BIO
-
 KAIST Research Team Proves How a Neurotransmitter may be the Key in Controlling Alzheimer’s Toxicity
With nearly 50 million dementia patients worldwide, and Alzheimers’s disease is the most common neurodegenerative disease. Its main symptom is the impairment of general cognitive abilities, including the ability to speak or to remember. The importance of finding a cure is widely understood with increasingly aging population and the life expectancy being ever-extended. However, even the cause of the grim disease is yet to be given a clear definition.
A KAIST research team in the Department of Chemistry led by professor Mi Hee Lim took on a lead to discovered a new role for somatostatin, a protein-based neurotransmitter, in reducing the toxicity caused in the pathogenic mechanism taken towards development of Alzheimer’s disease. The study was published in the July issue of Nature Chemistry under the title, “Conformational and functional changes of the native neuropeptide somatostatin occur in the presence of copper and amyloid-β”.
According to the amyloid hypothesis, the abnormal deposition of Aβ proteins causes death of neuronal cells. While Aβ agglomerations make up most of the aged plaques through fibrosis, in recent studies, high concentrations of transitional metal were found in the plaques from Alzheimer’s patients.
This suggests a close interaction between metallic ions and Aβ, which accelerates the fibrosis of proteins. Copper in particular is a redox-activating transition metal that can produce large amounts of oxygen and cause serious oxidative stress on cell organelles. Aβ proteins and transition metals can closely interact with neurotransmitters at synapses, but the direct effects of such abnormalities on the structure and function of neurotransmitters are yet to be understood.
Figure 1. Functional shift of somatostatin (SST) by factors in the pathogenesis of Alzheimer's disease.
Figure 2. Somatostatin’s loss-of-function as neurotransmitter. a. Schematic diagram of SST auto-aggregation due to Alzheimer's pathological factors. b. SST’s aggregation by copper ions. c. Coordination-prediction structure and N-terminal folding of copper-SST. d. Inhibition of SST receptor binding specificity by metals.
In their research, Professor Lim’s team discovered that when somatostatin, the protein-based neurotransmitter, is met with copper, Aβ, and metal-Aβ complexes, self-aggregates and ceases to perform its innate function of transmitting neural signals, but begins to attenuate the toxicity and agglomeration of metal-Aβ complexes.
Figure 3. Gain-of-function of somatostatin (SST) in the dementia setting. a. Prediction of docking of SST and amyloid beta. b. SST making metal-amyloid beta aggregates into an amorphous form. c. Cytotoxic mitigation effect of SST. d. SST mitigating the interaction between amyloid beta protein with the cell membrane.
This research, by Dr. Jiyeon Han et al. from the KAIST Department of Chemistry, revealed the coordination structure between copper and somatostatin at a molecular level through which it suggested the agglomeration mechanism, and discovered the effects of somatostatin on Aβ agglomeration path depending on the presence or absence of metals. The team has further confirmed somatostatin’s receptor binding, interactions with cell membranes, and effects on cell toxicity for the first time to receive international attention.
Professor Mi Hee Lim said, “This research has great significance in having discovered a new role of neurotransmitters in the pathogenesis of Alzheimer’s disease.” “We expect this research to contribute to defining the pathogenic network of neurodegenerative diseases caused by aging, and to the development of future biomarkers and medicine,” she added.
This research was conducted jointly by Professor Seung-Hee Lee’s team of KAIST Department of Biological Sciences, Professor Kiyoung Park’s Team of KAIST Department of Chemistry, and Professor Yulong Li’s team of Peking University.
The research was funded by Basic Science Research Program of the National Research Foundation of Korea and KAIST.
For more information about the research team, visit the website: https://sites.google.com/site/miheelimlab/1-professor-mi-hee-lim.
2022.07.29 View 15865
KAIST Research Team Proves How a Neurotransmitter may be the Key in Controlling Alzheimer’s Toxicity
With nearly 50 million dementia patients worldwide, and Alzheimers’s disease is the most common neurodegenerative disease. Its main symptom is the impairment of general cognitive abilities, including the ability to speak or to remember. The importance of finding a cure is widely understood with increasingly aging population and the life expectancy being ever-extended. However, even the cause of the grim disease is yet to be given a clear definition.
A KAIST research team in the Department of Chemistry led by professor Mi Hee Lim took on a lead to discovered a new role for somatostatin, a protein-based neurotransmitter, in reducing the toxicity caused in the pathogenic mechanism taken towards development of Alzheimer’s disease. The study was published in the July issue of Nature Chemistry under the title, “Conformational and functional changes of the native neuropeptide somatostatin occur in the presence of copper and amyloid-β”.
According to the amyloid hypothesis, the abnormal deposition of Aβ proteins causes death of neuronal cells. While Aβ agglomerations make up most of the aged plaques through fibrosis, in recent studies, high concentrations of transitional metal were found in the plaques from Alzheimer’s patients.
