Bioengineering
-
 A Korean research team develops a new clinical candidate for fatty liver disease
A team of Korean researchers have succeeded in developing a new drug candidate for the treatment of non-alcoholic fatty liver disease (NAFLD) acting on peripheral tissues. To date, there has not been an optimal treatment for non-alcoholic steatohepatitis (NASH), and this discovery is expected to set the grounds for the development of new drugs that can safely suppress both liver fat accumulation and liver fibrosis at the same time.
A joint research team led by Professor Jin Hee Ahn from Gwangju Institute of Science and Technology (GIST) and Professor Hail Kim from the KAIST Graduate School of Medical Science and Engineering developed a new chemical that can suppress disease-specific protein (HTR2A) through years of basic research. The team also revealed to have verified its efficacy and safety through preclinical tests (animal tests) at JD Bioscience Inc., a start-up company founded by Professor Ahn.
Although NAFLD has a prevalence rate as high as 20-30%, and about 5% of the global adult population suffers from NASH, there are no commercial drugs targeting them to date. NAFLD is a chronic disease that starts from the fatty liver and progresses into steatohepatitis, fibrosis, cirrhosis, and liver cancer. The mortality rate of patients increases with accompanied cardiovascular diseases and liver-related complications, and appropriate treatment in the early stage is hence necessary.
< Figure 1. Strategy and history of 5HT2A antagonists. Library and rational design for the development of compound 11c as a potent 5HT2A antagonist. Previous research efforts were discontinued due to limited oral absorption and safety. A therapeutic candidate to overcome this problem was identified and phase 1 clinical trials are currently in progress. >
The new synthetic chemical developed by the joint GIST-KAIST research is an innovative drug candidate that shows therapeutic effects on NASH based on a dual action mechanism that inhibits the accumulation of fat in the liver and liver fibrosis by suppressing the serotonin receptor protein 5HT2A.
The research team confirmed its therapeutic effects in animal models for NAFLD and NASH, in which hepatic steatosis and liver fibrosis* caused by fat accumulation in the liver were suppressed simultaneously by 50-70%.
*fibrosis: stiffening of parts of the liver, also used as a major indicator to track the prognosis of steatosis
The research team explained that the material was designed with optimal polarity and lipid affinity to minimize its permeability across the blood-brain barrier. It therefore does not affect the brain, and causes little side effects in the central nervous system (CNS) such as depression and suicidal ideations, while demonstrating excellent inhibition on its target protein present in tissues outside brain (IC50* = 14 nM). The team also demonstrated its superior efficacy in improving liver fibrosis when compared to similar drugs in the phase 3 clinical trial.
*IC50 (half maximal inhibitory concentration): the concentration at which a chemical suppresses 50% of a particular biological function
< Figure 2. GM-60106 (11c)'s effect on obesity: When GM-60106 was administered to an obese animal model (mice) for 2 months, body weight, body fat mass, and blood sugar were significantly reduced (a-d). In addition, the steatohepatitis level (NAFLD Activity Score) and the expression of genes of the treated mice involved in adipogenesis along with blood/liver fat decreased (e-h) >
Based on the pharmacological data obtained through preclinical trials, the team evaluated the effects of the drug on 88 healthy adults as part of their phase 1 clinical trial, where the side effects and the safe dosage of a drug are tested against healthy adults. Results showed no serious side effects and a good level of drug safety.
In addition, a preliminary efficacy evaluation on eight adults with steatohepatitis is currently underway.
Professor Jin Hee Ahn said, “The aim of this research is to develop a treatment for NASH with little side effects and guaranteed safety by developing a new target. The developed chemical is currently going through phase 1 of the global clinical trial in Australia through JD Bioscience Inc., a bio venture company for innovative drug development.” he added, “The candidate material the research team is currently developing shows not only a high level of safety and preventative effects by suppressing fat accumulation in the liver, but also a direct therapeutic effect on liver fibrosis. This is a strength that distinguishes our material from other competing drugs.”
< Figure 3. Efficacy of GM-60106 (11c) on liver fibrosis: When GM-60106 was administered to a steatohepatitis model (mice) for 3 months, the expression of genes associated with tissue fibrosis was significantly reduced (b-c). As a result of a detailed analysis of the tissues of the animal model, it was confirmed that the rate of tissue fibrosis was reduced and the expression rate of genes related to tissue fibrosis and inflammation was also significantly reduced (e-h). >
Professor Hail Kim from KAIST said, “Until now, this disease did not have a method of treatment other than weight control, and there has been no attempt to develop a drug that can be used for non-obese patients.” He added, “Through this research, we look forward to the development of various treatment techniques targeting a range of metabolic diseases including NASH that do not affect the weight of the patient.”
This study, conducted together by the research teams led by Professor Ahn from GIST and Professor Kim from KAIST, as well as the research team from JD Bioscience Inc., was supported by the Ministry of Science and ICT, and the National New Drug Development Project. The results of this research were published by Nature Communications on January 20.
The team also presented the results of their clinical study on the candidate material coded GM-60106 targeting metabolic abnormality-related MASH* at NASH-TAG Conference 2024, which was held in Utah for three days starting on January 4, which was selected as an excellent abstract.
*MASH (Metabolic Dysfunction-Associated Steatohepatitis): new replacement term for NASH
2024.02.21 View 8615
A Korean research team develops a new clinical candidate for fatty liver disease
A team of Korean researchers have succeeded in developing a new drug candidate for the treatment of non-alcoholic fatty liver disease (NAFLD) acting on peripheral tissues. To date, there has not been an optimal treatment for non-alcoholic steatohepatitis (NASH), and this discovery is expected to set the grounds for the development of new drugs that can safely suppress both liver fat accumulation and liver fibrosis at the same time.
A joint research team led by Professor Jin Hee Ahn from Gwangju Institute of Science and Technology (GIST) and Professor Hail Kim from the KAIST Graduate School of Medical Science and Engineering developed a new chemical that can suppress disease-specific protein (HTR2A) through years of basic research. The team also revealed to have verified its efficacy and safety through preclinical tests (animal tests) at JD Bioscience Inc., a start-up company founded by Professor Ahn.
Although NAFLD has a prevalence rate as high as 20-30%, and about 5% of the global adult population suffers from NASH, there are no commercial drugs targeting them to date. NAFLD is a chronic disease that starts from the fatty liver and progresses into steatohepatitis, fibrosis, cirrhosis, and liver cancer. The mortality rate of patients increases with accompanied cardiovascular diseases and liver-related complications, and appropriate treatment in the early stage is hence necessary.
< Figure 1. Strategy and history of 5HT2A antagonists. Library and rational design for the development of compound 11c as a potent 5HT2A antagonist. Previous research efforts were discontinued due to limited oral absorption and safety. A therapeutic candidate to overcome this problem was identified and phase 1 clinical trials are currently in progress. >
The new synthetic chemical developed by the joint GIST-KAIST research is an innovative drug candidate that shows therapeutic effects on NASH based on a dual action mechanism that inhibits the accumulation of fat in the liver and liver fibrosis by suppressing the serotonin receptor protein 5HT2A.
The research team confirmed its therapeutic effects in animal models for NAFLD and NASH, in which hepatic steatosis and liver fibrosis* caused by fat accumulation in the liver were suppressed simultaneously by 50-70%.
*fibrosis: stiffening of parts of the liver, also used as a major indicator to track the prognosis of steatosis
The research team explained that the material was designed with optimal polarity and lipid affinity to minimize its permeability across the blood-brain barrier. It therefore does not affect the brain, and causes little side effects in the central nervous system (CNS) such as depression and suicidal ideations, while demonstrating excellent inhibition on its target protein present in tissues outside brain (IC50* = 14 nM). The team also demonstrated its superior efficacy in improving liver fibrosis when compared to similar drugs in the phase 3 clinical trial.
*IC50 (half maximal inhibitory concentration): the concentration at which a chemical suppresses 50% of a particular biological function
< Figure 2. GM-60106 (11c)'s effect on obesity: When GM-60106 was administered to an obese animal model (mice) for 2 months, body weight, body fat mass, and blood sugar were significantly reduced (a-d). In addition, the steatohepatitis level (NAFLD Activity Score) and the expression of genes of the treated mice involved in adipogenesis along with blood/liver fat decreased (e-h) >
Based on the pharmacological data obtained through preclinical trials, the team evaluated the effects of the drug on 88 healthy adults as part of their phase 1 clinical trial, where the side effects and the safe dosage of a drug are tested against healthy adults. Results showed no serious side effects and a good level of drug safety.
