ACT
-
 KAIST Develops Healthcare Device Tracking Chronic Diabetic Wounds
A KAIST research team has developed an effective wireless system that monitors the wound healing process by tracking the spatiotemporal temperature changes and heat transfer characteristics of damaged areas such as diabetic wounds.
On the 5th of March, KAIST (represented by President Kwang Hyung Lee) announced that the research team led by Professor Kyeongha Kwon from KAIST’s School of Electrical Engineering, in association with Chung-Ang University professor Hanjun Ryu, developed digital healthcare technology that tracks the wound healing process in real time, which allows appropriate treatments to be administered.
< Figure 1. Schematic illustrations and diagrams of real-time wound monitoring systems. >
The skin serves as a barrier protecting the body from harmful substances, therefore damage to the skin may cause severe health risks to patients in need of intensive care. Especially in the case of diabetic patients, chronic wounds are easily formed due to complications in normal blood circulation and the wound healing process. In the United States alone, hundreds of billions of dollars of medical costs stem from regenerating the skin from such wounds. While various methods exist to promote wound healing, personalized management is essential depending on the condition of each patient's wounds.
Accordingly, the research team tracked the heating response within the wound by utilizing the differences in temperature between the damaged area and the surrounding healthy skin. They then measured heat transfer characteristics to observe moisture changes near the skin surface, ultimately establishing a basis for understanding the formation process of scar tissue. The team conducted experiments using diabetic mice models regarding the delay in wound healing under pathological conditions, and it was demonstrated that the collected data accurately tracks the wound healing process and the formation of scar tissue.
To minimize the tissue damage that may occur in the process of removing the tracking device after healing, the system integrates biodegradable sensor modules capable of natural decomposition within the body. These biodegradable modules disintegrate within the body after use, thus reducing the risk of additional discomfort or tissue damage upon device removal. Furthermore, the device could one day be used for monitoring inside the wound area as there is no need for removal.
Professor Kyeongha Kwon, who led the research, anticipates that continuous monitoring of wound temperature and heat transfer characteristics will enable medical professionals to more accurately assess the status of diabetic patients' wounds and provide appropriate treatment. He further predicted that the implementation of biodegradable sensors allows for the safe decomposition of the device after wound healing without the need for removal, making live monitoring possible not only in hospitals but also at home.
The research team plans to integrate antimicrobial materials into this device, aiming to expand its technological capabilities to enable the observation and prevention of inflammatory responses, bacterial infections, and other complications. The goal is to provide a multi-purpose wound monitoring platform capable of real-time antimicrobial monitoring in hospitals or homes by detecting changes in temperature and heat transfer characteristics indicative of infection levels.
< Image 1. Image of the bioresorbable temperature sensor >
The results of this study were published on February 19th in the international journal Advanced Healthcare Materials and selected as the inside back cover article, titled "Materials and Device Designs for Wireless Monitoring of Temperature and Thermal Transport Properties of Wound Beds during Healing."
This research was conducted with support from the Basic Research Program, the Regional Innovation Center Program, and the BK21 Program.
2024.03.11 View 6007
KAIST Develops Healthcare Device Tracking Chronic Diabetic Wounds
A KAIST research team has developed an effective wireless system that monitors the wound healing process by tracking the spatiotemporal temperature changes and heat transfer characteristics of damaged areas such as diabetic wounds.
On the 5th of March, KAIST (represented by President Kwang Hyung Lee) announced that the research team led by Professor Kyeongha Kwon from KAIST’s School of Electrical Engineering, in association with Chung-Ang University professor Hanjun Ryu, developed digital healthcare technology that tracks the wound healing process in real time, which allows appropriate treatments to be administered.
< Figure 1. Schematic illustrations and diagrams of real-time wound monitoring systems. >
The skin serves as a barrier protecting the body from harmful substances, therefore damage to the skin may cause severe health risks to patients in need of intensive care. Especially in the case of diabetic patients, chronic wounds are easily formed due to complications in normal blood circulation and the wound healing process. In the United States alone, hundreds of billions of dollars of medical costs stem from regenerating the skin from such wounds. While various methods exist to promote wound healing, personalized management is essential depending on the condition of each patient's wounds.
