Sang+Yup+Lee
-
 A KAIST Research Team Produces Eco-Friendly Nylon with Engineered Bacterium
With worsening climate change and environmental issues, in recent years, there has been increased interest in the eco-friendly production of polymers like nylon.
On August 10, Dr. Taehee Han from a KAIST research team led by Distinguished Professor Sang Yup Lee in the Department of Chemical and Biomolecular Engineering revealed the successful development of a microbial strain that produces valerolactam, a monomer of nylon-5.
Valerolactam is an important monomer that constitutes nylon-5 and nylon-6,5. Nylon is the oldest synthetic polymer, and nylon-5 is one of its derivatives composed of monomers with five carbons, while nylon-5,6 is composed of two types of monomers with either five or six carbons. They not only have excellent processability, but are also light and tough, which allows them to be applied in a wide range of industrial sectors including clothing, badminton rackets, fishing nets, tents, and gear parts. Monomers are materials that can be built into polymers, and synthetic processes are what connects them into a polymer.
The chemical production of valerolactam, however, is based on petrochemistry, where extreme reaction conditions are required and toxic waste is produced. To solve these problems, efforts are being made to develop environmentally friendly and highly efficient microbial cell factories for lactam production. Systems metabolic engineering, a key strategy for effective microbial strain development, is a research field pioneered by Professor Sang Yup Lee.
Professor Lee’s team used metabolic engineering, a technique for manipulating microbial metabolic pathways, to construct a synthetic metabolic pathway for valerolactam production in Corynebacteriam glutamicum, a bacterium commonly used for amino acid production. With this, they successfully developed a microbial strain that utilizes biomass-derived glucose as a carbon source to produce high-value valerolactam.
In 2017, the team suggested a novel method that metabolically manipulates Escherichia coli to produce valerolactam. However, there were several limitations at the time including low producibility and the generation of harmful byproducts.
< Figure 1. Schematic graphical representation of the development of microorganisms that produce valerolactam, a nylon-5 monomer >
In this research, the team improved valerolactam producibility and incorporated an additional systems metabolic strategy to the developed microbial strain while eliminating the harmful byproducts. By removing the gene involved in the production of the main byproduct and through gene screening, the team successfully converted 5-aminovaleric acid, a byproduct and a precursor, into valerolactam.
Furthermore, by employing a strategy where the 5-aminovaleric acid-converting gene is inserted multiple times into the genome, the team strengthened the metabolic flux for valerolactam production. As a result, they reached a world-record concentration of 76.1 g/L, which is 6.17 times greater than what was previously reported.
This study was published in Metabolic Engineering on July 12, under the title, “Metabolic engineering of Corynebacterium glutamicum for the high-level production of valerolactam, a nylon-5 monomer”.
Dr. Taehee Han, the first author of the paper, said, “The significance of this research lies in our development of an environmentally friendly technology that efficiently produces monomer lactam for nylon production using microorganisms.” She added, “Through this technology, we will be able to take a step forward in replacing the petrochemical industry with a microorganism-based biopolymer industry.”
This work was supported by the “Development of Next-Generation Biofinery Platform Technologies for Leading Bio-based Chemicals Industry Project” funded by the Korean Ministry of Science and ICT.
2023.08.24 View 5017
A KAIST Research Team Produces Eco-Friendly Nylon with Engineered Bacterium
With worsening climate change and environmental issues, in recent years, there has been increased interest in the eco-friendly production of polymers like nylon.
On August 10, Dr. Taehee Han from a KAIST research team led by Distinguished Professor Sang Yup Lee in the Department of Chemical and Biomolecular Engineering revealed the successful development of a microbial strain that produces valerolactam, a monomer of nylon-5.
Valerolactam is an important monomer that constitutes nylon-5 and nylon-6,5. Nylon is the oldest synthetic polymer, and nylon-5 is one of its derivatives composed of monomers with five carbons, while nylon-5,6 is composed of two types of monomers with either five or six carbons. They not only have excellent processability, but are also light and tough, which allows them to be applied in a wide range of industrial sectors including clothing, badminton rackets, fishing nets, tents, and gear parts. Monomers are materials that can be built into polymers, and synthetic processes are what connects them into a polymer.
The chemical production of valerolactam, however, is based on petrochemistry, where extreme reaction conditions are required and toxic waste is produced. To solve these problems, efforts are being made to develop environmentally friendly and highly efficient microbial cell factories for lactam production. Systems metabolic engineering, a key strategy for effective microbial strain development, is a research field pioneered by Professor Sang Yup Lee.