This suggests a close interaction between metallic ions and Aβ, which accelerates the fibrosis of proteins. Copper in particular is a redox-activating transition metal that can produce large amounts of oxygen and cause serious oxidative stress on cell organelles. Aβ proteins and transition metals can closely interact with neurotransmitters at synapses, but the direct effects of such abnormalities on the structure and function of neurotransmitters are yet to be understood.
Figure 1. Functional shift of somatostatin (SST) by factors in the pathogenesis of Alzheimer's disease.
Figure 2. Somatostatin’s loss-of-function as neurotransmitter. a. Schematic diagram of SST auto-aggregation due to Alzheimer's pathological factors. b. SST’s aggregation by copper ions. c. Coordination-prediction structure and N-terminal folding of copper-SST. d. Inhibition of SST receptor binding specificity by metals.
In their research, Professor Lim’s team discovered that when somatostatin, the protein-based neurotransmitter, is met with copper, Aβ, and metal-Aβ complexes, self-aggregates and ceases to perform its innate function of transmitting neural signals, but begins to attenuate the toxicity and agglomeration of metal-Aβ complexes.
Figure 3. Gain-of-function of somatostatin (SST) in the dementia setting. a. Prediction of docking of SST and amyloid beta. b. SST making metal-amyloid beta aggregates into an amorphous form. c. Cytotoxic mitigation effect of SST. d. SST mitigating the interaction between amyloid beta protein with the cell membrane.
This research, by Dr. Jiyeon Han et al. from the KAIST Department of Chemistry, revealed the coordination structure between copper and somatostatin at a molecular level through which it suggested the agglomeration mechanism, and discovered the effects of somatostatin on Aβ agglomeration path depending on the presence or absence of metals. The team has further confirmed somatostatin’s receptor binding, interactions with cell membranes, and effects on cell toxicity for the first time to receive international attention.
Professor Mi Hee Lim said, “This research has great significance in having discovered a new role of neurotransmitters in the pathogenesis of Alzheimer’s disease.” “We expect this research to contribute to defining the pathogenic network of neurodegenerative diseases caused by aging, and to the development of future biomarkers and medicine,” she added.
This research was conducted jointly by Professor Seung-Hee Lee’s team of KAIST Department of Biological Sciences, Professor Kiyoung Park’s Team of KAIST Department of Chemistry, and Professor Yulong Li’s team of Peking University.
The research was funded by Basic Science Research Program of the National Research Foundation of Korea and KAIST.
For more information about the research team, visit the website: https://sites.google.com/site/miheelimlab/1-professor-mi-hee-lim.
2022.07.29 View 15865 -
 PICASSO Technique Drives Biological Molecules into Technicolor
The new imaging approach brings current imaging colors from four to more than 15 for mapping overlapping proteins
Pablo Picasso’s surreal cubist artistic style shifted common features into unrecognizable scenes, but a new imaging approach bearing his namesake may elucidate the most complicated subject: the brain. Employing artificial intelligence to clarify spectral color blending of tiny molecules used to stain specific proteins and other items of research interest, the PICASSO technique, allows researchers to use more than 15 colors to image and parse our overlapping proteins.
The PICASSO developers, based in Korea, published their approach on May 5 in Nature Communications.
Fluorophores — the staining molecules — emit specific colors when excited by a light, but if more than four fluorophores are used, their emitted colors overlap and blend. Researchers previously developed techniques to correct this spectral overlap by precisely defining the matrix of mixed and unmixed images. This measurement depends on reference spectra, found by identifying clear images of only one fluorophore-stained specimen or of multiple, identically prepared specimens that only contain a single fluorophore each.
“Such reference spectra measurement could be complicated to perform in highly heterogeneous specimens, such as the brain, due to the highly varied emission spectra of fluorophores depending on the subregions from which the spectra were measured,” said co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST. He explained that the subregions would each need their own spectra reference measurements, making for an inefficient, time-consuming process. “To address this problem, we developed an approach that does not require reference spectra measurements.”
The approach is the “Process of ultra-multiplexed Imaging of biomolecules viA the unmixing of the Signals of Spectrally Overlapping fluorophores,” also known as PICASSO. Ultra-multiplexed imaging refers to visualizing the numerous individual components of a unit. Like a cinema multiplex in which each theater plays a different movie, each protein in a cell has a different role. By staining with fluorophores, researchers can begin to understand those roles.
“We devised a strategy based on information theory; unmixing is performed by iteratively minimizing the mutual information between mixed images,” said co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering, KAIST. “This allows us to get away with the assumption that the spatial distribution of different proteins is mutually exclusive and enables accurate information unmixing.”
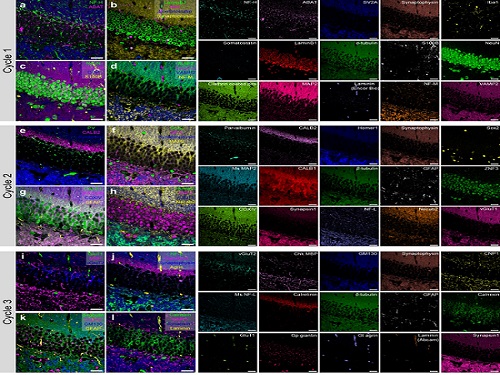
To demonstrate PICASSO’s capabilities, the researchers applied the technique to imaging a mouse brain. With a single round of staining, they performed 15-color multiplexed imaging of a mouse brain. Although small, mouse brains are still complex, multifaceted organs that can take significant resources to map. According to the researchers, PICASSO can improve the capabilities of other imaging techniques and allow for the use of even more fluorophore colors.
