research
A deep learning-based tool predicts transcription factors using protein sequences as inputs
A joint research team from KAIST and UCSD has developed a deep neural network named DeepTFactor that predicts transcription factors from protein sequences. DeepTFactor will serve as a useful tool for understanding the regulatory systems of organisms, accelerating the use of deep learning for solving biological problems.
A transcription factor is a protein that specifically binds to DNA sequences to control the transcription initiation. Analyzing transcriptional regulation enables the understanding of how organisms control gene expression in response to genetic or environmental changes. In this regard, finding the transcription factor of an organism is the first step in the analysis of the transcriptional regulatory system of an organism.
Previously, transcription factors have been predicted by analyzing sequence homology with already characterized transcription factors or by data-driven approaches such as machine learning. Conventional machine learning models require a rigorous feature selection process that relies on domain expertise such as calculating the physicochemical properties of molecules or analyzing the homology of biological sequences. Meanwhile, deep learning can inherently learn latent features for the specific task.
A joint research team comprised of Ph.D. candidate Gi Bae Kim and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Ye Gao and Professor Bernhard O. Palsson of the Department of Biochemical Engineering at UCSD reported a deep learning-based tool for the prediction of transcription factors. Their research paper “DeepTFactor: A deep learning-based tool for the prediction of transcription factors” was published online in PNAS.
Their article reports the development of DeepTFactor, a deep learning-based tool that predicts whether a given protein sequence is a transcription factor using three parallel convolutional neural networks. The joint research team predicted 332 transcription factors of Escherichia coli K-12 MG1655 using DeepTFactor and the performance of DeepTFactor by experimentally confirming the genome-wide binding sites of three predicted transcription factors (YqhC, YiaU, and YahB).
The joint research team further used a saliency method to understand the reasoning process of DeepTFactor. The researchers confirmed that even though information on the DNA binding domains of the transcription factor was not explicitly given the training process, DeepTFactor implicitly learned and used them for prediction. Unlike previous transcription factor prediction tools that were developed only for protein sequences of specific organisms, DeepTFactor is expected to be used in the analysis of the transcription systems of all organisms at a high level of performance.
Distinguished Professor Sang Yup Lee said, “DeepTFactor can be used to discover unknown transcription factors from numerous protein sequences that have not yet been characterized. It is expected that DeepTFactor will serve as an important tool for analyzing the regulatory systems of organisms of interest.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea.
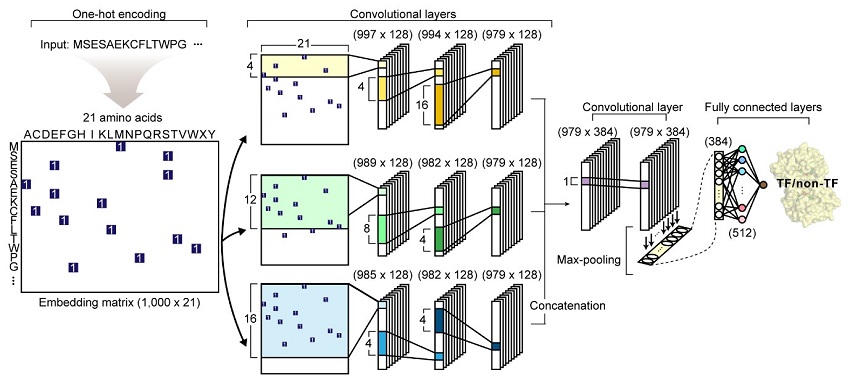
< Figure: The network architecture of DeepTFactor. An input protein sequence is processed using three parallel subnetworks. >
-Publication
Gi Bae Kim, Ye Gao, Bernhard O. Palsson, and Sang Yup Lee. DeepTFactor: A deep learning-based tool for the prediction of transcription factors. (https://doi.org/10.1073/pnas202117118)
-Profile
Distinguished Professor Sang Yup Lee
leesy@kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
Department of Chemical and Biomolecular Engineering
KAIST
-
research 'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecula
2022-03-04 -
research Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents. Professor YongKeum Park’s team and the startup Tomocube encoded 3D refrac
2022-02-09 -
research AI Light-Field Camera Reads 3D Facial Expressions
Machine-learned, light-field camera reads facial expressions from high-contrast illumination invariant 3D facial images A joint research team led by Professors Ki-Hun Jeong and Doheon Lee from the KAIST Department of Bio and Brain Engineering reported the development of a technique for facial expression detection by merging near-infrared light-field camera techniques with artificial intelligence (AI) technology. Unlike a conventional camera, the light-field camera contains micro-lens arrays
2022-01-21 -
research Deep Learning Framework to Enable Material Design in Unseen Domain
Researchers propose a deep neural network-based forward design space exploration using active transfer learning and data augmentation A new study proposed a deep neural network-based forward design approach that enables an efficient search for superior materials far beyond the domain of the initial training set. This approach compensates for the weak predictive power of neural networks on an unseen domain through gradual updates of the neural network with active transfer learning and data
2021-09-29 -
research Deep-Learning and 3D Holographic Microscopy Beats Scientists at Analyzing Cancer Immunotherapy
Live tracking and analyzing of the dynamics of chimeric antigen receptor (CAR) T-cells targeting cancer cells can open new avenues for the development of cancer immunotherapy. However, imaging via conventional microscopy approaches can result in cellular damage, and assessments of cell-to-cell interactions are extremely difficult and labor-intensive. When researchers applied deep learning and 3D holographic microscopy to the task, however, they not only avoided these difficultues but found tha
2021-02-24