In addition, a preliminary efficacy evaluation on eight adults with steatohepatitis is currently underway.
Professor Jin Hee Ahn said, “The aim of this research is to develop a treatment for NASH with little side effects and guaranteed safety by developing a new target. The developed chemical is currently going through phase 1 of the global clinical trial in Australia through JD Bioscience Inc., a bio venture company for innovative drug development.” he added, “The candidate material the research team is currently developing shows not only a high level of safety and preventative effects by suppressing fat accumulation in the liver, but also a direct therapeutic effect on liver fibrosis. This is a strength that distinguishes our material from other competing drugs.”
< Figure 3. Efficacy of GM-60106 (11c) on liver fibrosis: When GM-60106 was administered to a steatohepatitis model (mice) for 3 months, the expression of genes associated with tissue fibrosis was significantly reduced (b-c). As a result of a detailed analysis of the tissues of the animal model, it was confirmed that the rate of tissue fibrosis was reduced and the expression rate of genes related to tissue fibrosis and inflammation was also significantly reduced (e-h). >
Professor Hail Kim from KAIST said, “Until now, this disease did not have a method of treatment other than weight control, and there has been no attempt to develop a drug that can be used for non-obese patients.” He added, “Through this research, we look forward to the development of various treatment techniques targeting a range of metabolic diseases including NASH that do not affect the weight of the patient.”
This study, conducted together by the research teams led by Professor Ahn from GIST and Professor Kim from KAIST, as well as the research team from JD Bioscience Inc., was supported by the Ministry of Science and ICT, and the National New Drug Development Project. The results of this research were published by Nature Communications on January 20.
The team also presented the results of their clinical study on the candidate material coded GM-60106 targeting metabolic abnormality-related MASH* at NASH-TAG Conference 2024, which was held in Utah for three days starting on January 4, which was selected as an excellent abstract.
*MASH (Metabolic Dysfunction-Associated Steatohepatitis): new replacement term for NASH
2024.02.21 View 8615 -
 A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time.
On January 9, a joint research team led by Professor Won Do Heo from the KAIST Department of Biological Sciences, Professor Hyung-Bae Kwon from Johns Hopkins School of Medicine, and Professor Sangkyu Lee from the Institute for Basic Science (IBS) revealed that they have developed the world’s first technique to allow a real-time observation of synapse formation, extinction, and alterations.
Professor Heo’s team conjugated dimerization-dependent fluorescent proteins (ddFP) to synapses in order to observe the process in which synapses create connections between neurons in real time. The team named this technique SynapShot, by combining the words ‘synapse’ and snapshot’, and successfully tracked and observed the live formation and extinction processes of synapses as well as their dynamic changes.
< Figure 1. To observe dynamically changing synapses, dimerization-dependent fluorescent protein (ddFP) was expressed to observe flourescent signals upon synapse formation as ddFP enables fluorescence detection through reversible binding to pre- and postsynaptic terminals. >
Through a joint research project, the teams led by Professor Heo and Professor Sangkyu Lee at IBS together designed a SynapShot with green and red fluorescence, and were able to easily distinguish the synapse connecting two different neurons. Additionally, by combining an optogenetic technique that can control the function of a molecule using light, the team was able to observe the changes in the synapses while simultaneously inducing certain functions of the neurons using light.
Through more joint research with the team led by Professor Hyung-Bae Kwon at the Johns Hopkins School of Medicine, Professor Heo’s team induced several situations on live mice, including visual discrimination training, exercise, and anaesthesia, and used SynapShot to observe the changes in the synapses during each situation in real time. The observations revealed that each synapse could change fairly quickly and dynamically. This was the first-ever case in which the changes in synapses were observed in a live mammal.
< Figure 2. Microscopic photos observed through changes of the flourescence of the synapse sensor (SynapShot) by cultivating the neurons of an experimental rat and expressing the SynapShot. The changes in the synapse that is created when the pre- and post-synaptic terminals come into contact and the synapse that disappears after a certain period of time are measured by the fluorescence of the SynapShot. >
Professor Heo said, “Our group developed SynapShot through a collaboration with domestic and international research teams, and have opened up the possibility for first-hand live observations of the quick and dynamic changes of synapses, which was previously difficult to do. We expect this technique to revolutionize research methodology in the neurological field, and play an important role in brightening the future of brain science.”
This research, conducted by co-first authors Seungkyu Son (Ph.D. candidate), Jinsu Lee (Ph.D. candidate) and Dr. Kanghoon Jung from Johns Hopkins, was published in the online edition of Nature Methods on January 8 under the title “Real-time visualization of structural dynamics of synapses in live cells in vivo”, and will be printed in the February volume.
< Figure 3. Simultaneous use of green-SynapShot and red-SynapShot to distinguish and observe synapses with one post-terminal and different pre-terminals. >
< Figure 4. Dimer-dependent fluorescent protein (ddFP) exists as a green fluorescent protein as well as a red fluorescent protein, and can be applied together with blue light-activated optogenetic technology. After activating Tropomyosin receptor kinase B (TrkB) by blue light using optogenetic technology, the strengthening of synaptic connections through signals of brain-derived neurotrophic factor is observed using red-SynapShot. >
< Figure 5. Micrographs showing real-time changing synapses in the visual cortex of mice trained through visual training using in vivo imaging techniques such as two-photon microscopy as well as at the cellular level. >
This research was supported by Mid-Sized Research Funds and the Singularity Project from KAIST, and by IBS.
2024.01.18 View 6589
A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time.
On January 9, a joint research team led by Professor Won Do Heo from the KAIST Department of Biological Sciences, Professor Hyung-Bae Kwon from Johns Hopkins School of Medicine, and Professor Sangkyu Lee from the Institute for Basic Science (IBS) revealed that they have developed the world’s first technique to allow a real-time observation of synapse formation, extinction, and alterations.
Professor Heo’s team conjugated dimerization-dependent fluorescent proteins (ddFP) to synapses in order to observe the process in which synapses create connections between neurons in real time. The team named this technique SynapShot, by combining the words ‘synapse’ and snapshot’, and successfully tracked and observed the live formation and extinction processes of synapses as well as their dynamic changes.
< Figure 1. To observe dynamically changing synapses, dimerization-dependent fluorescent protein (ddFP) was expressed to observe flourescent signals upon synapse formation as ddFP enables fluorescence detection through reversible binding to pre- and postsynaptic terminals. >
Through a joint research project, the teams led by Professor Heo and Professor Sangkyu Lee at IBS together designed a SynapShot with green and red fluorescence, and were able to easily distinguish the synapse connecting two different neurons. Additionally, by combining an optogenetic technique that can control the function of a molecule using light, the team was able to observe the changes in the synapses while simultaneously inducing certain functions of the neurons using light.
Through more joint research with the team led by Professor Hyung-Bae Kwon at the Johns Hopkins School of Medicine, Professor Heo’s team induced several situations on live mice, including visual discrimination training, exercise, and anaesthesia, and used SynapShot to observe the changes in the synapses during each situation in real time. The observations revealed that each synapse could change fairly quickly and dynamically. This was the first-ever case in which the changes in synapses were observed in a live mammal.
< Figure 2. Microscopic photos observed through changes of the flourescence of the synapse sensor (SynapShot) by cultivating the neurons of an experimental rat and expressing the SynapShot. The changes in the synapse that is created when the pre- and post-synaptic terminals come into contact and the synapse that disappears after a certain period of time are measured by the fluorescence of the SynapShot. >
Professor Heo said, “Our group developed SynapShot through a collaboration with domestic and international research teams, and have opened up the possibility for first-hand live observations of the quick and dynamic changes of synapses, which was previously difficult to do. We expect this technique to revolutionize research methodology in the neurological field, and play an important role in brightening the future of brain science.”
This research, conducted by co-first authors Seungkyu Son (Ph.D. candidate), Jinsu Lee (Ph.D. candidate) and Dr. Kanghoon Jung from Johns Hopkins, was published in the online edition of Nature Methods on January 8 under the title “Real-time visualization of structural dynamics of synapses in live cells in vivo”, and will be printed in the February volume.
< Figure 3. Simultaneous use of green-SynapShot and red-SynapShot to distinguish and observe synapses with one post-terminal and different pre-terminals. >
< Figure 4. Dimer-dependent fluorescent protein (ddFP) exists as a green fluorescent protein as well as a red fluorescent protein, and can be applied together with blue light-activated optogenetic technology. After activating Tropomyosin receptor kinase B (TrkB) by blue light using optogenetic technology, the strengthening of synaptic connections through signals of brain-derived neurotrophic factor is observed using red-SynapShot. >
< Figure 5. Micrographs showing real-time changing synapses in the visual cortex of mice trained through visual training using in vivo imaging techniques such as two-photon microscopy as well as at the cellular level. >
This research was supported by Mid-Sized Research Funds and the Singularity Project from KAIST, and by IBS.