Accordingly, the research team tracked the heating response within the wound by utilizing the differences in temperature between the damaged area and the surrounding healthy skin. They then measured heat transfer characteristics to observe moisture changes near the skin surface, ultimately establishing a basis for understanding the formation process of scar tissue. The team conducted experiments using diabetic mice models regarding the delay in wound healing under pathological conditions, and it was demonstrated that the collected data accurately tracks the wound healing process and the formation of scar tissue.
To minimize the tissue damage that may occur in the process of removing the tracking device after healing, the system integrates biodegradable sensor modules capable of natural decomposition within the body. These biodegradable modules disintegrate within the body after use, thus reducing the risk of additional discomfort or tissue damage upon device removal. Furthermore, the device could one day be used for monitoring inside the wound area as there is no need for removal.
Professor Kyeongha Kwon, who led the research, anticipates that continuous monitoring of wound temperature and heat transfer characteristics will enable medical professionals to more accurately assess the status of diabetic patients' wounds and provide appropriate treatment. He further predicted that the implementation of biodegradable sensors allows for the safe decomposition of the device after wound healing without the need for removal, making live monitoring possible not only in hospitals but also at home.
The research team plans to integrate antimicrobial materials into this device, aiming to expand its technological capabilities to enable the observation and prevention of inflammatory responses, bacterial infections, and other complications. The goal is to provide a multi-purpose wound monitoring platform capable of real-time antimicrobial monitoring in hospitals or homes by detecting changes in temperature and heat transfer characteristics indicative of infection levels.
< Image 1. Image of the bioresorbable temperature sensor >
The results of this study were published on February 19th in the international journal Advanced Healthcare Materials and selected as the inside back cover article, titled "Materials and Device Designs for Wireless Monitoring of Temperature and Thermal Transport Properties of Wound Beds during Healing."
This research was conducted with support from the Basic Research Program, the Regional Innovation Center Program, and the BK21 Program.
2024.03.11 View 6007 -
 Genome Sequencing Unveils Mutational Impacts of Radiation on Mammalian Cells
Recent release of the waste water from Japan's Fukushima nuclear disaster stirred apprehension regarding the health implications of radiation exposure. Classified as a Group 1 carcinogen, ionizing radiation has long been associated with various cancers and genetic disorders, as evidenced by survivors and descendants of atomic bombings and the Chernobyl disaster. Despite much smaller amount, we remain consistently exposed to low levels of radiation in everyday life and medical procedures.
Radiation, whether in the form of high-energy particles or electromagnetic waves, is conventionally known to break our cellular DNA, leading to cancer and genetic disorders. Yet, our understanding of the quantitative and qualitative mutational impacts of ionizing radiation has been incomplete.
On the 14th, Professor Young Seok Ju and his research team from KAIST, in collaboration with Dr. Tae Gen Son from the Dongnam Institute of Radiological and Medical Science, and Professors Kyung Su Kim and Ji Hyun Chang from Seoul National University, unveiled a breakthrough. Their study, led by joint first authors Drs. Jeonghwan Youk, Hyun Woo Kwon, Joonoh Lim, Eunji Kim and Tae-Woo Kim, titled "Quantitative and qualitative mutational impact of ionizing radiation on normal cells," was published in Cell Genomics.
Employing meticulous techniques, the research team comprehensively analyzed the whole-genome sequences of cells pre- and post-radiation exposure, pinpointing radiation-induced DNA mutations. Experiments involving cells from different organs of humans and mice exposed to varying radiation doses revealed mutation patterns correlating with exposure levels. (Figure 1)
Notably, exposure to 1 Gray (Gy) of radiation resulted in on average 14 mutations in every post-exposure cell. (Figure 2) Unlike other carcinogens, radiation-induced mutations primarily comprised short base deletions and a set of structural variations including inversions, translocations, and various complex genomic rearrangements. (Figure 3) Interestingly, experiments subjecting cells to low radiation dose rate over 100 days demonstrated that mutation quantities, under equivalent total radiation doses, mirrored those of high-dose exposure.
"Through this study, we have clearly elucidated the effects of radiation on cells at the molecular level," said Prof. Ju at KAIST. "Now we understand better how radiation changes the DNA of our cells," he added.
Dr. Son from the Dongnam Institute of Radiological and Medical Science stated, "Based on this study, we will continue to research the effects of very low and very high doses of radiation on the human body," and further remarked, "We will advance the development of safe and effective radiation therapy techniques."