Using one such imaging technique in combination with PICASSO, the team achieved 45-color multiplexed imaging of the mouse brain in only three staining and imaging cycles, according to Yoon.
“PICASSO is a versatile tool for the multiplexed biomolecule imaging of cultured cells, tissue slices and clinical specimens,” Chang said. “We anticipate that PICASSO will be useful for a broad range of applications for which biomolecules’ spatial information is important. One such application the tool would be useful for is revealing the cellular heterogeneities of tumor microenvironments, especially the heterogeneous populations of immune cells, which are closely related to cancer prognoses and the efficacy of cancer therapies.”
The Samsung Research Funding & Incubation Center for Future Technology supported this work. Spectral imaging was performed at the Korea Basic Science Institute Western Seoul Center.
-PublicationJunyoung Seo, Yeonbo Sim, Jeewon Kim, Hyunwoo Kim, In Cho, Hoyeon Nam, Yong-Gyu Yoon, Jae-Byum Chang, “PICASSO allows ultra-multiplexed fluorescence imaging of spatiallyoverlapping proteins without reference spectra measurements,” May 5, Nature Communications (doi.org/10.1038/s41467-022-30168-z)
-ProfileProfessor Jae-Byum ChangDepartment of Materials Science and EngineeringCollege of EngineeringKAIST
Professor Young-Gyu YoonSchool of Electrical EngineeringCollege of EngineeringKAIST
2022.06.22 View 11379
PICASSO Technique Drives Biological Molecules into Technicolor
The new imaging approach brings current imaging colors from four to more than 15 for mapping overlapping proteins
Pablo Picasso’s surreal cubist artistic style shifted common features into unrecognizable scenes, but a new imaging approach bearing his namesake may elucidate the most complicated subject: the brain. Employing artificial intelligence to clarify spectral color blending of tiny molecules used to stain specific proteins and other items of research interest, the PICASSO technique, allows researchers to use more than 15 colors to image and parse our overlapping proteins.
The PICASSO developers, based in Korea, published their approach on May 5 in Nature Communications.
Fluorophores — the staining molecules — emit specific colors when excited by a light, but if more than four fluorophores are used, their emitted colors overlap and blend. Researchers previously developed techniques to correct this spectral overlap by precisely defining the matrix of mixed and unmixed images. This measurement depends on reference spectra, found by identifying clear images of only one fluorophore-stained specimen or of multiple, identically prepared specimens that only contain a single fluorophore each.
“Such reference spectra measurement could be complicated to perform in highly heterogeneous specimens, such as the brain, due to the highly varied emission spectra of fluorophores depending on the subregions from which the spectra were measured,” said co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST. He explained that the subregions would each need their own spectra reference measurements, making for an inefficient, time-consuming process. “To address this problem, we developed an approach that does not require reference spectra measurements.”
The approach is the “Process of ultra-multiplexed Imaging of biomolecules viA the unmixing of the Signals of Spectrally Overlapping fluorophores,” also known as PICASSO. Ultra-multiplexed imaging refers to visualizing the numerous individual components of a unit. Like a cinema multiplex in which each theater plays a different movie, each protein in a cell has a different role. By staining with fluorophores, researchers can begin to understand those roles.
“We devised a strategy based on information theory; unmixing is performed by iteratively minimizing the mutual information between mixed images,” said co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering, KAIST. “This allows us to get away with the assumption that the spatial distribution of different proteins is mutually exclusive and enables accurate information unmixing.”
To demonstrate PICASSO’s capabilities, the researchers applied the technique to imaging a mouse brain. With a single round of staining, they performed 15-color multiplexed imaging of a mouse brain. Although small, mouse brains are still complex, multifaceted organs that can take significant resources to map. According to the researchers, PICASSO can improve the capabilities of other imaging techniques and allow for the use of even more fluorophore colors.
Using one such imaging technique in combination with PICASSO, the team achieved 45-color multiplexed imaging of the mouse brain in only three staining and imaging cycles, according to Yoon.
“PICASSO is a versatile tool for the multiplexed biomolecule imaging of cultured cells, tissue slices and clinical specimens,” Chang said. “We anticipate that PICASSO will be useful for a broad range of applications for which biomolecules’ spatial information is important. One such application the tool would be useful for is revealing the cellular heterogeneities of tumor microenvironments, especially the heterogeneous populations of immune cells, which are closely related to cancer prognoses and the efficacy of cancer therapies.”
The Samsung Research Funding & Incubation Center for Future Technology supported this work. Spectral imaging was performed at the Korea Basic Science Institute Western Seoul Center.