2024.01.18 View 6589 -
 KAIST-UCSD researchers build an enzyme discovering AI
- A joint research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Bernhard Palsson of UCSD developed ‘DeepECtransformer’, an artificial intelligence that can predict Enzyme Commission (EC) number of proteins.
- The AI is tasked to discover new enzymes that have not been discovered yet, which would allow prediction for a total of 5,360 types of Enzyme Commission (EC) numbers
- It is expected to be used in the development of microbial cell factories that produce environmentally friendly chemicals as a core technology for analyzing the metabolic network of a genome.
While E. coli is one of the most studied organisms, the function of 30% of proteins that make up E. coli has not yet been clearly revealed. For this, an artificial intelligence was used to discover 464 types of enzymes from the proteins that were unknown, and the researchers went on to verify the predictions of 3 types of proteins were successfully identified through in vitro enzyme assay.
KAIST (President Kwang-Hyung Lee) announced on the 24th that a joint research team comprised of Gi Bae Kim, Ji Yeon Kim, Dr. Jong An Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Dr. Charles J. Norsigian and Professor Bernhard O. Palsson of the Department of Bioengineering at UCSD has developed DeepECtransformer, an artificial intelligence that can predict the enzyme functions from the protein sequence, and has established a prediction system by utilizing the AI to quickly and accurately identify the enzyme function.
Enzymes are proteins that catalyze biological reactions, and identifying the function of each enzyme is essential to understanding the various chemical reactions that exist in living organisms and the metabolic characteristics of those organisms. Enzyme Commission (EC) number is an enzyme function classification system designed by the International Union of Biochemistry and Molecular Biology, and in order to understand the metabolic characteristics of various organisms, it is necessary to develop a technology that can quickly analyze enzymes and EC numbers of the enzymes present in the genome.
Various methodologies based on deep learning have been developed to analyze the features of biological sequences, including protein function prediction, but most of them have a problem of a black box, where the inference process of AI cannot be interpreted. Various prediction systems that utilize AI for enzyme function prediction have also been reported, but they do not solve this black box problem, or cannot interpret the reasoning process in fine-grained level (e.g., the level of amino acid residues in the enzyme sequence).
The joint team developed DeepECtransformer, an AI that utilizes deep learning and a protein homology analysis module to predict the enzyme function of a given protein sequence. To better understand the features of protein sequences, the transformer architecture, which is commonly used in natural language processing, was additionally used to extract important features about enzyme functions in the context of the entire protein sequence, which enabled the team to accurately predict the EC number of the enzyme. The developed DeepECtransformer can predict a total of 5360 EC numbers.
The joint team further analyzed the transformer architecture to understand the inference process of DeepECtransformer, and found that in the inference process, the AI utilizes information on catalytic active sites and/or the cofactor binding sites which are important for enzyme function. By analyzing the black box of DeepECtransformer, it was confirmed that the AI was able to identify the features that are important for enzyme function on its own during the learning process.
"By utilizing the prediction system we developed, we were able to predict the functions of enzymes that had not yet been identified and verify them experimentally," said Gi Bae Kim, the first author of the paper. "By using DeepECtransformer to identify previously unknown enzymes in living organisms, we will be able to more accurately analyze various facets involved in the metabolic processes of organisms, such as the enzymes needed to biosynthesize various useful compounds or the enzymes needed to biodegrade plastics." he added.
"DeepECtransformer, which quickly and accurately predicts enzyme functions, is a key technology in functional genomics, enabling us to analyze the function of entire enzymes at the systems level," said Professor Sang Yup Lee. He added, “We will be able to use it to develop eco-friendly microbial factories based on comprehensive genome-scale metabolic models, potentially minimizing missing information of metabolism.”
The joint team’s work on DeepECtransformer is described in the paper titled "Functional annotation of enzyme-encoding genes using deep learning with transformer layers" written by Gi Bae Kim, Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST and their colleagues. The paper was published via peer-review on the 14th of November on “Nature Communications”.
This research was conducted with the support by “the Development of next-generation biorefinery platform technologies for leading bio-based chemicals industry project (2022M3J5A1056072)” and by “Development of platform technologies of microbial cell factories for the next-generation biorefineries project (2022M3J5A1056117)” from National Research Foundation supported by the Korean Ministry of Science and ICT (Project Leader: Distinguished Professor Sang Yup Lee, KAIST).
< Figure 1. The structure of DeepECtransformer's artificial neural network >
2023.11.24 View 5824
KAIST-UCSD researchers build an enzyme discovering AI
- A joint research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Bernhard Palsson of UCSD developed ‘DeepECtransformer’, an artificial intelligence that can predict Enzyme Commission (EC) number of proteins.
- The AI is tasked to discover new enzymes that have not been discovered yet, which would allow prediction for a total of 5,360 types of Enzyme Commission (EC) numbers
- It is expected to be used in the development of microbial cell factories that produce environmentally friendly chemicals as a core technology for analyzing the metabolic network of a genome.
While E. coli is one of the most studied organisms, the function of 30% of proteins that make up E. coli has not yet been clearly revealed. For this, an artificial intelligence was used to discover 464 types of enzymes from the proteins that were unknown, and the researchers went on to verify the predictions of 3 types of proteins were successfully identified through in vitro enzyme assay.
KAIST (President Kwang-Hyung Lee) announced on the 24th that a joint research team comprised of Gi Bae Kim, Ji Yeon Kim, Dr. Jong An Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Dr. Charles J. Norsigian and Professor Bernhard O. Palsson of the Department of Bioengineering at UCSD has developed DeepECtransformer, an artificial intelligence that can predict the enzyme functions from the protein sequence, and has established a prediction system by utilizing the AI to quickly and accurately identify the enzyme function.
Enzymes are proteins that catalyze biological reactions, and identifying the function of each enzyme is essential to understanding the various chemical reactions that exist in living organisms and the metabolic characteristics of those organisms. Enzyme Commission (EC) number is an enzyme function classification system designed by the International Union of Biochemistry and Molecular Biology, and in order to understand the metabolic characteristics of various organisms, it is necessary to develop a technology that can quickly analyze enzymes and EC numbers of the enzymes present in the genome.
Various methodologies based on deep learning have been developed to analyze the features of biological sequences, including protein function prediction, but most of them have a problem of a black box, where the inference process of AI cannot be interpreted. Various prediction systems that utilize AI for enzyme function prediction have also been reported, but they do not solve this black box problem, or cannot interpret the reasoning process in fine-grained level (e.g., the level of amino acid residues in the enzyme sequence).
The joint team developed DeepECtransformer, an AI that utilizes deep learning and a protein homology analysis module to predict the enzyme function of a given protein sequence. To better understand the features of protein sequences, the transformer architecture, which is commonly used in natural language processing, was additionally used to extract important features about enzyme functions in the context of the entire protein sequence, which enabled the team to accurately predict the EC number of the enzyme. The developed DeepECtransformer can predict a total of 5360 EC numbers.
The joint team further analyzed the transformer architecture to understand the inference process of DeepECtransformer, and found that in the inference process, the AI utilizes information on catalytic active sites and/or the cofactor binding sites which are important for enzyme function. By analyzing the black box of DeepECtransformer, it was confirmed that the AI was able to identify the features that are important for enzyme function on its own during the learning process.
"By utilizing the prediction system we developed, we were able to predict the functions of enzymes that had not yet been identified and verify them experimentally," said Gi Bae Kim, the first author of the paper. "By using DeepECtransformer to identify previously unknown enzymes in living organisms, we will be able to more accurately analyze various facets involved in the metabolic processes of organisms, such as the enzymes needed to biosynthesize various useful compounds or the enzymes needed to biodegrade plastics." he added.
"DeepECtransformer, which quickly and accurately predicts enzyme functions, is a key technology in functional genomics, enabling us to analyze the function of entire enzymes at the systems level," said Professor Sang Yup Lee. He added, “We will be able to use it to develop eco-friendly microbial factories based on comprehensive genome-scale metabolic models, potentially minimizing missing information of metabolism.”
The joint team’s work on DeepECtransformer is described in the paper titled "Functional annotation of enzyme-encoding genes using deep learning with transformer layers" written by Gi Bae Kim, Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST and their colleagues. The paper was published via peer-review on the 14th of November on “Nature Communications”.