Professors Kim and Chang from Seoul National University College of Medicine expressed their views, saying, "Through this study, we believe we now have a tool to accurately understand the impact of radiation on human DNA," and added, "We hope that many subsequent studies will emerge using the research methodologies employed in this study."
This research represents a significant leap forward in radiation studies, made possible through collaborative efforts and interdisciplinary approaches. This pioneering research engaged scholars from diverse backgrounds, spanning from the Genetic Engineering Research Institute at Seoul National University, the Cambridge Stem Cell Institute in the UK, the Institute for Molecular Biotechnology in Austria (IMBA), and the Genome Insight Inc. (a KAIST spin-off start-up). This study was supported by various institutions including the National Research Foundation of Korea, Dongnam Institute of Radiological and Medical Science (supported by Ministry of Science and ICT, the government of South Korea), the Suh Kyungbae Foundation, the Human Frontier Science Program (HFSP), and the Korea University Anam Hospital Korea Foundation for the Advancement of Science and Creativity, the Ministry of Science and ICT, and the National R&D Program.
2024.02.15 View 7099
Genome Sequencing Unveils Mutational Impacts of Radiation on Mammalian Cells
Recent release of the waste water from Japan's Fukushima nuclear disaster stirred apprehension regarding the health implications of radiation exposure. Classified as a Group 1 carcinogen, ionizing radiation has long been associated with various cancers and genetic disorders, as evidenced by survivors and descendants of atomic bombings and the Chernobyl disaster. Despite much smaller amount, we remain consistently exposed to low levels of radiation in everyday life and medical procedures.
Radiation, whether in the form of high-energy particles or electromagnetic waves, is conventionally known to break our cellular DNA, leading to cancer and genetic disorders. Yet, our understanding of the quantitative and qualitative mutational impacts of ionizing radiation has been incomplete.
On the 14th, Professor Young Seok Ju and his research team from KAIST, in collaboration with Dr. Tae Gen Son from the Dongnam Institute of Radiological and Medical Science, and Professors Kyung Su Kim and Ji Hyun Chang from Seoul National University, unveiled a breakthrough. Their study, led by joint first authors Drs. Jeonghwan Youk, Hyun Woo Kwon, Joonoh Lim, Eunji Kim and Tae-Woo Kim, titled "Quantitative and qualitative mutational impact of ionizing radiation on normal cells," was published in Cell Genomics.
Employing meticulous techniques, the research team comprehensively analyzed the whole-genome sequences of cells pre- and post-radiation exposure, pinpointing radiation-induced DNA mutations. Experiments involving cells from different organs of humans and mice exposed to varying radiation doses revealed mutation patterns correlating with exposure levels. (Figure 1)
Notably, exposure to 1 Gray (Gy) of radiation resulted in on average 14 mutations in every post-exposure cell. (Figure 2) Unlike other carcinogens, radiation-induced mutations primarily comprised short base deletions and a set of structural variations including inversions, translocations, and various complex genomic rearrangements. (Figure 3) Interestingly, experiments subjecting cells to low radiation dose rate over 100 days demonstrated that mutation quantities, under equivalent total radiation doses, mirrored those of high-dose exposure.
"Through this study, we have clearly elucidated the effects of radiation on cells at the molecular level," said Prof. Ju at KAIST. "Now we understand better how radiation changes the DNA of our cells," he added.
Dr. Son from the Dongnam Institute of Radiological and Medical Science stated, "Based on this study, we will continue to research the effects of very low and very high doses of radiation on the human body," and further remarked, "We will advance the development of safe and effective radiation therapy techniques."
Professors Kim and Chang from Seoul National University College of Medicine expressed their views, saying, "Through this study, we believe we now have a tool to accurately understand the impact of radiation on human DNA," and added, "We hope that many subsequent studies will emerge using the research methodologies employed in this study."
This research represents a significant leap forward in radiation studies, made possible through collaborative efforts and interdisciplinary approaches. This pioneering research engaged scholars from diverse backgrounds, spanning from the Genetic Engineering Research Institute at Seoul National University, the Cambridge Stem Cell Institute in the UK, the Institute for Molecular Biotechnology in Austria (IMBA), and the Genome Insight Inc. (a KAIST spin-off start-up). This study was supported by various institutions including the National Research Foundation of Korea, Dongnam Institute of Radiological and Medical Science (supported by Ministry of Science and ICT, the government of South Korea), the Suh Kyungbae Foundation, the Human Frontier Science Program (HFSP), and the Korea University Anam Hospital Korea Foundation for the Advancement of Science and Creativity, the Ministry of Science and ICT, and the National R&D Program.