-PublicationJunyoung Seo, Yeonbo Sim, Jeewon Kim, Hyunwoo Kim, In Cho, Hoyeon Nam, Yong-Gyu Yoon, Jae-Byum Chang, “PICASSO allows ultra-multiplexed fluorescence imaging of spatiallyoverlapping proteins without reference spectra measurements,” May 5, Nature Communications (doi.org/10.1038/s41467-022-30168-z)
-ProfileProfessor Jae-Byum ChangDepartment of Materials Science and EngineeringCollege of EngineeringKAIST
Professor Young-Gyu YoonSchool of Electrical EngineeringCollege of EngineeringKAIST
2022.06.22 View 11379 -
 New Polymer Mesophase Structure Discovered
Bilayer-folded lamellar mesophase induced by random polymer sequence
Polymers, large molecules made up of repeating smaller molecules called monomers, are found in nearly everything we use in our day-to-day lives. Polymers can be natural or created synthetically. Natural polymers, also called biopolymers, include DNA, proteins, and materials like silk, gelatin, and collagen. Synthetic polymers make up many different kinds of materials, including plastic, that are used in constructing everything from toys to industrial fiber cables to brake pads.
As polymers are formed through a process called polymerization, the monomers are connected through a chain. As the chain develops, the structure of the polymer determines its unique physical and chemical properties. Researchers are continually studying polymers, how they form, how they are structured, and how they develop these unique properties. By understanding this information, scientists can develop new uses for polymers and create new materials that can be used in a wide variety of industries.
In a paper published in Nature Communications on May 4, researchers describe a new structure found in an aqueous solution of an amphiphilic copolymer, called a bilayer-folded lamellar mesophase, that has been discovered through a random copolymer sequence.
“A new mesophase is an important discovery as it shows a new way for molecules to self-organize,” said Professor Myungeun Seo at the Department of Chemistry at KAIST. “We were particularly thrilled to identify this bilayer-folded lamellar phase because pure bilayer membranes are difficult to fold thermodynamically.”
Researchers think that this mesophase structure comes from the sequence of the monomers within the copolymer. The way the different monomers arrange themselves in the chain that makes up a copolymer is important and can have implications for what the copolymer can do. Many copolymers are random, which means that their structure relies on how the monomers interact with each other. In this case, the interaction between the hydrophobic monomers associates the copolymer chains to conceal the hydrophobic domain from water. As the structure gets more complex, researchers have found that a visible order develops so that monomers can be matched up with the right pair.
“While we tend to think random means disorder, here we showed that a periodic order can spontaneously arise from the random copolymer sequence based on their collective behavior,” said Professor Seo. “We believe this comes from the sequence matching problem: finding a perfectly complementary pair for a long sequence is nearly impossible.”
This is what creates the unique structure of this newly discovered mesophase. The copolymer spontaneously folds and creates a multilamellar structure that is separated by water. A multilamellar structure refers to plate-like folds and the folded layers stack on top of each other. The resulting mesophase is birefringent, meaning light refracts through it, it is similar to liquid crystalline, and viscoelastic, which means that it is both viscous and elastic at the same time.
Looking ahead, researchers hope to learn more about this new mesophase and figure out how to control the outcome. Once more is understood about the mesophase and how it is formed, it’s possible that new mesophases could be discovered as more sequences are researched. “One of the obvious questions for us is how to control the folding frequency and adjust the folded height, which we are currently working to address. Ultimately, we want to understand how different multinary sequences can associate with another to create order and apply the knowledge to develop new materials,” said Professor Seo. The National Research Foundation, the Ministry of Education, and the Ministry of Science and ICT of Korea funded this research.
-PublicationMinjoong Shin, Hayeon Kim, Geonhyeong Park, Jongmin Park, Hyungju Ahn, Dong Ki Yoon, Eunji Lee, Myungeun Seo, “Bilayer-folded lamellar mesophase induced by random polymersequence,” May 4, 2022, Nature Communications (https://doi.org/10.1038/s41467-022-30122-z)
-ProfileProfessor Myungeun SeoMacromolecular Materials Chemistry Lab (https://nanopsg.kaist.ac.kr/)Department of ChemistryCollege of Natural SciencesKAIST
2022.06.17 View 9834
New Polymer Mesophase Structure Discovered
Bilayer-folded lamellar mesophase induced by random polymer sequence
Polymers, large molecules made up of repeating smaller molecules called monomers, are found in nearly everything we use in our day-to-day lives. Polymers can be natural or created synthetically. Natural polymers, also called biopolymers, include DNA, proteins, and materials like silk, gelatin, and collagen. Synthetic polymers make up many different kinds of materials, including plastic, that are used in constructing everything from toys to industrial fiber cables to brake pads.
As polymers are formed through a process called polymerization, the monomers are connected through a chain. As the chain develops, the structure of the polymer determines its unique physical and chemical properties. Researchers are continually studying polymers, how they form, how they are structured, and how they develop these unique properties. By understanding this information, scientists can develop new uses for polymers and create new materials that can be used in a wide variety of industries.