Professor Lee’s team used metabolic engineering, a technique for manipulating microbial metabolic pathways, to construct a synthetic metabolic pathway for valerolactam production in Corynebacteriam glutamicum, a bacterium commonly used for amino acid production. With this, they successfully developed a microbial strain that utilizes biomass-derived glucose as a carbon source to produce high-value valerolactam.
In 2017, the team suggested a novel method that metabolically manipulates Escherichia coli to produce valerolactam. However, there were several limitations at the time including low producibility and the generation of harmful byproducts.
< Figure 1. Schematic graphical representation of the development of microorganisms that produce valerolactam, a nylon-5 monomer >
In this research, the team improved valerolactam producibility and incorporated an additional systems metabolic strategy to the developed microbial strain while eliminating the harmful byproducts. By removing the gene involved in the production of the main byproduct and through gene screening, the team successfully converted 5-aminovaleric acid, a byproduct and a precursor, into valerolactam.
Furthermore, by employing a strategy where the 5-aminovaleric acid-converting gene is inserted multiple times into the genome, the team strengthened the metabolic flux for valerolactam production. As a result, they reached a world-record concentration of 76.1 g/L, which is 6.17 times greater than what was previously reported.
This study was published in Metabolic Engineering on July 12, under the title, “Metabolic engineering of Corynebacterium glutamicum for the high-level production of valerolactam, a nylon-5 monomer”.
Dr. Taehee Han, the first author of the paper, said, “The significance of this research lies in our development of an environmentally friendly technology that efficiently produces monomer lactam for nylon production using microorganisms.” She added, “Through this technology, we will be able to take a step forward in replacing the petrochemical industry with a microorganism-based biopolymer industry.”
This work was supported by the “Development of Next-Generation Biofinery Platform Technologies for Leading Bio-based Chemicals Industry Project” funded by the Korean Ministry of Science and ICT.
2023.08.24 View 5017 -
 KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 7091
KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 7091 -
 Synthetic sRNAs to knockdown genes in medical and industrial bacteria
Bacteria are intimately involved in our daily lives. These microorganisms have been used in human history for food such as cheese, yogurt, and wine, In more recent years, through metabolic engineering, microorganisms been used extensively as microbial cell factories to manufacture plastics, feed for livestock, dietary supplements, and drugs. However, in addition to these bacteria that are beneficial to human lives, pathogens such as Pneumonia, Salmonella, and Staphylococcus that cause various infectious diseases are also ubiquitously present. It is important to be able to metabolically control these beneficial industrial bacteria for high value-added chemicals production and to manipulate harmful pathogens to suppress its pathogenic traits.
KAIST (President Kwang Hyung Lee) announced on the 10th that a research team led by Distinguished Professor Sang Yup Lee of the Department of Biochemical Engineering has developed a new sRNA tool that can effectively inhibit target genes in various bacteria, including both Gram-negative and Gram-positive bacteria. The research results were published online on April 24 in Nature Communications.
※ Thesis title: Targeted and high-throughput gene knockdown in diverse bacteria using synthetic sRNAs
※ Author information : Jae Sung Cho (co-1st), Dongsoo Yang (co-1st), Cindy Pricilia Surya Prabowo (co-author), Mohammad Rifqi Ghiffary (co-author), Taehee Han (co-author), Kyeong Rok Choi (co-author), Cheon Woo Moon (co-author), Hengrui Zhou (co-author), Jae Yong Ryu (co-author), Hyun Uk Kim (co-author) and Sang Yup Lee (corresponding author).
sRNA is an effective tool for synthesizing and regulating target genes in E. coli, but it has been difficult to apply to industrially useful Gram-positive bacteria such as Bacillus subtilis and Corynebacterium in addition to Gram-negative bacteria such as E. coli.
To address this issue, a research team led by Distinguished Professor Lee Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST developed a new sRNA platform that can effectively suppress target genes in various bacteria, including both Gram-negative and positive bacteria. The research team surveyed thousands of microbial-derived sRNA systems in the microbial database, and eventually designated the sRNA system derived from 'Bacillus subtilis' that showed the highest gene knockdown efficiency, and designated it as “Broad-Host-Range sRNA”, or BHR-sRNA.