2024.02.15 View 7099 -
 KAIST Demonstrates AI and sustainable technologies at CES 2024
On January 2, KAIST announced it will be participating in the Consumer Electronics Show (CES) 2024, held between January 9 and 12.
CES 2024 is one of the world’s largest tech conferences to take place in Las Vegas. Under the slogan “KAIST, the Global Value Creator” for its exhibition, KAIST has submitted technologies falling under one of following themes: “Expansion of Human Intelligence, Mobility, and Reality”, and “Pursuit of Human Security and Sustainable Development”.
24 startups and pre-startups whose technologies stand out in various fields including artificial intelligence (AI), mobility, virtual reality, healthcare and human security, and sustainable development, will welcome their visitors at an exclusive booth of 232 m2 prepared for KAIST at Eureka Park in Las Vegas.
12 businesses will participate in the first category, “Expansion of Human Intelligence, Mobility, and Reality”, including MicroPix, Panmnesia, DeepAuto, MGL, Reports, Narnia Labs, EL FACTORY, Korea Position Technology, AudAi, Planby Technologies, Movin, and Studio Lab.
In the “Pursuit of Human Security and Sustainable Development” category, 12 businesses including Aldaver, ADNC, Solve, Iris, Blue Device, Barreleye, TR, A2US, Greeners, Iron Boys, Shard Partners and Kingbot, will be introduced.
In particular, Aldaver is a startup that received the Korean Business Award 2023 as well as the presidential award at the Challenge K-Startup with its biomimetic material and printing technology. It has attracted 4.5 billion KRW of investment thus far.
Narnia Labs, with its AI design solution for manufacturing, won the grand prize for K-tech Startups 2022, and has so far attracted 3.5 billion KRW of investments.
Panmnesia is a startup that won the 2024 CES Innovation Award, recognized for their fab-less AI semiconductor technology. They attracted 16 billion KRW of investment through seed round alone.
Meanwhile, student startups will also be presented during the exhibition. Studio Lab received a CES 2024 Best of Innovation Award in the AI category. The team developed the software Seller Canvas, which automatically generates a page for product details when a user uploads an image of a product.
The central stage at the KAIST exhibition booth will be used to interview members of the participating startups between Jan 9 to 11, as well as a networking site for businesses and invited investors during KAIST NIGHT on the evening of 10th, between 5 and 7 PM.
Director Sung-Yool Choi of the KAIST Institute of Technology Value Creation said, “Through CES 2024, KAIST will overcome the limits of human intelligence, mobility, and space with the deep-tech based technologies developed by its startups, and will demonstrate its achievements for realizing its vision as a global value-creating university through the solutions for human security and sustainable development.”
2024.01.05 View 8996
KAIST Demonstrates AI and sustainable technologies at CES 2024
On January 2, KAIST announced it will be participating in the Consumer Electronics Show (CES) 2024, held between January 9 and 12.
CES 2024 is one of the world’s largest tech conferences to take place in Las Vegas. Under the slogan “KAIST, the Global Value Creator” for its exhibition, KAIST has submitted technologies falling under one of following themes: “Expansion of Human Intelligence, Mobility, and Reality”, and “Pursuit of Human Security and Sustainable Development”.
24 startups and pre-startups whose technologies stand out in various fields including artificial intelligence (AI), mobility, virtual reality, healthcare and human security, and sustainable development, will welcome their visitors at an exclusive booth of 232 m2 prepared for KAIST at Eureka Park in Las Vegas.
12 businesses will participate in the first category, “Expansion of Human Intelligence, Mobility, and Reality”, including MicroPix, Panmnesia, DeepAuto, MGL, Reports, Narnia Labs, EL FACTORY, Korea Position Technology, AudAi, Planby Technologies, Movin, and Studio Lab.
In the “Pursuit of Human Security and Sustainable Development” category, 12 businesses including Aldaver, ADNC, Solve, Iris, Blue Device, Barreleye, TR, A2US, Greeners, Iron Boys, Shard Partners and Kingbot, will be introduced.
In particular, Aldaver is a startup that received the Korean Business Award 2023 as well as the presidential award at the Challenge K-Startup with its biomimetic material and printing technology. It has attracted 4.5 billion KRW of investment thus far.