In a paper published in Nature Communications on May 4, researchers describe a new structure found in an aqueous solution of an amphiphilic copolymer, called a bilayer-folded lamellar mesophase, that has been discovered through a random copolymer sequence.
“A new mesophase is an important discovery as it shows a new way for molecules to self-organize,” said Professor Myungeun Seo at the Department of Chemistry at KAIST. “We were particularly thrilled to identify this bilayer-folded lamellar phase because pure bilayer membranes are difficult to fold thermodynamically.”
Researchers think that this mesophase structure comes from the sequence of the monomers within the copolymer. The way the different monomers arrange themselves in the chain that makes up a copolymer is important and can have implications for what the copolymer can do. Many copolymers are random, which means that their structure relies on how the monomers interact with each other. In this case, the interaction between the hydrophobic monomers associates the copolymer chains to conceal the hydrophobic domain from water. As the structure gets more complex, researchers have found that a visible order develops so that monomers can be matched up with the right pair.
“While we tend to think random means disorder, here we showed that a periodic order can spontaneously arise from the random copolymer sequence based on their collective behavior,” said Professor Seo. “We believe this comes from the sequence matching problem: finding a perfectly complementary pair for a long sequence is nearly impossible.”
This is what creates the unique structure of this newly discovered mesophase. The copolymer spontaneously folds and creates a multilamellar structure that is separated by water. A multilamellar structure refers to plate-like folds and the folded layers stack on top of each other. The resulting mesophase is birefringent, meaning light refracts through it, it is similar to liquid crystalline, and viscoelastic, which means that it is both viscous and elastic at the same time.
Looking ahead, researchers hope to learn more about this new mesophase and figure out how to control the outcome. Once more is understood about the mesophase and how it is formed, it’s possible that new mesophases could be discovered as more sequences are researched. “One of the obvious questions for us is how to control the folding frequency and adjust the folded height, which we are currently working to address. Ultimately, we want to understand how different multinary sequences can associate with another to create order and apply the knowledge to develop new materials,” said Professor Seo. The National Research Foundation, the Ministry of Education, and the Ministry of Science and ICT of Korea funded this research.
-PublicationMinjoong Shin, Hayeon Kim, Geonhyeong Park, Jongmin Park, Hyungju Ahn, Dong Ki Yoon, Eunji Lee, Myungeun Seo, “Bilayer-folded lamellar mesophase induced by random polymersequence,” May 4, 2022, Nature Communications (https://doi.org/10.1038/s41467-022-30122-z)
-ProfileProfessor Myungeun SeoMacromolecular Materials Chemistry Lab (https://nanopsg.kaist.ac.kr/)Department of ChemistryCollege of Natural SciencesKAIST
2022.06.17 View 9834 -
 Machine Learning-Based Algorithm to Speed up DNA Sequencing
The algorithm presents the first full-fledged, short-read alignment software that leverages learned indices for solving the exact match search problem for efficient seeding
The human genome consists of a complete set of DNA, which is about 6.4 billion letters long. Because of its size, reading the whole genome sequence at once is challenging. So scientists use DNA sequencers to produce hundreds of millions of DNA sequence fragments, or short reads, up to 300 letters long. Then the DNA sequencer assembles all the short reads like a giant jigsaw puzzle to reconstruct the entire genome sequence. Even with very fast computers, this job can take hours to complete.
A research team at KAIST has achieved up to 3.45x faster speeds by developing the first short-read alignment software that uses a recent advance in machine-learning called a learned index.
The research team reported their findings on March 7, 2022 in the journal Bioinformatics. The software has been released as open source and can be found on github (https://github.com/kaist-ina/BWA-MEME).
Next-generation sequencing (NGS) is a state-of-the-art DNA sequencing method. Projects are underway with the goal of producing genome sequencing at population scale. Modern NGS hardware is capable of generating billions of short reads in a single run. Then the short reads have to be aligned with the reference DNA sequence. With large-scale DNA sequencing operations running hundreds of next-generation sequences, the need for an efficient short read alignment tool has become even more critical. Accelerating the DNA sequence alignment would be a step toward achieving the goal of population-scale sequencing. However, existing algorithms are limited in their performance because of their frequent memory accesses.
BWA-MEM2 is a popular short-read alignment software package currently used to sequence the DNA. However, it has its limitations. The state-of-the-art alignment has two phases – seeding and extending. During the seeding phase, searches find exact matches of short reads in the reference DNA sequence. During the extending phase, the short reads from the seeding phase are extended. In the current process, bottlenecks occur in the seeding phase. Finding the exact matches slows the process.
The researchers set out to solve the problem of accelerating the DNA sequence alignment. To speed the process, they applied machine learning techniques to create an algorithmic improvement. Their algorithm, BWA-MEME (BWA-MEM emulated) leverages learned indices to solve the exact match search problem. The original software compared one character at a time for an exact match search. The team’s new algorithm achieves up to 3.45x faster speeds in seeding throughput over BWA-MEM2 by reducing the number of instructions by 4.60x and memory accesses by 8.77x. “Through this study, it has been shown that full genome big data analysis can be performed faster and less costly than conventional methods by applying machine learning technology,” said Professor Dongsu Han from the School of Electrical Engineering at KAIST.