A similar well-known system is the CRISPR interference (CRISPRi) system, which is a modified CRISPR system that knocks down gene expression by suppressing the gene transcription process. However, the Cas9 protein in the CRISPRi system has a very high molecular weight, and there have been reports growth inhibition in bacteria. The BHR-sRNA system developed in this study did not affect bacterial growth while showing similar gene knockdown efficiencies to CRISPRi.
< Figure 1. a) Schematic illustration demonstrating the mechanism of syntetic sRNA b) Phylogenetic tree of the 16 Gram-negative and Gram-positive bacterial species tested for gene knockdown by the BHR-sRNA system. >
To validate the versatility of the BHR-sRNA system, 16 different gram-negative and gram-positive bacteria were selected and tested, where the BHR-sRNA system worked successfully in 15 of them. In addition, it was demonstrated that the gene knockdown capability was more effective than that of the existing E. coli-based sRNA system in 10 bacteria. The BHR-sRNA system proved to be a universal tool capable of effectively inhibiting gene expression in various bacteria.
In order to address the problem of antibiotic-resistant pathogens that have recently become more serious, the BHR-sRNA was demonstrated to suppress the pathogenicity by suppressing the gene producing the virulence factor. By using BHR-sRNA, biofilm formation, one of the factors resulting in antibiotic resistance, was inhibited by 73% in Staphylococcus epidermidis a pathogen that can cause hospital-acquired infections. Antibiotic resistance was also weakened by 58% in the pneumonia causing bacteria Klebsiella pneumoniae. In addition, BHR-sRNA was applied to industrial bacteria to develop microbial cell factories to produce high value-added chemicals with better production performance. Notably, superior industrial strains were constructed with the aid of BHR-sRNA to produce the following chemicals: valerolactam, a raw material for polyamide polymers, methyl-anthranilate, a grape-flavor food additive, and indigoidine, a blue-toned natural dye.
The BHR-sRNA developed through this study will help expedite the commercialization of bioprocesses to produce high value-added compounds and materials such as artificial meat, jet fuel, health supplements, pharmaceuticals, and plastics. It is also anticipated that to help eradicating antibiotic-resistant pathogens in preparation for another upcoming pandemic. “In the past, we could only develop new tools for gene knockdown for each bacterium, but now we have developed a tool that works for a variety of bacteria” said Distinguished Professor Sang Yup Lee.
This work was supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project from NRF supported by the Korean MSIT.
2023.05.10 View 6459
Synthetic sRNAs to knockdown genes in medical and industrial bacteria
Bacteria are intimately involved in our daily lives. These microorganisms have been used in human history for food such as cheese, yogurt, and wine, In more recent years, through metabolic engineering, microorganisms been used extensively as microbial cell factories to manufacture plastics, feed for livestock, dietary supplements, and drugs. However, in addition to these bacteria that are beneficial to human lives, pathogens such as Pneumonia, Salmonella, and Staphylococcus that cause various infectious diseases are also ubiquitously present. It is important to be able to metabolically control these beneficial industrial bacteria for high value-added chemicals production and to manipulate harmful pathogens to suppress its pathogenic traits.
KAIST (President Kwang Hyung Lee) announced on the 10th that a research team led by Distinguished Professor Sang Yup Lee of the Department of Biochemical Engineering has developed a new sRNA tool that can effectively inhibit target genes in various bacteria, including both Gram-negative and Gram-positive bacteria. The research results were published online on April 24 in Nature Communications.
※ Thesis title: Targeted and high-throughput gene knockdown in diverse bacteria using synthetic sRNAs
※ Author information : Jae Sung Cho (co-1st), Dongsoo Yang (co-1st), Cindy Pricilia Surya Prabowo (co-author), Mohammad Rifqi Ghiffary (co-author), Taehee Han (co-author), Kyeong Rok Choi (co-author), Cheon Woo Moon (co-author), Hengrui Zhou (co-author), Jae Yong Ryu (co-author), Hyun Uk Kim (co-author) and Sang Yup Lee (corresponding author).
sRNA is an effective tool for synthesizing and regulating target genes in E. coli, but it has been difficult to apply to industrially useful Gram-positive bacteria such as Bacillus subtilis and Corynebacterium in addition to Gram-negative bacteria such as E. coli.
To address this issue, a research team led by Distinguished Professor Lee Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST developed a new sRNA platform that can effectively suppress target genes in various bacteria, including both Gram-negative and positive bacteria. The research team surveyed thousands of microbial-derived sRNA systems in the microbial database, and eventually designated the sRNA system derived from 'Bacillus subtilis' that showed the highest gene knockdown efficiency, and designated it as “Broad-Host-Range sRNA”, or BHR-sRNA.