Narnia Labs, with its AI design solution for manufacturing, won the grand prize for K-tech Startups 2022, and has so far attracted 3.5 billion KRW of investments.
Panmnesia is a startup that won the 2024 CES Innovation Award, recognized for their fab-less AI semiconductor technology. They attracted 16 billion KRW of investment through seed round alone.
Meanwhile, student startups will also be presented during the exhibition. Studio Lab received a CES 2024 Best of Innovation Award in the AI category. The team developed the software Seller Canvas, which automatically generates a page for product details when a user uploads an image of a product.
The central stage at the KAIST exhibition booth will be used to interview members of the participating startups between Jan 9 to 11, as well as a networking site for businesses and invited investors during KAIST NIGHT on the evening of 10th, between 5 and 7 PM.
Director Sung-Yool Choi of the KAIST Institute of Technology Value Creation said, “Through CES 2024, KAIST will overcome the limits of human intelligence, mobility, and space with the deep-tech based technologies developed by its startups, and will demonstrate its achievements for realizing its vision as a global value-creating university through the solutions for human security and sustainable development.”
2024.01.05 View 8996 -
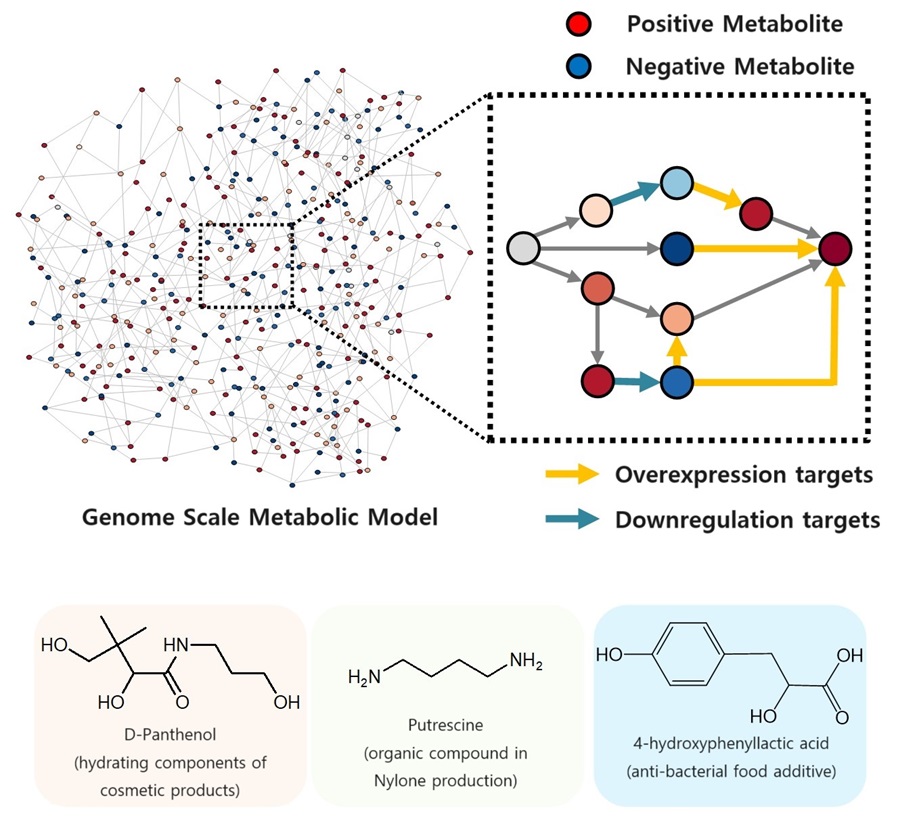 KAIST proposes alternatives to chemical factories through “iBridge”
- A computer simulation program “iBridge” was developed at KAIST that can put together microbial cell factories quickly and efficiently to produce cosmetics and food additives, and raw materials for nylons
- Eco-friendly and sustainable fermentation process to establish an alternative to chemical plants
As climate change and environmental concerns intensify, sustainable microbial cell factories garner significant attention as candidates to replace chemical plants. To develop microorganisms to be used in the microbial cell factories, it is crucial to modify their metabolic processes to induce efficient target chemical production by modulating its gene expressions. Yet, the challenge persists in determining which gene expressions to amplify and suppress, and the experimental verification of these modification targets is a time- and resource-intensive process even for experts. The challenges were addressed by a team of researchers at KAIST (President Kwang-Hyung Lee) led by Distinguished Professor Sang Yup Lee.