This research was conducted with the support by “the Development of next-generation biorefinery platform technologies for leading bio-based chemicals industry project (2022M3J5A1056072)” and by “Development of platform technologies of microbial cell factories for the next-generation biorefineries project (2022M3J5A1056117)” from National Research Foundation supported by the Korean Ministry of Science and ICT (Project Leader: Distinguished Professor Sang Yup Lee, KAIST).
< Figure 1. The structure of DeepECtransformer's artificial neural network >
2023.11.24 View 5824 -
 KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 8698
KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 8698 -
 KAIST researchers discovers the neural circuit that reacts to alarm clock
KAIST (President Kwang Hyung Lee) announced on the 20th that a research team led by Professor Daesoo Kim of the Department of Brain and Cognitive Sciences and Dr. Jeongjin Kim 's team from the Korea Institute of Science and Technology (KIST) have identified the principle of awakening animals by responding to sounds even while sleeping.
Sleep is a very important physiological process that organizes brain activity and maintains health. During sleep, the function of sensory nerves is blocked, so the ability to detect danger in the proximity is reduced. However, many animals detect approaching predators and respond even while sleeping. Scientists thought that animals ready for danger by alternating between deep sleep and light sleep.
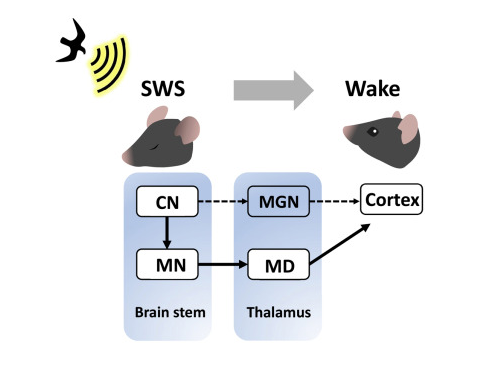
A research team led by Professor Daesoo Kim at KAIST discovered that animals have neural circuits that respond to sounds even during deep sleep. While awake, the medial geniculate thalamus responds to sounds, but during deep sleep, or Non-REM sleep, the Mediodorsal thalamus responds to sounds to wake up the brain.
As a result of the study, when the rats fell into deep sleep, the nerves of the medial geniculate thalamus were also sleeping, but the nerves of mediodorsal thalamus were awake and responded immediately to sounds. In addition, it was observed that when mediodorsal thalamus was inhibited, the rats could not wake up even when a sound was heard, and when the mediodorsal thalamus was stimulated, the rats woke up within a few seconds without sound.
This is the first study to show that sleep and wakefulness can transmit auditory signals through different neural circuits, and was reported in the international journal, Current Biology on February 7, and was highlighted by Nature. (https://www.nature.com/articles/d41586-023-00354-0)
Professor Daesoo Kim explained, “The findings of this study can used in developing digital healthcare technologies to be used to improve understanding of disorders of senses and wakefulness seen in various brain diseases and to control the senses in the future.”
This research was carried out with the support from the National Research Foundation of Korea's Mid-Career Research Foundation Program.
Figure 1. Traditionally, sound signals were thought to be propagated from the auditory nerve to the auditory thalamus. However, while in slow-wave sleep, the auditory nerve sends sound signals to the mediodorsal thalamic neurons via the brainstem nerve to induce arousal in the brain.
Figure 2. GRIK4 dorsomedial nerve in response to sound stimulation. The awakening effect is induced as the activity of the GRIK4 dorsal medial nerve increases based on the time when sound stimulation is given.
2023.03.03 View 6039
KAIST researchers discovers the neural circuit that reacts to alarm clock
KAIST (President Kwang Hyung Lee) announced on the 20th that a research team led by Professor Daesoo Kim of the Department of Brain and Cognitive Sciences and Dr. Jeongjin Kim 's team from the Korea Institute of Science and Technology (KIST) have identified the principle of awakening animals by responding to sounds even while sleeping.
Sleep is a very important physiological process that organizes brain activity and maintains health. During sleep, the function of sensory nerves is blocked, so the ability to detect danger in the proximity is reduced. However, many animals detect approaching predators and respond even while sleeping. Scientists thought that animals ready for danger by alternating between deep sleep and light sleep.
A research team led by Professor Daesoo Kim at KAIST discovered that animals have neural circuits that respond to sounds even during deep sleep. While awake, the medial geniculate thalamus responds to sounds, but during deep sleep, or Non-REM sleep, the Mediodorsal thalamus responds to sounds to wake up the brain.
As a result of the study, when the rats fell into deep sleep, the nerves of the medial geniculate thalamus were also sleeping, but the nerves of mediodorsal thalamus were awake and responded immediately to sounds. In addition, it was observed that when mediodorsal thalamus was inhibited, the rats could not wake up even when a sound was heard, and when the mediodorsal thalamus was stimulated, the rats woke up within a few seconds without sound.
This is the first study to show that sleep and wakefulness can transmit auditory signals through different neural circuits, and was reported in the international journal, Current Biology on February 7, and was highlighted by Nature. (https://www.nature.com/articles/d41586-023-00354-0)
Professor Daesoo Kim explained, “The findings of this study can used in developing digital healthcare technologies to be used to improve understanding of disorders of senses and wakefulness seen in various brain diseases and to control the senses in the future.”
This research was carried out with the support from the National Research Foundation of Korea's Mid-Career Research Foundation Program.
Figure 1. Traditionally, sound signals were thought to be propagated from the auditory nerve to the auditory thalamus. However, while in slow-wave sleep, the auditory nerve sends sound signals to the mediodorsal thalamic neurons via the brainstem nerve to induce arousal in the brain.
Figure 2. GRIK4 dorsomedial nerve in response to sound stimulation. The awakening effect is induced as the activity of the GRIK4 dorsal medial nerve increases based on the time when sound stimulation is given.
2023.03.03 View 6039 -
 KAIST Offers Hope to Musicians with Dystonia
< Photo 1. Conductor and Pianist João Carlos Martins before the Recital at the Carnegie Hall preparing with his bionic gloves >
KAIST’s neuroscientist and professor, Dr. Daesoo Kim attended the “Conference for Musicians with Dystonia” supported by the World Health Organization (WHO) and the Carnegie Hall concert of legendary pianist João Carlos Martins, who is also a dystonia patient, to announce his team’s recent advancements toward finding a cure for dystonia.
On November 19, 2022, a “miracle concert” was held in Carnegie Hall. João Carlos Martins was a renowned world-class pianist in the 70s and 80s, but he had to put an end to his musical career due to focal dystonia in his fingers. But in 2020, he began using a bionic glove developed by industrial designer Ubiratã Bizarro Costa and after years of hard work he was back in Carnegie Hall as an 82-year-old man.
During the concert, he conducted the NOVUS NY orchestra in a performance of Bach, and later even played the piano himself. In particular, between his performances, he gave shout-outs to scientists studying dystonia including KAIST Professor Daesoo Kim, asking them to continue working towards curing rare diseases for musicians.
< Photo 2. Professor Daesoo Kim with Conductor and Pianist João Carlos Martins >
Musician’s dystonia affects 1-3% of musicians around the world and musicians make up approximately 5% of the total number of dystonia patients. Musicians who are no longer able to practice music due to the disease often experience stress and depression, which may even lead to suicide in extreme cases. Musicians are known to be particularly prone to such diseases due to excessive practice regimens, perfectionism, and even genetics. Currently, botulinum toxin (Botox) is used to suppress abnormal muscles, but muscle function suppression ultimately means that the musician is no longer able to play the instrument. João Carlos Martins himself underwent several Botox procedures and three brain surgeries, but saw no therapeutic results. This is why a new treatment was necessary.
Professor Daesoo Kim’s research team at KAIST took note of the fact that abnormal muscle tension is caused by excessive stress, and developed NT-1, a treatment that blocks the development of the symptoms of dystonia from the brain, allowing patients to use their muscles as they normally would. The research team published their findings in Science Advances in 2021, and João Carlos Martins invited Professor Daesoo Kim to the UN conference and his concert after reading this paper.
< Photo 3. Professor Daesoo Kim (3rd from the left) photographed with other guests at the recital including Dr. Dévora Kestel, the Director of the Mental Health and Substance Use at WHO, sharing the center with Conductor and Pianist João Carlos Martins >
During the UN conference held the day prior to the Carnegie Hall concert, Dr. Dévora Kestel, Director of the Mental Health and Substance Use at WHO, said, “Although dystonia is not as well-known, it is a common disease around the world, and needs our society’s attention and the devotion of many researchers.” Professor Daesoo Kim said, “NT-1 is a drug that blocks the cause of dystonia in the brain, and will allow musicians to continue practicing music. We aim to attain clinical approval in Korea by 2024.”