A similar well-known system is the CRISPR interference (CRISPRi) system, which is a modified CRISPR system that knocks down gene expression by suppressing the gene transcription process. However, the Cas9 protein in the CRISPRi system has a very high molecular weight, and there have been reports growth inhibition in bacteria. The BHR-sRNA system developed in this study did not affect bacterial growth while showing similar gene knockdown efficiencies to CRISPRi.
< Figure 1. a) Schematic illustration demonstrating the mechanism of syntetic sRNA b) Phylogenetic tree of the 16 Gram-negative and Gram-positive bacterial species tested for gene knockdown by the BHR-sRNA system. >
To validate the versatility of the BHR-sRNA system, 16 different gram-negative and gram-positive bacteria were selected and tested, where the BHR-sRNA system worked successfully in 15 of them. In addition, it was demonstrated that the gene knockdown capability was more effective than that of the existing E. coli-based sRNA system in 10 bacteria. The BHR-sRNA system proved to be a universal tool capable of effectively inhibiting gene expression in various bacteria.
In order to address the problem of antibiotic-resistant pathogens that have recently become more serious, the BHR-sRNA was demonstrated to suppress the pathogenicity by suppressing the gene producing the virulence factor. By using BHR-sRNA, biofilm formation, one of the factors resulting in antibiotic resistance, was inhibited by 73% in Staphylococcus epidermidis a pathogen that can cause hospital-acquired infections. Antibiotic resistance was also weakened by 58% in the pneumonia causing bacteria Klebsiella pneumoniae. In addition, BHR-sRNA was applied to industrial bacteria to develop microbial cell factories to produce high value-added chemicals with better production performance. Notably, superior industrial strains were constructed with the aid of BHR-sRNA to produce the following chemicals: valerolactam, a raw material for polyamide polymers, methyl-anthranilate, a grape-flavor food additive, and indigoidine, a blue-toned natural dye.
The BHR-sRNA developed through this study will help expedite the commercialization of bioprocesses to produce high value-added compounds and materials such as artificial meat, jet fuel, health supplements, pharmaceuticals, and plastics. It is also anticipated that to help eradicating antibiotic-resistant pathogens in preparation for another upcoming pandemic. “In the past, we could only develop new tools for gene knockdown for each bacterium, but now we have developed a tool that works for a variety of bacteria” said Distinguished Professor Sang Yup Lee.
This work was supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project from NRF supported by the Korean MSIT.
2023.05.10 View 6459 -
 A biohybrid system to extract 20 times more bioplastic from CO2 developed by KAIST researchers
As the issues surrounding global climate change intensify, more attention and determined efforts are required to re-grasp the issue as a state of “crisis” and respond to it properly. Among the various methods of recycling CO2, the electrochemical CO2 conversion technology is a technology that can convert CO2 into useful chemical substances using electrical energy. Since it is easy to operate facilities and can use the electricity from renewable sources like the solar cells or the wind power, it has received a lot of attention as an eco-friendly technology can contribute to reducing greenhouse gases and achieve carbon neutrality.
KAIST (President Kwang Hyung Lee) announced on the 30th that the joint research team led by Professor Hyunjoo Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering succeeded in developing a technology that produces bioplastics from CO2 with high efficiency by developing a hybrid system that interlinked the electrochemical CO2 conversion and microbial bio conversion methods together. The results of the research, which showed the world's highest productivity by more than 20 times compared to similar systems, were published online on March 27th in the "Proceedings of the National Academy of Sciences (PNAS)".
※ Paper title: Biohybrid CO2 electrolysis for the direct synthesis of polyesters from CO2
※ Author information: Jinkyu Lim (currently at Stanford Linear Accelerator Center, co-first author), So Young Choi (KAIST, co-first author), Jae Won Lee (KAIST, co-first author), Hyunjoo Lee (KAIST, corresponding author), Sang Yup Lee (KAIST, corresponding author)
For the efficient conversion of CO2, high-efficiency electrode catalysts and systems are actively being developed. As conversion products, only compounds containing one or up to three carbon atoms are produced on a limited basis. Compounds of one carbon, such as CO, formic acid, and ethylene, are produced with relatively high efficiency. Liquid compounds of several carbons, such as ethanol, acetic acid, and propanol, can also be produced by these systems, but due to the nature of the chemical reaction that requires more electrons, there are limitations involving the conversion efficiency and the product selection.
