers has developed a stretchable display that can expand by 25% while maintaining clear image quality without distortion. It can also stretch and contract up to 5,000 times at 15% expansion without any performance degradation, making it the first deformation-free stretchable display with a negative Poisson's rat
2024-09-20