NT-1 is currently under development by NeuroTobe, a faculty-led start-up company at KAIST, headed by Professor Daesoo Kim as the CEO. The synthesis of the drug for clinical testing has been successfully completed, and it has shown excellent efficacy and safety through various rounds of animal testing. Unlike Botox, which takes a few days to show its therapeutic effects after receiving the procedure from a hospital, NT-1 shows its therapeutic effects within an hour after taking it. As a so-called “edible Botox”, it is expected to help treat various muscular diseases and ailments.
2022.12.27 View 11599
KAIST Offers Hope to Musicians with Dystonia
< Photo 1. Conductor and Pianist João Carlos Martins before the Recital at the Carnegie Hall preparing with his bionic gloves >
KAIST’s neuroscientist and professor, Dr. Daesoo Kim attended the “Conference for Musicians with Dystonia” supported by the World Health Organization (WHO) and the Carnegie Hall concert of legendary pianist João Carlos Martins, who is also a dystonia patient, to announce his team’s recent advancements toward finding a cure for dystonia.
On November 19, 2022, a “miracle concert” was held in Carnegie Hall. João Carlos Martins was a renowned world-class pianist in the 70s and 80s, but he had to put an end to his musical career due to focal dystonia in his fingers. But in 2020, he began using a bionic glove developed by industrial designer Ubiratã Bizarro Costa and after years of hard work he was back in Carnegie Hall as an 82-year-old man.
During the concert, he conducted the NOVUS NY orchestra in a performance of Bach, and later even played the piano himself. In particular, between his performances, he gave shout-outs to scientists studying dystonia including KAIST Professor Daesoo Kim, asking them to continue working towards curing rare diseases for musicians.
< Photo 2. Professor Daesoo Kim with Conductor and Pianist João Carlos Martins >
Musician’s dystonia affects 1-3% of musicians around the world and musicians make up approximately 5% of the total number of dystonia patients. Musicians who are no longer able to practice music due to the disease often experience stress and depression, which may even lead to suicide in extreme cases. Musicians are known to be particularly prone to such diseases due to excessive practice regimens, perfectionism, and even genetics. Currently, botulinum toxin (Botox) is used to suppress abnormal muscles, but muscle function suppression ultimately means that the musician is no longer able to play the instrument. João Carlos Martins himself underwent several Botox procedures and three brain surgeries, but saw no therapeutic results. This is why a new treatment was necessary.
Professor Daesoo Kim’s research team at KAIST took note of the fact that abnormal muscle tension is caused by excessive stress, and developed NT-1, a treatment that blocks the development of the symptoms of dystonia from the brain, allowing patients to use their muscles as they normally would. The research team published their findings in Science Advances in 2021, and João Carlos Martins invited Professor Daesoo Kim to the UN conference and his concert after reading this paper.
< Photo 3. Professor Daesoo Kim (3rd from the left) photographed with other guests at the recital including Dr. Dévora Kestel, the Director of the Mental Health and Substance Use at WHO, sharing the center with Conductor and Pianist João Carlos Martins >
During the UN conference held the day prior to the Carnegie Hall concert, Dr. Dévora Kestel, Director of the Mental Health and Substance Use at WHO, said, “Although dystonia is not as well-known, it is a common disease around the world, and needs our society’s attention and the devotion of many researchers.” Professor Daesoo Kim said, “NT-1 is a drug that blocks the cause of dystonia in the brain, and will allow musicians to continue practicing music. We aim to attain clinical approval in Korea by 2024.”
NT-1 is currently under development by NeuroTobe, a faculty-led start-up company at KAIST, headed by Professor Daesoo Kim as the CEO. The synthesis of the drug for clinical testing has been successfully completed, and it has shown excellent efficacy and safety through various rounds of animal testing. Unlike Botox, which takes a few days to show its therapeutic effects after receiving the procedure from a hospital, NT-1 shows its therapeutic effects within an hour after taking it. As a so-called “edible Botox”, it is expected to help treat various muscular diseases and ailments.
2022.12.27 View 11599 -
 KAIST develops biocompatible adhesive applicable to hair transplants
Aside from being used as a new medical adhesive, the new material can be applied to developing a new method of hair transplants, which cannot be repeated multiple times using current method of implanting the wholly intact follicles into the skin.
Medical adhesives are materials that can be applied to various uses such as wound healing, hemostasis, vascular anastomosis, and tissue engineering, and is expected to contribute greatly to the development of minimally invasive surgery and organ transplants. However, adhesives with high adhesion, low toxicity, and capable of decomposing in the body are rare. Adhesives based on natural proteins, such as fibrin and collagen, have high biocompatibility but insufficient adhesive strength. Synthetic polymer adhesives based on urethane or acrylic have greater adhesion but do not decompose well and may cause an inflammatory reaction in the body.
A joint research team led by Professor Myungeun Seo and Professor Haeshin Lee from the KAIST Department of Chemistry developed a bio-friendly adhesive from biocompatible polymers using tannic acid, the source of astringency in wine.
The research team focused on tannic acid, a natural polyphenolic product. Tannic acid is a polyphenol present in large amounts in fruit peels, nuts, and cacao. It has a high affinity and coating ability on other substances, and we sense the astringent taste in wine when tannic acid sticks to the surface of our tongue. When tannic acid is mixed with hydrophilic polymers, they form coacervates, or small droplets of jelly-like fluids that sink. If the polymers used are biocompatible, the mixture can be applied as a medical adhesive with low toxicity. However, coacervates are fundamentally fluid-like and cannot withstand large forces, which limits their adhesive capabilities. Thus, while research to utilize it as an adhesive has been actively discussed, a biodegradable material exhibiting strong adhesion due to its high shear strength has not yet been developed.
The research team figured out a way to enhance adhesion by mixing two biocompatible FDA-approved polymers, polyethylene glycol (PEG) and polylactic acid (PLA). While PEG, which is used widely in eyedrops and cream, is hydrophilic, PLA, a well-known bioplastic derived from lactic acid, is insoluble in water. The team combined the two into a block copolymer, which forms hydrophilic PLA aggregates in water with PEG blocks surrounding them. A coacervate created by mixing the micelles and tannic acid would behave like a solid due to the hard PLA components, and show an elastic modulus improved by a thousand times compared to PEG, enabling it to withstand much greater force as an adhesive.
Figure 1. (Above) Principle of biodegradable adhesive made by mixing poly(ethylene glycol)-poly(lactic acid) diblock copolymer and tannic acid in water. Yellow coacervate is precipitated through hydrogen bonding between the block copolymer micelles and tannic acid, and exhibits adhesion. After heat treatment, hydrogen bonds are rearranged to further improve adhesion. (Bottom) Adhesion comparison. Compared to using poly(ethylene glycol) polymer (d), it can support 10 times more weight when using block copolymer (e) and 60 times more weight after heat treatment (f). The indicated G' values represent the elastic modulus of the material.
Furthermore, the research team observed that the material’s mechanical properties can be improved by over a hundred times through a heating and cooling process that is used to heat-treat metals. They also discovered that this is due to the enforced interactions between micelle and tannic acid arrays.
The research team used the fact that the material shows minimal irritation to the skin and decomposes well in the body to demonstrate its possible application as an adhesive for hair transplantation through an animal experiment. Professor Haeshin Lee, who has pioneered various application fields including medical adhesives, hemostatic agents, and browning shampoo, focused on the adhesive capacities and low toxicity of polyphenols like tannic acid, and now looks forward to it improving the limitations of current hair transplant methods, which still involve follicle transfer and are difficult to be repeated multiple times.
Figure 2. (a) Overview of a hair transplantation method using a biodegradable adhesive (right) compared to a conventional hair transplantation method (left) that transplants hair containing hair follicles. After applying an adhesive to the tip of the hair, it is fixed to the skin by implanting it through a subcutaneous injection, and repeated treatment is possible. (b) Initial animal test results. One day after 15 hair transplantation, 12 strands of hair remain. If you pull the 3 strands of hair, you can see that the whole body is pulled up, indicating that it is firmly implanted into the skin. All strands of hair applied without the new adhesive material fell off, and in the case of adhesive without heat treatment, the efficiency was 1/7.