The researchers’ ultimate goal was to develop efficient software that scientists from academia and industry could use on a daily basis for analyzing big data in genomics. “With the recent advances in artificial intelligence and machine learning, we see so many opportunities for designing better software for genomic data analysis. The potential is there for accelerating existing analysis as well as enabling new types of analysis, and our goal is to develop such software,” added Han.
Whole genome sequencing has traditionally been used for discovering genomic mutations and identifying the root causes of diseases, which leads to the discovery and development of new drugs and cures. There could be many potential applications. Whole genome sequencing is used not only for research, but also for clinical purposes. “The science and technology for analyzing genomic data is making rapid progress to make it more accessible for scientists and patients. This will enhance our understanding about diseases and develop a better cure for patients of various diseases.”
The research was funded by the National Research Foundation of the Korean government’s Ministry of Science and ICT.
-PublicationYoungmok Jung, Dongsu Han, “BWA-MEME:BWA-MEM emulated with a machine learning approach,” Bioinformatics, Volume 38, Issue 9, May 2022
(https://doi.org/10.1093/bioinformatics/btac137)
-ProfileProfessor Dongsu HanSchool of Electrical EngineeringKAIST
2022.05.10 View 10382
Machine Learning-Based Algorithm to Speed up DNA Sequencing
The algorithm presents the first full-fledged, short-read alignment software that leverages learned indices for solving the exact match search problem for efficient seeding
The human genome consists of a complete set of DNA, which is about 6.4 billion letters long. Because of its size, reading the whole genome sequence at once is challenging. So scientists use DNA sequencers to produce hundreds of millions of DNA sequence fragments, or short reads, up to 300 letters long. Then the DNA sequencer assembles all the short reads like a giant jigsaw puzzle to reconstruct the entire genome sequence. Even with very fast computers, this job can take hours to complete.
A research team at KAIST has achieved up to 3.45x faster speeds by developing the first short-read alignment software that uses a recent advance in machine-learning called a learned index.
The research team reported their findings on March 7, 2022 in the journal Bioinformatics. The software has been released as open source and can be found on github (https://github.com/kaist-ina/BWA-MEME).
Next-generation sequencing (NGS) is a state-of-the-art DNA sequencing method. Projects are underway with the goal of producing genome sequencing at population scale. Modern NGS hardware is capable of generating billions of short reads in a single run. Then the short reads have to be aligned with the reference DNA sequence. With large-scale DNA sequencing operations running hundreds of next-generation sequences, the need for an efficient short read alignment tool has become even more critical. Accelerating the DNA sequence alignment would be a step toward achieving the goal of population-scale sequencing. However, existing algorithms are limited in their performance because of their frequent memory accesses.
BWA-MEM2 is a popular short-read alignment software package currently used to sequence the DNA. However, it has its limitations. The state-of-the-art alignment has two phases – seeding and extending. During the seeding phase, searches find exact matches of short reads in the reference DNA sequence. During the extending phase, the short reads from the seeding phase are extended. In the current process, bottlenecks occur in the seeding phase. Finding the exact matches slows the process.
The researchers set out to solve the problem of accelerating the DNA sequence alignment. To speed the process, they applied machine learning techniques to create an algorithmic improvement. Their algorithm, BWA-MEME (BWA-MEM emulated) leverages learned indices to solve the exact match search problem. The original software compared one character at a time for an exact match search. The team’s new algorithm achieves up to 3.45x faster speeds in seeding throughput over BWA-MEM2 by reducing the number of instructions by 4.60x and memory accesses by 8.77x. “Through this study, it has been shown that full genome big data analysis can be performed faster and less costly than conventional methods by applying machine learning technology,” said Professor Dongsu Han from the School of Electrical Engineering at KAIST.
The researchers’ ultimate goal was to develop efficient software that scientists from academia and industry could use on a daily basis for analyzing big data in genomics. “With the recent advances in artificial intelligence and machine learning, we see so many opportunities for designing better software for genomic data analysis. The potential is there for accelerating existing analysis as well as enabling new types of analysis, and our goal is to develop such software,” added Han.
Whole genome sequencing has traditionally been used for discovering genomic mutations and identifying the root causes of diseases, which leads to the discovery and development of new drugs and cures. There could be many potential applications. Whole genome sequencing is used not only for research, but also for clinical purposes. “The science and technology for analyzing genomic data is making rapid progress to make it more accessible for scientists and patients. This will enhance our understanding about diseases and develop a better cure for patients of various diseases.”
The research was funded by the National Research Foundation of the Korean government’s Ministry of Science and ICT.