Accordingly, a joint research team led by Professor Hyunjoo Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST developed a technology to produce bioplastics from CO2 by linking electrochemical conversion technology with bioconversion method that uses microorganisms.
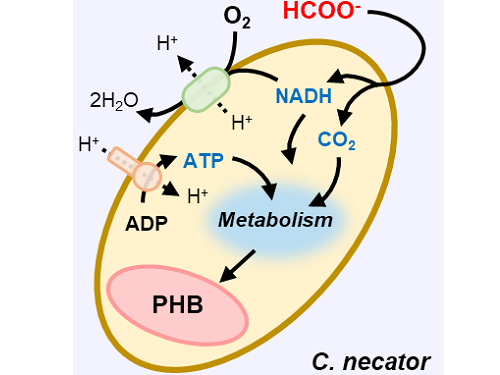
This electrochemical-bio hybrid system is in the form of having an electrolyzer, in which electrochemical conversion reactions occur, connected to a fermenter, in which microorganisms are cultured. When CO2 is converted to formic acid in the electrolyzer, and it is fed into the fermenter in which the microbes like the Cupriavidus necator, in this case, consumes the carbon source to produce polyhydroxyalkanoate (PHA), a microbial-derived bioplastic.
According to the research results of the existing hybrid concepts, there was a disadvantage of having low productivity or stopping at a non-continuous process due to problems of low efficiency of the electrolysis and irregular results arising from the culturing conditions of the microbes.
In order to overcome these problems, the joint research team made formic acid with a gas diffusion electrode using gaseous CO2. In addition, the team developed a 'physiologically compatible catholyte' that can be used as a culture medium for microorganisms as well as an electrolyte that allows the electrolysis to occur sufficiently without inhibiting the growth of microorganisms, without having to have a additional separation and purification process, which allowed the acide to be supplied directly to microorganisms.
Through this, the electrolyte solution containing formic acid made from CO2 enters the fermentation tank, is used for microbial culture, and enters the electrolyzer to be circulated, maximizing the utilization of the electrolyte solution and remaining formic acid. In addition, a filter was installed to ensure that only the electrolyte solution with any and all microorganisms that can affect the electrosis filtered out is supplied back to the electrolyzer, and that the microorganisms exist only in the fermenter, designing the two system to work well together with utmost efficiency.
Through the developed hybrid system, the produced bioplastic, poly-3-hydroxybutyrate (PHB), of up to 83% of the cell dry weight was produced from CO2, which produced 1.38g of PHB from a 4 cm2 electrode, which is the world's first gram(g) level production and is more than 20 times more productive than previous research. In addition, the hybrid system is expected to be applied to various industrial processes in the future as it shows promises of the continuous culture system.
The corresponding authors, Professor Hyunjoo Lee and Distinguished Professor Sang Yup Lee noted that “The results of this research are technologies that can be applied to the production of various chemical substances as well as bioplastics, and are expected to be used as key parts needed in achieving carbon neutrality in the future.”
This research was received and performed with the supports from the CO2 Reduction Catalyst and Energy Device Technology Development Project, the Heterogeneous Atomic Catalyst Control Project, and the Next-generation Biorefinery Source Technology Development Project to lead the Biochemical Industry of the Oil-replacement Eco-friendly Chemical Technology Development Program by the Ministry of Science and ICT.
Figure 1. Schematic diagram and photo of the biohybrid CO2 electrolysis system.
(A) A conceptual scheme and (B) a photograph of the biohybrid CO2 electrolysis system. (C) A detailed scheme of reaction inside the system. Gaseous CO2 was converted to formate in the electrolyzer, and the formate was converted to PHB by the cells in the fermenter. The catholyte was developed so that it is compatible with both CO2 electrolysis and fermentation and was continuously circulated.
2023.03.30 View 8953
A biohybrid system to extract 20 times more bioplastic from CO2 developed by KAIST researchers
As the issues surrounding global climate change intensify, more attention and determined efforts are required to re-grasp the issue as a state of “crisis” and respond to it properly. Among the various methods of recycling CO2, the electrochemical CO2 conversion technology is a technology that can convert CO2 into useful chemical substances using electrical energy. Since it is easy to operate facilities and can use the electricity from renewable sources like the solar cells or the wind power, it has received a lot of attention as an eco-friendly technology can contribute to reducing greenhouse gases and achieve carbon neutrality.