This research was conducted by first co-authors Dr. Jongmin Park (currently a senior researcher at the Korea Research Institute of Chemical Technology) from Professor Myeongeun Seo’s team and Dr. Eunsook Park from Professor Haeshin Lee’s team in the KAIST Department of Chemistry, and through joint research with the teams led by Professor Hyungjun Kim from the KAIST Department of Chemistry and Professor Siyoung Choi from the Department of Chemical and Biomolecular Engineering. The research was published online on August 22 in the international journal Au (JACS Au) under the title Biodegradable Block Copolymer-Tannic Acid Glue.
This study was funded by the Support Research Under Protection Project of the National Research Foundation (NRF), Leading Research Center Support Project (Research Center for Multiscale Chiral Structure), Biodegradable Plastics Commercialization and Demonstration Project by the Ministry of Trade and Industry, and institutional funding from the Korea Research Institute of Chemical Technology.
2022.10.07 View 11672
KAIST develops biocompatible adhesive applicable to hair transplants
Aside from being used as a new medical adhesive, the new material can be applied to developing a new method of hair transplants, which cannot be repeated multiple times using current method of implanting the wholly intact follicles into the skin.
Medical adhesives are materials that can be applied to various uses such as wound healing, hemostasis, vascular anastomosis, and tissue engineering, and is expected to contribute greatly to the development of minimally invasive surgery and organ transplants. However, adhesives with high adhesion, low toxicity, and capable of decomposing in the body are rare. Adhesives based on natural proteins, such as fibrin and collagen, have high biocompatibility but insufficient adhesive strength. Synthetic polymer adhesives based on urethane or acrylic have greater adhesion but do not decompose well and may cause an inflammatory reaction in the body.
A joint research team led by Professor Myungeun Seo and Professor Haeshin Lee from the KAIST Department of Chemistry developed a bio-friendly adhesive from biocompatible polymers using tannic acid, the source of astringency in wine.
The research team focused on tannic acid, a natural polyphenolic product. Tannic acid is a polyphenol present in large amounts in fruit peels, nuts, and cacao. It has a high affinity and coating ability on other substances, and we sense the astringent taste in wine when tannic acid sticks to the surface of our tongue. When tannic acid is mixed with hydrophilic polymers, they form coacervates, or small droplets of jelly-like fluids that sink. If the polymers used are biocompatible, the mixture can be applied as a medical adhesive with low toxicity. However, coacervates are fundamentally fluid-like and cannot withstand large forces, which limits their adhesive capabilities. Thus, while research to utilize it as an adhesive has been actively discussed, a biodegradable material exhibiting strong adhesion due to its high shear strength has not yet been developed.
The research team figured out a way to enhance adhesion by mixing two biocompatible FDA-approved polymers, polyethylene glycol (PEG) and polylactic acid (PLA). While PEG, which is used widely in eyedrops and cream, is hydrophilic, PLA, a well-known bioplastic derived from lactic acid, is insoluble in water. The team combined the two into a block copolymer, which forms hydrophilic PLA aggregates in water with PEG blocks surrounding them. A coacervate created by mixing the micelles and tannic acid would behave like a solid due to the hard PLA components, and show an elastic modulus improved by a thousand times compared to PEG, enabling it to withstand much greater force as an adhesive.
Figure 1. (Above) Principle of biodegradable adhesive made by mixing poly(ethylene glycol)-poly(lactic acid) diblock copolymer and tannic acid in water. Yellow coacervate is precipitated through hydrogen bonding between the block copolymer micelles and tannic acid, and exhibits adhesion. After heat treatment, hydrogen bonds are rearranged to further improve adhesion. (Bottom) Adhesion comparison. Compared to using poly(ethylene glycol) polymer (d), it can support 10 times more weight when using block copolymer (e) and 60 times more weight after heat treatment (f). The indicated G' values represent the elastic modulus of the material.
Furthermore, the research team observed that the material’s mechanical properties can be improved by over a hundred times through a heating and cooling process that is used to heat-treat metals. They also discovered that this is due to the enforced interactions between micelle and tannic acid arrays.
The research team used the fact that the material shows minimal irritation to the skin and decomposes well in the body to demonstrate its possible application as an adhesive for hair transplantation through an animal experiment. Professor Haeshin Lee, who has pioneered various application fields including medical adhesives, hemostatic agents, and browning shampoo, focused on the adhesive capacities and low toxicity of polyphenols like tannic acid, and now looks forward to it improving the limitations of current hair transplant methods, which still involve follicle transfer and are difficult to be repeated multiple times.
Figure 2. (a) Overview of a hair transplantation method using a biodegradable adhesive (right) compared to a conventional hair transplantation method (left) that transplants hair containing hair follicles. After applying an adhesive to the tip of the hair, it is fixed to the skin by implanting it through a subcutaneous injection, and repeated treatment is possible. (b) Initial animal test results. One day after 15 hair transplantation, 12 strands of hair remain. If you pull the 3 strands of hair, you can see that the whole body is pulled up, indicating that it is firmly implanted into the skin. All strands of hair applied without the new adhesive material fell off, and in the case of adhesive without heat treatment, the efficiency was 1/7.
This research was conducted by first co-authors Dr. Jongmin Park (currently a senior researcher at the Korea Research Institute of Chemical Technology) from Professor Myeongeun Seo’s team and Dr. Eunsook Park from Professor Haeshin Lee’s team in the KAIST Department of Chemistry, and through joint research with the teams led by Professor Hyungjun Kim from the KAIST Department of Chemistry and Professor Siyoung Choi from the Department of Chemical and Biomolecular Engineering. The research was published online on August 22 in the international journal Au (JACS Au) under the title Biodegradable Block Copolymer-Tannic Acid Glue.
This study was funded by the Support Research Under Protection Project of the National Research Foundation (NRF), Leading Research Center Support Project (Research Center for Multiscale Chiral Structure), Biodegradable Plastics Commercialization and Demonstration Project by the Ministry of Trade and Industry, and institutional funding from the Korea Research Institute of Chemical Technology.
2022.10.07 View 11672 -
 Establishing a novel strategy to tackle Huntington’s disease
A platform to take on the Huntington’s disease via an innovative approach established by KAIST’s researchers through international collaboration with scientists in the Netherlands, France, and Sweden.
Through an international joint research effort involving ProQR Therapeutics of the Netherlands, Université Grenoble Alpes of France, and KTH Royal Institute of Technology of Sweden, Professor Ji-Soon Song's research team in the Department of Biological Sciences and KAIST Institute for BioCentury of KAIST, established a noble strategy to treat Huntington's disease. The new works showed that the protein converted from disease form to its disease-free form maintains its original function, providing new roadblocks to approach Huntington’s disease.
This research, titled, “A pathogenic-proteolysis resistant huntingtin isoform induced by an antisense oligonucleotide maintains huntingtin function”, co-authored by Hyeongju Kim, was published in the online edition of 'Journal of Clinical Investigation Insight' on August 9, 2022.
Huntington's disease is a dominantly inherited neurodegenerative disease and is caused by a mutation in a protein called ‘huntingtin’, which adds a distinctive feature of an expanded stretch of glutamine amino acids called polyglutamine to the protein. It is estimated that one in every 10,000 have Huntington's disease in United States. The patients would suffer a decade of regression before death, and, thus far, there is no known cure for the disease.
The cleavage near the stretched polyglutamine in mutated huntingtin is known to be the cause of the Huntington’s disease. However, as huntingtin protein is required for the development and normal function of the brain, it is critical to specifically eliminate the disease-causing protein while maintaining the ones that are still normally functioning.
The research team showed that huntingtin delta 12, the converted form of huntingtin that is resistant to developing cleavages at the ends of the protein, the known cause of the Huntington’s disease (HD), alleviated the disease’s symptoms while maintaining the functions of normal huntingtin.
Figure. Huntington's disease resistance huntingtin protein induced by antisense oligonucleotide (AON) is resistant to Caspase-6 cleavage, therefore, does not cause Huntington’s disease while maintaining normal functions of huntingtin.
The research was welcomed as it is sure to fuel innovate strategies to tackle Huntington’s disease without altering the essential function of huntingtin.
This work was supported by a Global Research Lab grant from the National Research Foundation of Korea (NRF) and by a EUREKA Eurostars 2 grant from European Union Horizon 2020.
2022.09.02 View 7641
Establishing a novel strategy to tackle Huntington’s disease
A platform to take on the Huntington’s disease via an innovative approach established by KAIST’s researchers through international collaboration with scientists in the Netherlands, France, and Sweden.