-PublicationYoungmok Jung, Dongsu Han, “BWA-MEME:BWA-MEM emulated with a machine learning approach,” Bioinformatics, Volume 38, Issue 9, May 2022
(https://doi.org/10.1093/bioinformatics/btac137)
-ProfileProfessor Dongsu HanSchool of Electrical EngineeringKAIST
2022.05.10 View 10382 -
 VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 11760
VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 11760 -
 Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 11387
Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 11387 -
 Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 9450
Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 9450 -
 Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
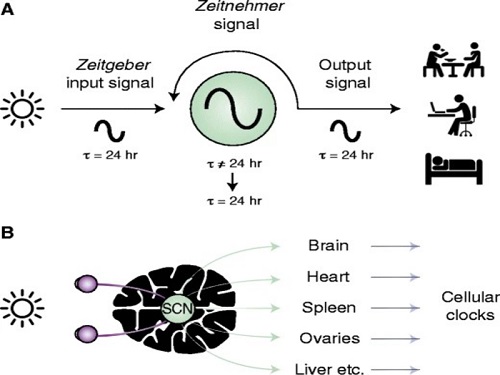
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 11366
Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 11366 -
 Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 12475
Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 12475 -
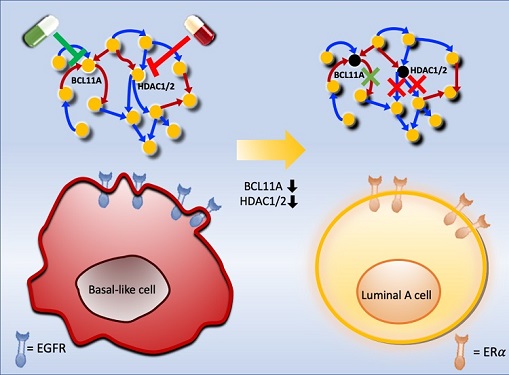 Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 12079
Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 12079 -
 Scientists Develop Wireless Networks that Allow Brain Circuits to Be Controlled Remotely through the Internet
Wireless implantable devices and IoT could manipulate the brains of animals from anywhere around the world due to their minimalistic hardware, low setup cost, ease of use, and customizable versatility
A new study shows that researchers can remotely control the brain circuits of numerous animals simultaneously and independently through the internet. The scientists believe this newly developed technology can speed up brain research and various neuroscience studies to uncover basic brain functions as well as the underpinnings of various neuropsychiatric and neurological disorders.
A multidisciplinary team of researchers at KAIST, Washington University in St. Louis, and the University of Colorado, Boulder, created a wireless ecosystem with its own wireless implantable devices and Internet of Things (IoT) infrastructure to enable high-throughput neuroscience experiments over the internet. This innovative technology could enable scientists to manipulate the brains of animals from anywhere around the world. The study was published in the journal Nature Biomedical Engineering on November 25
“This novel technology is highly versatile and adaptive. It can remotely control numerous neural implants and laboratory tools in real-time or in a scheduled way without direct human interactions,” said Professor Jae-Woong Jeong of the School of Electrical Engineering at KAIST and a senior author of the study. “These wireless neural devices and equipment integrated with IoT technology have enormous potential for science and medicine.”
The wireless ecosystem only requires a mini-computer that can be purchased for under $45, which connects to the internet and communicates with wireless multifunctional brain probes or other types of conventional laboratory equipment using IoT control modules. By optimally integrating the versatility and modular construction of both unique IoT hardware and software within a single ecosystem, this wireless technology offers new applications that have not been demonstrated before by a single standalone technology. This includes, but is not limited to minimalistic hardware, global remote access, selective and scheduled experiments, customizable automation, and high-throughput scalability.
“As long as researchers have internet access, they are able to trigger, customize, stop, validate, and store the outcomes of large experiments at any time and from anywhere in the world. They can remotely perform large-scale neuroscience experiments in animals deployed in multiple countries,” said one of the lead authors, Dr. Raza Qazi, a researcher with KAIST and the University of Colorado, Boulder. “The low cost of this system allows it to be easily adopted and can further fuel innovation across many laboratories,” Dr. Qazi added.
One of the significant advantages of this IoT neurotechnology is its ability to be mass deployed across the globe due to its minimalistic hardware, low setup cost, ease of use, and customizable versatility. Scientists across the world can quickly implement this technology within their existing laboratories with minimal budget concerns to achieve globally remote access, scalable experimental automation, or both, thus potentially reducing the time needed to unravel various neuroscientific challenges such as those associated with intractable neurological conditions.
Another senior author on the study, Professor Jordan McCall from the Department of Anesthesiology and Center for Clinical Pharmacology at Washington University in St. Louis, said this technology has the potential to change how basic neuroscience studies are performed. “One of the biggest limitations when trying to understand how the mammalian brain works is that we have to study these functions in unnatural conditions. This technology brings us one step closer to performing important studies without direct human interaction with the study subjects.”
The ability to remotely schedule experiments moves toward automating these types of experiments. Dr. Kyle Parker, an instructor at Washington University in St. Louis and another lead author on the study added, “This experimental automation can potentially help us reduce the number of animals used in biomedical research by reducing the variability introduced by various experimenters. This is especially important given our moral imperative to seek research designs that enable this reduction.”
The researchers believe this wireless technology may open new opportunities for many applications including brain research, pharmaceuticals, and telemedicine to treat diseases in the brain and other organs remotely. This remote automation technology could become even more valuable when many labs need to shut down, such as during the height of the COVID-19 pandemic.