KAIST (President Kwang Hyung Lee) announced on the 30th that the joint research team led by Professor Hyunjoo Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering succeeded in developing a technology that produces bioplastics from CO2 with high efficiency by developing a hybrid system that interlinked the electrochemical CO2 conversion and microbial bio conversion methods together. The results of the research, which showed the world's highest productivity by more than 20 times compared to similar systems, were published online on March 27th in the "Proceedings of the National Academy of Sciences (PNAS)".
※ Paper title: Biohybrid CO2 electrolysis for the direct synthesis of polyesters from CO2
※ Author information: Jinkyu Lim (currently at Stanford Linear Accelerator Center, co-first author), So Young Choi (KAIST, co-first author), Jae Won Lee (KAIST, co-first author), Hyunjoo Lee (KAIST, corresponding author), Sang Yup Lee (KAIST, corresponding author)
For the efficient conversion of CO2, high-efficiency electrode catalysts and systems are actively being developed. As conversion products, only compounds containing one or up to three carbon atoms are produced on a limited basis. Compounds of one carbon, such as CO, formic acid, and ethylene, are produced with relatively high efficiency. Liquid compounds of several carbons, such as ethanol, acetic acid, and propanol, can also be produced by these systems, but due to the nature of the chemical reaction that requires more electrons, there are limitations involving the conversion efficiency and the product selection.
Accordingly, a joint research team led by Professor Hyunjoo Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST developed a technology to produce bioplastics from CO2 by linking electrochemical conversion technology with bioconversion method that uses microorganisms.
This electrochemical-bio hybrid system is in the form of having an electrolyzer, in which electrochemical conversion reactions occur, connected to a fermenter, in which microorganisms are cultured. When CO2 is converted to formic acid in the electrolyzer, and it is fed into the fermenter in which the microbes like the Cupriavidus necator, in this case, consumes the carbon source to produce polyhydroxyalkanoate (PHA), a microbial-derived bioplastic.
According to the research results of the existing hybrid concepts, there was a disadvantage of having low productivity or stopping at a non-continuous process due to problems of low efficiency of the electrolysis and irregular results arising from the culturing conditions of the microbes.
In order to overcome these problems, the joint research team made formic acid with a gas diffusion electrode using gaseous CO2. In addition, the team developed a 'physiologically compatible catholyte' that can be used as a culture medium for microorganisms as well as an electrolyte that allows the electrolysis to occur sufficiently without inhibiting the growth of microorganisms, without having to have a additional separation and purification process, which allowed the acide to be supplied directly to microorganisms.
Through this, the electrolyte solution containing formic acid made from CO2 enters the fermentation tank, is used for microbial culture, and enters the electrolyzer to be circulated, maximizing the utilization of the electrolyte solution and remaining formic acid. In addition, a filter was installed to ensure that only the electrolyte solution with any and all microorganisms that can affect the electrosis filtered out is supplied back to the electrolyzer, and that the microorganisms exist only in the fermenter, designing the two system to work well together with utmost efficiency.
Through the developed hybrid system, the produced bioplastic, poly-3-hydroxybutyrate (PHB), of up to 83% of the cell dry weight was produced from CO2, which produced 1.38g of PHB from a 4 cm2 electrode, which is the world's first gram(g) level production and is more than 20 times more productive than previous research. In addition, the hybrid system is expected to be applied to various industrial processes in the future as it shows promises of the continuous culture system.
The corresponding authors, Professor Hyunjoo Lee and Distinguished Professor Sang Yup Lee noted that “The results of this research are technologies that can be applied to the production of various chemical substances as well as bioplastics, and are expected to be used as key parts needed in achieving carbon neutrality in the future.”
This research was received and performed with the supports from the CO2 Reduction Catalyst and Energy Device Technology Development Project, the Heterogeneous Atomic Catalyst Control Project, and the Next-generation Biorefinery Source Technology Development Project to lead the Biochemical Industry of the Oil-replacement Eco-friendly Chemical Technology Development Program by the Ministry of Science and ICT.
Figure 1. Schematic diagram and photo of the biohybrid CO2 electrolysis system.
(A) A conceptual scheme and (B) a photograph of the biohybrid CO2 electrolysis system. (C) A detailed scheme of reaction inside the system. Gaseous CO2 was converted to formate in the electrolyzer, and the formate was converted to PHB by the cells in the fermenter. The catholyte was developed so that it is compatible with both CO2 electrolysis and fermentation and was continuously circulated.