Through an international joint research effort involving ProQR Therapeutics of the Netherlands, Université Grenoble Alpes of France, and KTH Royal Institute of Technology of Sweden, Professor Ji-Soon Song's research team in the Department of Biological Sciences and KAIST Institute for BioCentury of KAIST, established a noble strategy to treat Huntington's disease. The new works showed that the protein converted from disease form to its disease-free form maintains its original function, providing new roadblocks to approach Huntington’s disease.
This research, titled, “A pathogenic-proteolysis resistant huntingtin isoform induced by an antisense oligonucleotide maintains huntingtin function”, co-authored by Hyeongju Kim, was published in the online edition of 'Journal of Clinical Investigation Insight' on August 9, 2022.
Huntington's disease is a dominantly inherited neurodegenerative disease and is caused by a mutation in a protein called ‘huntingtin’, which adds a distinctive feature of an expanded stretch of glutamine amino acids called polyglutamine to the protein. It is estimated that one in every 10,000 have Huntington's disease in United States. The patients would suffer a decade of regression before death, and, thus far, there is no known cure for the disease.
The cleavage near the stretched polyglutamine in mutated huntingtin is known to be the cause of the Huntington’s disease. However, as huntingtin protein is required for the development and normal function of the brain, it is critical to specifically eliminate the disease-causing protein while maintaining the ones that are still normally functioning.
The research team showed that huntingtin delta 12, the converted form of huntingtin that is resistant to developing cleavages at the ends of the protein, the known cause of the Huntington’s disease (HD), alleviated the disease’s symptoms while maintaining the functions of normal huntingtin.
Figure. Huntington's disease resistance huntingtin protein induced by antisense oligonucleotide (AON) is resistant to Caspase-6 cleavage, therefore, does not cause Huntington’s disease while maintaining normal functions of huntingtin.
The research was welcomed as it is sure to fuel innovate strategies to tackle Huntington’s disease without altering the essential function of huntingtin.
This work was supported by a Global Research Lab grant from the National Research Foundation of Korea (NRF) and by a EUREKA Eurostars 2 grant from European Union Horizon 2020.
2022.09.02 View 7641 -
 Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 15158
Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 15158 -
 A New Therapeutic Drug for Alzheimer’s Disease without Inflammatory Side Effects
Although Aduhelm, a monoclonal antibody targeting amyloid beta (Aβ), recently became the first US FDA approved drug for Alzheimer’s disease (AD) based on its ability to decrease Aβ plaque burden in AD patients, its effect on cognitive improvement is still controversial. Moreover, about 40% of the patients treated with this antibody experienced serious side effects including cerebral edemas (ARIA-E) and hemorrhages (ARIA-H) that are likely related to inflammatory responses in the brain when the Aβ antibody binds Fc receptors (FCR) of immune cells such as microglia and macrophages.
These inflammatory side effects can cause neuronal cell death and synapse elimination by activated microglia, and even have the potential to exacerbate cognitive impairment in AD patients. Thus, current Aβ antibody-based immunotherapy holds the inherent risk of doing more harm than good due to their inflammatory side effects.
To overcome these problems, a team of researchers at KAIST in South Korea has developed a novel fusion protein drug, αAβ-Gas6, which efficiently eliminates Aβ via an entirely different mechanism than Aβ antibody-based immunotherapy. In a mouse model of AD, αAβ-Gas6 not only removed Aβ with higher potency, but also circumvented the neurotoxic inflammatory side effects associated with conventional antibody treatments.
Their findings were published on August 4 in Nature Medicine.
Schematic of a chimeric Gas6 fusion protein. A single chain variable fragment (scFv) of
an Amyloid β (Aβ)-targeting monoclonal antibody is fused with a truncated receptor binding
domain of Gas6, a bridging molecule for the clearance of dead cells via TAM (TYRO3, AXL,
and MERTK) receptors, which are expressed by microglia and astrocytes.
“FcR activation by Aβ targeting antibodies induces microglia-mediated Aβ phagocytosis, but it also produces inflammatory signals, inevitably damaging brain tissues,” said paper authors Chan Hyuk Kim and Won-Suk Chung, associate professors in the Department of Biological Sciences at KAIST.
“Therefore, we utilized efferocytosis, a cellular process by which dead cells are removed by phagocytes as an alternative pathway for the clearance of Aβ in the brain,” Prof. Kim and Chung said. “Efferocytosis is accompanied by anti-inflammatory responses to maintain tissue homeostasis. To exploit this process, we engineered Gas6, a soluble adaptor protein that mediates efferocytosis via TAM phagocytic receptors in such a way that its target specificity was redirected from dead cells to Aβ plaques.”
The professors and their team demonstrated that the resulting αAβ-Gas6 induced Aβ engulfment by activating not only microglial but also astrocytic phagocytosis since TAM phagocytic receptors are highly expressed by these two major phagocytes in the brain. Importantly, αAβ-Gas6 promoted the robust uptake of Aβ without showing any signs of inflammation and neurotoxicity, which contrasts sharply with the treatment using an Aβ monoclonal antibody. Moreover, they showed that αAβ-Gas6 substantially reduced excessive synapse elimination by microglia, consequently leading to better behavioral rescues in AD model mice.
“By using a mouse model of cerebral amyloid angiopathy (CAA), a cerebrovascular disorder caused by the deposition of Aβ within the walls of the brain’s blood vessels, we also showed that the intrathecal administration of Gas6 fusion protein significantly eliminated cerebrovascular amyloids, along with a reduction of microhemorrhages. These data demonstrate that aAb-Gas6 is a potent therapeutic agent in eliminating Aβ without exacerbating CAA-related microhemorrhages.”
The resulting αAβ-Gas6 clears Aβ oligomers and fibrils without causing neurotoxicity (a-b, neurons: red, and fragmented axons: yellow) and proinflammatory responses (c, TNF release), which are conversely exacerbated by the treatment of an Aβ-targeting monoclonal antibody (Aducanumab).
Professors Kim and Chung noted, “We believe our approach can be a breakthrough in treating AD without causing inflammatory side effects and synapse loss. Our approach holds promise as a novel therapeutic platform that is applicable to more than AD. By modifying the target-specificity of the fusion protein, the Gas6-fusion protein can be applied to various neurological disorders as well as autoimmune diseases affected by toxic molecules that should be removed without causing inflammatory responses.”
The number and total area of Aβ plaques (Thioflavin-T, green) were significantly reduced in αAβ-Gas6-treated AD mouse brains compared to Aducanumab-treated ones (a, b). The cognitive functions of AD model mice were significantly rescued by αAβ-Gas6 treatment, whereas Aducanumab-treated AD mice showed partial rescue in these cognitive tests (c-e).
Professors Kim and Chung founded “Illimis Therapeutics” based on this strategy of designing chimeric Gas6 fusion proteins that would remove toxic aggregates from the nervous system. Through this company, they are planning to further develop various Gas6-fusion proteins not only for Ab but also for Tau to treat AD symptoms.
This work was supported by KAIST and the Korea Health Technology R&D Project that was administered by the Korea Health Industry Development Institute (KHIDI) and the Korea Dementia Research Center (KDRC) funded by the Ministry of Health & Welfare (MOHW) and the Ministry of Science and ICT (MSIT), and KAIST.
Other contributors include Hyuncheol Jung and Se Young Lee, Sungjoon Lim, Hyeong Ryeol Choi, Yeseong Choi, Minjin Kim, Segi Kim, the Department of Biological Sciences, and the Korea Advanced Institute of Science and Technology (KAIST).
To receive more up-to-date information on this new development, follow “Illimis Therapeutics” on twitter @Illimistx.
2022.08.05 View 12648
A New Therapeutic Drug for Alzheimer’s Disease without Inflammatory Side Effects
Although Aduhelm, a monoclonal antibody targeting amyloid beta (Aβ), recently became the first US FDA approved drug for Alzheimer’s disease (AD) based on its ability to decrease Aβ plaque burden in AD patients, its effect on cognitive improvement is still controversial. Moreover, about 40% of the patients treated with this antibody experienced serious side effects including cerebral edemas (ARIA-E) and hemorrhages (ARIA-H) that are likely related to inflammatory responses in the brain when the Aβ antibody binds Fc receptors (FCR) of immune cells such as microglia and macrophages.
These inflammatory side effects can cause neuronal cell death and synapse elimination by activated microglia, and even have the potential to exacerbate cognitive impairment in AD patients. Thus, current Aβ antibody-based immunotherapy holds the inherent risk of doing more harm than good due to their inflammatory side effects.