This work was supported by grants from the KAIST Global Singularity Research Program, the National Research Foundation of Korea, the United States National Institute of Health, and Oak Ridge Associated Universities.
-PublicationRaza Qazi, Kyle Parker, Choong Yeon Kim, Jordan McCall, Jae-Woong Jeong et al. “Scalable and modular wireless-network infrastructure for large-scale behavioral neuroscience,” Nature Biomedical Engineering, November 25 2021 (doi.org/10.1038/s41551-021-00814-w)
-ProfileProfessor Jae-Woong JeongBio-Integrated Electronics and Systems LabSchool of Electrical EngineeringKAIST
2021.11.29 View 16906
Scientists Develop Wireless Networks that Allow Brain Circuits to Be Controlled Remotely through the Internet
Wireless implantable devices and IoT could manipulate the brains of animals from anywhere around the world due to their minimalistic hardware, low setup cost, ease of use, and customizable versatility
A new study shows that researchers can remotely control the brain circuits of numerous animals simultaneously and independently through the internet. The scientists believe this newly developed technology can speed up brain research and various neuroscience studies to uncover basic brain functions as well as the underpinnings of various neuropsychiatric and neurological disorders.
A multidisciplinary team of researchers at KAIST, Washington University in St. Louis, and the University of Colorado, Boulder, created a wireless ecosystem with its own wireless implantable devices and Internet of Things (IoT) infrastructure to enable high-throughput neuroscience experiments over the internet. This innovative technology could enable scientists to manipulate the brains of animals from anywhere around the world. The study was published in the journal Nature Biomedical Engineering on November 25
“This novel technology is highly versatile and adaptive. It can remotely control numerous neural implants and laboratory tools in real-time or in a scheduled way without direct human interactions,” said Professor Jae-Woong Jeong of the School of Electrical Engineering at KAIST and a senior author of the study. “These wireless neural devices and equipment integrated with IoT technology have enormous potential for science and medicine.”
The wireless ecosystem only requires a mini-computer that can be purchased for under $45, which connects to the internet and communicates with wireless multifunctional brain probes or other types of conventional laboratory equipment using IoT control modules. By optimally integrating the versatility and modular construction of both unique IoT hardware and software within a single ecosystem, this wireless technology offers new applications that have not been demonstrated before by a single standalone technology. This includes, but is not limited to minimalistic hardware, global remote access, selective and scheduled experiments, customizable automation, and high-throughput scalability.
“As long as researchers have internet access, they are able to trigger, customize, stop, validate, and store the outcomes of large experiments at any time and from anywhere in the world. They can remotely perform large-scale neuroscience experiments in animals deployed in multiple countries,” said one of the lead authors, Dr. Raza Qazi, a researcher with KAIST and the University of Colorado, Boulder. “The low cost of this system allows it to be easily adopted and can further fuel innovation across many laboratories,” Dr. Qazi added.
One of the significant advantages of this IoT neurotechnology is its ability to be mass deployed across the globe due to its minimalistic hardware, low setup cost, ease of use, and customizable versatility. Scientists across the world can quickly implement this technology within their existing laboratories with minimal budget concerns to achieve globally remote access, scalable experimental automation, or both, thus potentially reducing the time needed to unravel various neuroscientific challenges such as those associated with intractable neurological conditions.
Another senior author on the study, Professor Jordan McCall from the Department of Anesthesiology and Center for Clinical Pharmacology at Washington University in St. Louis, said this technology has the potential to change how basic neuroscience studies are performed. “One of the biggest limitations when trying to understand how the mammalian brain works is that we have to study these functions in unnatural conditions. This technology brings us one step closer to performing important studies without direct human interaction with the study subjects.”
The ability to remotely schedule experiments moves toward automating these types of experiments. Dr. Kyle Parker, an instructor at Washington University in St. Louis and another lead author on the study added, “This experimental automation can potentially help us reduce the number of animals used in biomedical research by reducing the variability introduced by various experimenters. This is especially important given our moral imperative to seek research designs that enable this reduction.”
The researchers believe this wireless technology may open new opportunities for many applications including brain research, pharmaceuticals, and telemedicine to treat diseases in the brain and other organs remotely. This remote automation technology could become even more valuable when many labs need to shut down, such as during the height of the COVID-19 pandemic.
This work was supported by grants from the KAIST Global Singularity Research Program, the National Research Foundation of Korea, the United States National Institute of Health, and Oak Ridge Associated Universities.
-PublicationRaza Qazi, Kyle Parker, Choong Yeon Kim, Jordan McCall, Jae-Woong Jeong et al. “Scalable and modular wireless-network infrastructure for large-scale behavioral neuroscience,” Nature Biomedical Engineering, November 25 2021 (doi.org/10.1038/s41551-021-00814-w)
-ProfileProfessor Jae-Woong JeongBio-Integrated Electronics and Systems LabSchool of Electrical EngineeringKAIST
2021.11.29 View 16906 -
 The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 10391
The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 10391