2023.03.30 View 8953 -
 KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
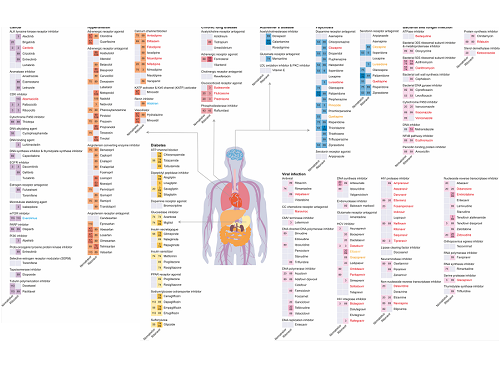
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 7102
KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 7102 -
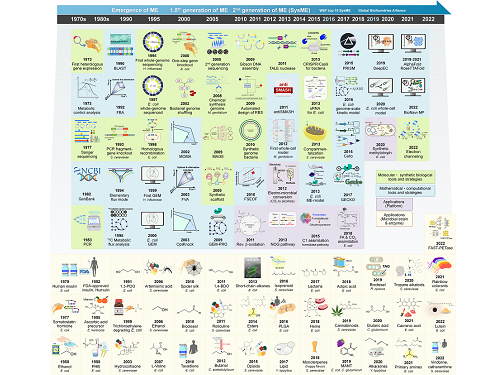 Overview of the 30-year history of metabolic engineering
< Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST >
A research team comprised of Gi Bae Kim, Dr. So Young Choi, Dr. In Jin Cho, Da-Hee Ahn, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST reported the 30-year history of metabolic engineering, highlighting examples of recent progress in the field and contributions to sustainability and health. Their paper “Metabolic engineering for sustainability and health” was published online in the 40th anniversary special issue of Trends in Biotechnology on January 10, 2023.
Metabolic engineering, a discipline of engineering that modifies cell phenotypes through molecular and genetic-level manipulations to improve cellular activities, has been studied since the early 1990s, and has progressed significantly over the past 30 years. In particular, metabolic engineering has enabled the engineering of microorganisms for the development of microbial cell factories capable of efficiently producing chemicals and materials as well as degrading recalcitrant contaminants.
This review article revisited how metabolic engineering has advanced over the past 30 years, from the advent of genetic engineering techniques such as recombinant DNA technologies to recent breakthroughs in systems metabolic engineering and data science aided by artificial intelligence. The research team highlighted momentous events and achievements in metabolic engineering, providing both trends and future directions in the field. Metabolic engineering’s contributions to bio-based sustainable chemicals and clean energy, health, and bioremediation were also reviewed. Finally, the research team shared their perspectives on the future challenges impacting metabolic engineering than must be overcome in order to achieve advancements in sustainability and health.
Distinguished Professor Sang Yup Lee said, “Replacing fossil resource-based chemical processes with bio-based sustainable processes for the production of chemicals, fuels, and materials using metabolic engineering has become our essential task for the future. By looking back on the 30+ years of metabolic engineering, we aimed to highlight the contributions of metabolic engineering to achieve sustainability and good health.” He added, “Metabolic engineering will play an increasingly important role as a key solution to the climate crisis, environmental pollution, food and energy shortages, and health problems in aging societies.”
< Figure: Metabolic Engineering Timeline >
2023.01.25 View 9317
Overview of the 30-year history of metabolic engineering
< Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST >
A research team comprised of Gi Bae Kim, Dr. So Young Choi, Dr. In Jin Cho, Da-Hee Ahn, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST reported the 30-year history of metabolic engineering, highlighting examples of recent progress in the field and contributions to sustainability and health. Their paper “Metabolic engineering for sustainability and health” was published online in the 40th anniversary special issue of Trends in Biotechnology on January 10, 2023.
Metabolic engineering, a discipline of engineering that modifies cell phenotypes through molecular and genetic-level manipulations to improve cellular activities, has been studied since the early 1990s, and has progressed significantly over the past 30 years. In particular, metabolic engineering has enabled the engineering of microorganisms for the development of microbial cell factories capable of efficiently producing chemicals and materials as well as degrading recalcitrant contaminants.