It was announced on the 9th by the school that a method for building a microbial factory at low cost, quickly and efficiently, was presented by a novel computer simulation program developed by the team under Professor Lee’s guidance, which is named “iBridge”. This innovative system is designed to predict gene targets to either overexpress or downregulate in the goal of producing a desired compound to enable the cost-effective and efficient construction of microbial cell factories specifically tailored for producing the chemical compound in demand from renewable biomass.
Systems metabolic engineering is a field of research and engineering pioneered by KAIST’s Distinguished Professor Sang Yup Lee that seeks to produce valuable compounds in industrial demands using microorganisms that are re-configured by a combination of methods including, but not limited to, metabolic engineering, synthetic biology, systems biology, and fermentation engineering.
In order to improve microorganisms’ capability to produce useful compounds, it is essential to delete, suppress, or overexpress microbial genes. However, it is difficult even for the experts to identify the gene targets to modify without experimental confirmations for each of them, which can take up immeasurable amount of time and resources.
The newly developed iBridge identifies positive and negative metabolites within cells, which exert positive and/or negative impact on formation of the products, by calculating the sum of covariances of their outgoing (consuming) reaction fluxes for a target chemical. Subsequently, it pinpoints "bridge" reactions responsible for converting negative metabolites into positive ones as candidates for overexpression, while identifying the opposites as targets for downregulation.
The research team successfully utilized the iBridge simulation to establish E. coli microbial cell factories each capable of producing three of the compounds that are in high demands at a production capacity that has not been reported around the world. They developed E. coli strains that can each produce panthenol, a moisturizing agent found in many cosmetics, putrescine, which is one of the key components in nylon production, and 4-hydroxyphenyllactic acid, an anti-bacterial food additive. In addition to these three compounds, the study presents predictions for overexpression and suppression genes to construct microbial factories for 298 other industrially valuable compounds.
Dr. Youngjoon Lee, the co-first author of this paper from KAIST, emphasized the accelerated construction of various microbial factories the newly developed simulation enabled. He stated, "With the use of this simulation, multiple microbial cell factories have been established significantly faster than it would have been using the conventional methods. Microbial cell factories producing a wider range of valuable compounds can now be constructed quickly using this technology."
Professor Sang Yup Lee said, "Systems metabolic engineering is a crucial technology for addressing the current climate change issues." He added, "This simulation could significantly expedite the transition from resorting to conventional chemical factories to utilizing environmentally friendly microbial factories."
< Figure. Conceptual diagram of the flow of iBridge simulation >
The team’s work on iBridge is described in a paper titled "Genome-Wide Identification of Overexpression and Downregulation Gene Targets Based on the Sum of Covariances of the Outgoing Reaction Fluxes" written by Dr. Won Jun Kim, and Dr. Youngjoon Lee of the Bioprocess Research Center and Professors Hyun Uk Kim and Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST. The paper was published via peer-review on the 6th of November on “Cell Systems” by Cell Press.
This research was conducted with the support from the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and Development of Platform Technology for the Production of Novel Aromatic Bioplastic using Microbial Cell Factories Project (Project Leader: Research Professor So Young Choi, KAIST) of the Korean Ministry of Science and ICT.
2023.11.09 View 7158
KAIST proposes alternatives to chemical factories through “iBridge”
- A computer simulation program “iBridge” was developed at KAIST that can put together microbial cell factories quickly and efficiently to produce cosmetics and food additives, and raw materials for nylons
- Eco-friendly and sustainable fermentation process to establish an alternative to chemical plants
As climate change and environmental concerns intensify, sustainable microbial cell factories garner significant attention as candidates to replace chemical plants. To develop microorganisms to be used in the microbial cell factories, it is crucial to modify their metabolic processes to induce efficient target chemical production by modulating its gene expressions. Yet, the challenge persists in determining which gene expressions to amplify and suppress, and the experimental verification of these modification targets is a time- and resource-intensive process even for experts. The challenges were addressed by a team of researchers at KAIST (President Kwang-Hyung Lee) led by Distinguished Professor Sang Yup Lee.