To overcome these problems, a team of researchers at KAIST in South Korea has developed a novel fusion protein drug, αAβ-Gas6, which efficiently eliminates Aβ via an entirely different mechanism than Aβ antibody-based immunotherapy. In a mouse model of AD, αAβ-Gas6 not only removed Aβ with higher potency, but also circumvented the neurotoxic inflammatory side effects associated with conventional antibody treatments.
Their findings were published on August 4 in Nature Medicine.
Schematic of a chimeric Gas6 fusion protein. A single chain variable fragment (scFv) of
an Amyloid β (Aβ)-targeting monoclonal antibody is fused with a truncated receptor binding
domain of Gas6, a bridging molecule for the clearance of dead cells via TAM (TYRO3, AXL,
and MERTK) receptors, which are expressed by microglia and astrocytes.
“FcR activation by Aβ targeting antibodies induces microglia-mediated Aβ phagocytosis, but it also produces inflammatory signals, inevitably damaging brain tissues,” said paper authors Chan Hyuk Kim and Won-Suk Chung, associate professors in the Department of Biological Sciences at KAIST.
“Therefore, we utilized efferocytosis, a cellular process by which dead cells are removed by phagocytes as an alternative pathway for the clearance of Aβ in the brain,” Prof. Kim and Chung said. “Efferocytosis is accompanied by anti-inflammatory responses to maintain tissue homeostasis. To exploit this process, we engineered Gas6, a soluble adaptor protein that mediates efferocytosis via TAM phagocytic receptors in such a way that its target specificity was redirected from dead cells to Aβ plaques.”
The professors and their team demonstrated that the resulting αAβ-Gas6 induced Aβ engulfment by activating not only microglial but also astrocytic phagocytosis since TAM phagocytic receptors are highly expressed by these two major phagocytes in the brain. Importantly, αAβ-Gas6 promoted the robust uptake of Aβ without showing any signs of inflammation and neurotoxicity, which contrasts sharply with the treatment using an Aβ monoclonal antibody. Moreover, they showed that αAβ-Gas6 substantially reduced excessive synapse elimination by microglia, consequently leading to better behavioral rescues in AD model mice.
“By using a mouse model of cerebral amyloid angiopathy (CAA), a cerebrovascular disorder caused by the deposition of Aβ within the walls of the brain’s blood vessels, we also showed that the intrathecal administration of Gas6 fusion protein significantly eliminated cerebrovascular amyloids, along with a reduction of microhemorrhages. These data demonstrate that aAb-Gas6 is a potent therapeutic agent in eliminating Aβ without exacerbating CAA-related microhemorrhages.”
The resulting αAβ-Gas6 clears Aβ oligomers and fibrils without causing neurotoxicity (a-b, neurons: red, and fragmented axons: yellow) and proinflammatory responses (c, TNF release), which are conversely exacerbated by the treatment of an Aβ-targeting monoclonal antibody (Aducanumab).
Professors Kim and Chung noted, “We believe our approach can be a breakthrough in treating AD without causing inflammatory side effects and synapse loss. Our approach holds promise as a novel therapeutic platform that is applicable to more than AD. By modifying the target-specificity of the fusion protein, the Gas6-fusion protein can be applied to various neurological disorders as well as autoimmune diseases affected by toxic molecules that should be removed without causing inflammatory responses.”
The number and total area of Aβ plaques (Thioflavin-T, green) were significantly reduced in αAβ-Gas6-treated AD mouse brains compared to Aducanumab-treated ones (a, b). The cognitive functions of AD model mice were significantly rescued by αAβ-Gas6 treatment, whereas Aducanumab-treated AD mice showed partial rescue in these cognitive tests (c-e).
Professors Kim and Chung founded “Illimis Therapeutics” based on this strategy of designing chimeric Gas6 fusion proteins that would remove toxic aggregates from the nervous system. Through this company, they are planning to further develop various Gas6-fusion proteins not only for Ab but also for Tau to treat AD symptoms.
This work was supported by KAIST and the Korea Health Technology R&D Project that was administered by the Korea Health Industry Development Institute (KHIDI) and the Korea Dementia Research Center (KDRC) funded by the Ministry of Health & Welfare (MOHW) and the Ministry of Science and ICT (MSIT), and KAIST.
Other contributors include Hyuncheol Jung and Se Young Lee, Sungjoon Lim, Hyeong Ryeol Choi, Yeseong Choi, Minjin Kim, Segi Kim, the Department of Biological Sciences, and the Korea Advanced Institute of Science and Technology (KAIST).
To receive more up-to-date information on this new development, follow “Illimis Therapeutics” on twitter @Illimistx.
2022.08.05 View 12648 -
 President-Elect Suk-Yeol Yoon Meets and Talks with KAIST Students
President-Elect Yoon stresses science and technology-powered economic growth during his visit to KAIST
Korean President-elect Suk-Yeol Yoon stressed that semiconductors are the key strategical industry that will take the lead during the fourth industrial revolution powered by AI and data during a meeting with KAIST graduate students on April 29. President-elect Yoon promised systemic policy support for making science and technology breakthroughs possible and better rewarding young researchers who are devoted to advances in R&D during his meeting at KAIST.
Before he met with the students, he toured the National Nanofab Center, which is affiliated with KAIST, and was briefed on the center’s role and responsibilities.
President-elect Yoon, who will take office on May 10, said that the best way to ensure prompt growth in Korea’s aging society hinges on advances in science and technology. “All-out investments in science and technology will help us move forward to improve people’s quality of life and lessen the social divide,” he explained.
Eight Master’s and PhD candidates majoring in nuclear engineering, AI robotics, semiconductors, electrical engineering, aerospace, and bioengineering attended the meeting with President-elect Yoon. The students asked for help dealing with the challenges they are experiencing while researching and called for deregulation in the process of forming startups.
PhD candidate Jae Wan Cho from the Department Nuclear and Quantum Engineering stressed the importance of energy security. He asked for the prompt development of new types of nuclear reactors such as small modular reactors, adding, “Korea has very excellent technologies in nuclear plant construction and parts manufacturing, but lags behind in the new types of nuclear reactors. This sector will develop new energy markets and create synergy along with the shipbuilding industry, which will emerge as new pillars of our export.”
Student entrepreneurs such as PhD candidate Kwang Min Kim from the Department of Bio and Brain Engineering and PhD candidate Dong Yoon Shin from the Department of Mechanical and Aerospace Engineering asked for more deregulation in the process of creating startups. PhD candidate Dong Hon Lee from the School of Electrical Engineering pointed out the insecure future caused by the ‘special research fellow system,’ where the number of fellows who have been designated alternative military service has drastically decreased.
2022.05.02 View 6429
President-Elect Suk-Yeol Yoon Meets and Talks with KAIST Students
President-Elect Yoon stresses science and technology-powered economic growth during his visit to KAIST
Korean President-elect Suk-Yeol Yoon stressed that semiconductors are the key strategical industry that will take the lead during the fourth industrial revolution powered by AI and data during a meeting with KAIST graduate students on April 29. President-elect Yoon promised systemic policy support for making science and technology breakthroughs possible and better rewarding young researchers who are devoted to advances in R&D during his meeting at KAIST.
Before he met with the students, he toured the National Nanofab Center, which is affiliated with KAIST, and was briefed on the center’s role and responsibilities.
President-elect Yoon, who will take office on May 10, said that the best way to ensure prompt growth in Korea’s aging society hinges on advances in science and technology. “All-out investments in science and technology will help us move forward to improve people’s quality of life and lessen the social divide,” he explained.
Eight Master’s and PhD candidates majoring in nuclear engineering, AI robotics, semiconductors, electrical engineering, aerospace, and bioengineering attended the meeting with President-elect Yoon. The students asked for help dealing with the challenges they are experiencing while researching and called for deregulation in the process of forming startups.
PhD candidate Jae Wan Cho from the Department Nuclear and Quantum Engineering stressed the importance of energy security. He asked for the prompt development of new types of nuclear reactors such as small modular reactors, adding, “Korea has very excellent technologies in nuclear plant construction and parts manufacturing, but lags behind in the new types of nuclear reactors. This sector will develop new energy markets and create synergy along with the shipbuilding industry, which will emerge as new pillars of our export.”
Student entrepreneurs such as PhD candidate Kwang Min Kim from the Department of Bio and Brain Engineering and PhD candidate Dong Yoon Shin from the Department of Mechanical and Aerospace Engineering asked for more deregulation in the process of creating startups. PhD candidate Dong Hon Lee from the School of Electrical Engineering pointed out the insecure future caused by the ‘special research fellow system,’ where the number of fellows who have been designated alternative military service has drastically decreased.
2022.05.02 View 6429 -
 Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 11652
Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 11652