This review article revisited how metabolic engineering has advanced over the past 30 years, from the advent of genetic engineering techniques such as recombinant DNA technologies to recent breakthroughs in systems metabolic engineering and data science aided by artificial intelligence. The research team highlighted momentous events and achievements in metabolic engineering, providing both trends and future directions in the field. Metabolic engineering’s contributions to bio-based sustainable chemicals and clean energy, health, and bioremediation were also reviewed. Finally, the research team shared their perspectives on the future challenges impacting metabolic engineering than must be overcome in order to achieve advancements in sustainability and health.
Distinguished Professor Sang Yup Lee said, “Replacing fossil resource-based chemical processes with bio-based sustainable processes for the production of chemicals, fuels, and materials using metabolic engineering has become our essential task for the future. By looking back on the 30+ years of metabolic engineering, we aimed to highlight the contributions of metabolic engineering to achieve sustainability and good health.” He added, “Metabolic engineering will play an increasingly important role as a key solution to the climate crisis, environmental pollution, food and energy shortages, and health problems in aging societies.”
< Figure: Metabolic Engineering Timeline >
2023.01.25 View 9317 -
 Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 11601
Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 11601 -
 Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 12747
Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 12747 -
 Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 9022
Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 9022 -
 VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 9417
VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 9417 -
 Five Projects Ranked in the Top 100 for National R&D Excellence
Five KAIST research projects were selected as the 2021 Top 100 for National R&D Excellence by the Ministry of Science and ICT and the Korea Institute of Science & Technology Evaluation and Planning.
The five projects are:-The development of E. coli that proliferates with only formic acid and carbon dioxide by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering
-An original reverse aging technology that restores an old human skin cell into a younger one by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering-The development of next-generation high-efficiency perovskite-silicon tandem solar cells by Professor Byungha Shin from the Department of Materials Science and Engineering-Research on the effects of ultrafine dust in the atmosphere has on energy consumption by Professor Jiyong Eom from the School of Business and Technology Management-Research on a molecular trigger that controls the phase transformation of bio materials by Professor Myungchul Kim from the Department of Bio and Brain Engineering
Started in 2006, an Evaluation Committee composed of experts in industries, universities, and research institutes has made the preliminary selections of the most outstanding research projects based on their significance as a scientific and technological development and their socioeconomic effects. The finalists went through an open public evaluation. The final 100 studies are from six fields: 18 from mechanics & materials, 26 from biology & marine sciences, 19 from ICT & electronics, 10 from interdisciplinary research, and nine from natural science and infrastructure.
The selected 100 studies will receive a certificate and an award plaque from the minister of MSIT as well as additional points for business and institutional evaluations according to appropriate regulations, and the selected researchers will be strongly recommended as candidates for national meritorious awards.
In particular, to help the 100 selected research projects become more accessible for the general public, their main contents will be provided in a free e-book ‘The Top 100 for National R&D Excellence of 2021’ that will be available from online booksellers.
2022.02.17 View 10045
Five Projects Ranked in the Top 100 for National R&D Excellence
Five KAIST research projects were selected as the 2021 Top 100 for National R&D Excellence by the Ministry of Science and ICT and the Korea Institute of Science & Technology Evaluation and Planning.
The five projects are:-The development of E. coli that proliferates with only formic acid and carbon dioxide by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering
-An original reverse aging technology that restores an old human skin cell into a younger one by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering-The development of next-generation high-efficiency perovskite-silicon tandem solar cells by Professor Byungha Shin from the Department of Materials Science and Engineering-Research on the effects of ultrafine dust in the atmosphere has on energy consumption by Professor Jiyong Eom from the School of Business and Technology Management-Research on a molecular trigger that controls the phase transformation of bio materials by Professor Myungchul Kim from the Department of Bio and Brain Engineering
Started in 2006, an Evaluation Committee composed of experts in industries, universities, and research institutes has made the preliminary selections of the most outstanding research projects based on their significance as a scientific and technological development and their socioeconomic effects. The finalists went through an open public evaluation. The final 100 studies are from six fields: 18 from mechanics & materials, 26 from biology & marine sciences, 19 from ICT & electronics, 10 from interdisciplinary research, and nine from natural science and infrastructure.
The selected 100 studies will receive a certificate and an award plaque from the minister of MSIT as well as additional points for business and institutional evaluations according to appropriate regulations, and the selected researchers will be strongly recommended as candidates for national meritorious awards.
In particular, to help the 100 selected research projects become more accessible for the general public, their main contents will be provided in a free e-book ‘The Top 100 for National R&D Excellence of 2021’ that will be available from online booksellers.
2022.02.17 View 10045 -
 3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 12305
3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 12305