research
Bacteria are intimately involved in our daily lives. These microorganisms have been used in human history for food such as cheese, yogurt, and wine, In more recent years, through metabolic engineering, microorganisms been used extensively as microbial cell factories to manufacture plastics, feed for livestock, dietary supplements, and drugs. However, in addition to these bacteria that are beneficial to human lives, pathogens such as Pneumonia, Salmonella, and Staphylococcus that cause various infectious diseases are also ubiquitously present. It is important to be able to metabolically control these beneficial industrial bacteria for high value-added chemicals production and to manipulate harmful pathogens to suppress its pathogenic traits.
KAIST (President Kwang Hyung Lee) announced on the 10th that a research team led by Distinguished Professor Sang Yup Lee of the Department of Biochemical Engineering has developed a new sRNA tool that can effectively inhibit target genes in various bacteria, including both Gram-negative and Gram-positive bacteria. The research results were published online on April 24 in Nature Communications.
※ Thesis title: Targeted and high-throughput gene knockdown in diverse bacteria using synthetic sRNAs
※ Author information : Jae Sung Cho (co-1st), Dongsoo Yang (co-1st), Cindy Pricilia Surya Prabowo (co-author), Mohammad Rifqi Ghiffary (co-author), Taehee Han (co-author), Kyeong Rok Choi (co-author), Cheon Woo Moon (co-author), Hengrui Zhou (co-author), Jae Yong Ryu (co-author), Hyun Uk Kim (co-author) and Sang Yup Lee (corresponding author).
sRNA is an effective tool for synthesizing and regulating target genes in E. coli, but it has been difficult to apply to industrially useful Gram-positive bacteria such as Bacillus subtilis and Corynebacterium in addition to Gram-negative bacteria such as E. coli.
To address this issue, a research team led by Distinguished Professor Lee Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST developed a new sRNA platform that can effectively suppress target genes in various bacteria, including both Gram-negative and positive bacteria. The research team surveyed thousands of microbial-derived sRNA systems in the microbial database, and eventually designated the sRNA system derived from 'Bacillus subtilis' that showed the highest gene knockdown efficiency, and designated it as “Broad-Host-Range sRNA”, or BHR-sRNA.
A similar well-known system is the CRISPR interference (CRISPRi) system, which is a modified CRISPR system that knocks down gene expression by suppressing the gene transcription process. However, the Cas9 protein in the CRISPRi system has a very high molecular weight, and there have been reports growth inhibition in bacteria. The BHR-sRNA system developed in this study did not affect bacterial growth while showing similar gene knockdown efficiencies to CRISPRi.
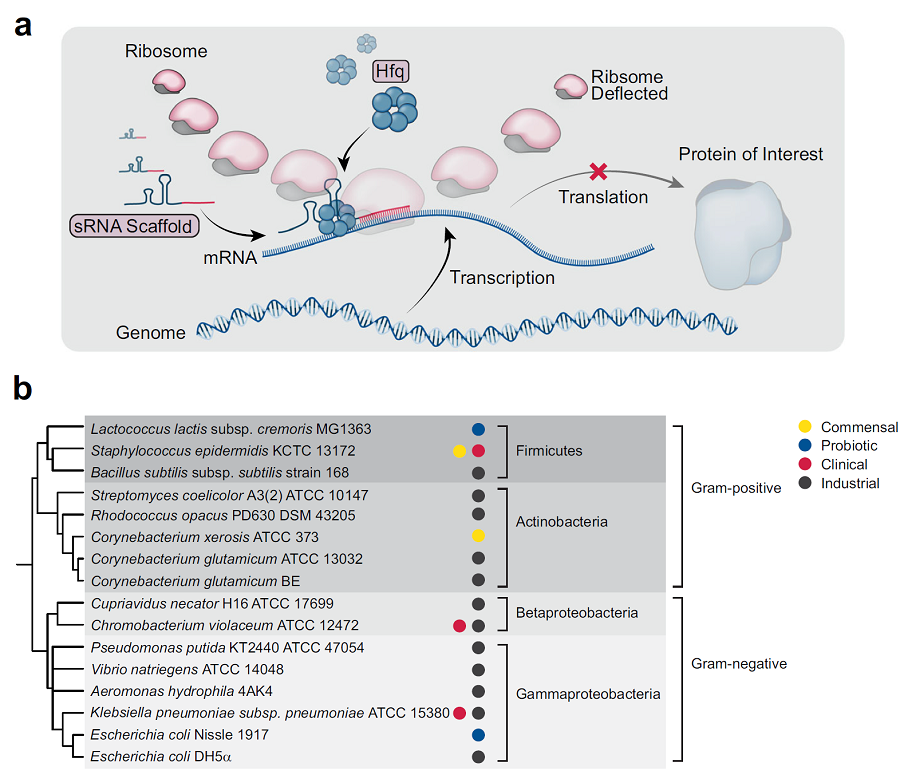
< Figure 1. a) Schematic illustration demonstrating the mechanism of syntetic sRNA b) Phylogenetic tree of the 16 Gram-negative and Gram-positive bacterial species tested for gene knockdown by the BHR-sRNA system. >
To validate the versatility of the BHR-sRNA system, 16 different gram-negative and gram-positive bacteria were selected and tested, where the BHR-sRNA system worked successfully in 15 of them. In addition, it was demonstrated that the gene knockdown capability was more effective than that of the existing E. coli-based sRNA system in 10 bacteria. The BHR-sRNA system proved to be a universal tool capable of effectively inhibiting gene expression in various bacteria.
In order to address the problem of antibiotic-resistant pathogens that have recently become more serious, the BHR-sRNA was demonstrated to suppress the pathogenicity by suppressing the gene producing the virulence factor. By using BHR-sRNA, biofilm formation, one of the factors resulting in antibiotic resistance, was inhibited by 73% in Staphylococcus epidermidis a pathogen that can cause hospital-acquired infections. Antibiotic resistance was also weakened by 58% in the pneumonia causing bacteria Klebsiella pneumoniae. In addition, BHR-sRNA was applied to industrial bacteria to develop microbial cell factories to produce high value-added chemicals with better production performance. Notably, superior industrial strains were constructed with the aid of BHR-sRNA to produce the following chemicals: valerolactam, a raw material for polyamide polymers, methyl-anthranilate, a grape-flavor food additive, and indigoidine, a blue-toned natural dye.
The BHR-sRNA developed through this study will help expedite the commercialization of bioprocesses to produce high value-added compounds and materials such as artificial meat, jet fuel, health supplements, pharmaceuticals, and plastics. It is also anticipated that to help eradicating antibiotic-resistant pathogens in preparation for another upcoming pandemic. “In the past, we could only develop new tools for gene knockdown for each bacterium, but now we have developed a tool that works for a variety of bacteria” said Distinguished Professor Sang Yup Lee.
This work was supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project from NRF supported by the Korean MSIT.
- No Data