It was announced on the 9th by the school that a method for building a microbial factory at low cost, quickly and efficiently, was presented by a novel computer simulation program developed by the team under Professor Lee’s guidance, which is named “iBridge”. This innovative system is designed to predict gene targets to either overexpress or downregulate in the goal of producing a desired compound to enable the cost-effective and efficient construction of microbial cell factories specifically tailored for producing the chemical compound in demand from renewable biomass.
Systems metabolic engineering is a field of research and engineering pioneered by KAIST’s Distinguished Professor Sang Yup Lee that seeks to produce valuable compounds in industrial demands using microorganisms that are re-configured by a combination of methods including, but not limited to, metabolic engineering, synthetic biology, systems biology, and fermentation engineering.
In order to improve microorganisms’ capability to produce useful compounds, it is essential to delete, suppress, or overexpress microbial genes. However, it is difficult even for the experts to identify the gene targets to modify without experimental confirmations for each of them, which can take up immeasurable amount of time and resources.
The newly developed iBridge identifies positive and negative metabolites within cells, which exert positive and/or negative impact on formation of the products, by calculating the sum of covariances of their outgoing (consuming) reaction fluxes for a target chemical. Subsequently, it pinpoints "bridge" reactions responsible for converting negative metabolites into positive ones as candidates for overexpression, while identifying the opposites as targets for downregulation.
The research team successfully utilized the iBridge simulation to establish E. coli microbial cell factories each capable of producing three of the compounds that are in high demands at a production capacity that has not been reported around the world. They developed E. coli strains that can each produce panthenol, a moisturizing agent found in many cosmetics, putrescine, which is one of the key components in nylon production, and 4-hydroxyphenyllactic acid, an anti-bacterial food additive. In addition to these three compounds, the study presents predictions for overexpression and suppression genes to construct microbial factories for 298 other industrially valuable compounds.
Dr. Youngjoon Lee, the co-first author of this paper from KAIST, emphasized the accelerated construction of various microbial factories the newly developed simulation enabled. He stated, "With the use of this simulation, multiple microbial cell factories have been established significantly faster than it would have been using the conventional methods. Microbial cell factories producing a wider range of valuable compounds can now be constructed quickly using this technology."
Professor Sang Yup Lee said, "Systems metabolic engineering is a crucial technology for addressing the current climate change issues." He added, "This simulation could significantly expedite the transition from resorting to conventional chemical factories to utilizing environmentally friendly microbial factories."
< Figure. Conceptual diagram of the flow of iBridge simulation >
The team’s work on iBridge is described in a paper titled "Genome-Wide Identification of Overexpression and Downregulation Gene Targets Based on the Sum of Covariances of the Outgoing Reaction Fluxes" written by Dr. Won Jun Kim, and Dr. Youngjoon Lee of the Bioprocess Research Center and Professors Hyun Uk Kim and Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST. The paper was published via peer-review on the 6th of November on “Cell Systems” by Cell Press.
This research was conducted with the support from the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and Development of Platform Technology for the Production of Novel Aromatic Bioplastic using Microbial Cell Factories Project (Project Leader: Research Professor So Young Choi, KAIST) of the Korean Ministry of Science and ICT.
2023.11.09 View 7158 -
 A KAIST Research Team Produces Eco-Friendly Nylon with Engineered Bacterium
With worsening climate change and environmental issues, in recent years, there has been increased interest in the eco-friendly production of polymers like nylon.
On August 10, Dr. Taehee Han from a KAIST research team led by Distinguished Professor Sang Yup Lee in the Department of Chemical and Biomolecular Engineering revealed the successful development of a microbial strain that produces valerolactam, a monomer of nylon-5.
Valerolactam is an important monomer that constitutes nylon-5 and nylon-6,5. Nylon is the oldest synthetic polymer, and nylon-5 is one of its derivatives composed of monomers with five carbons, while nylon-5,6 is composed of two types of monomers with either five or six carbons. They not only have excellent processability, but are also light and tough, which allows them to be applied in a wide range of industrial sectors including clothing, badminton rackets, fishing nets, tents, and gear parts. Monomers are materials that can be built into polymers, and synthetic processes are what connects them into a polymer.
The chemical production of valerolactam, however, is based on petrochemistry, where extreme reaction conditions are required and toxic waste is produced. To solve these problems, efforts are being made to develop environmentally friendly and highly efficient microbial cell factories for lactam production. Systems metabolic engineering, a key strategy for effective microbial strain development, is a research field pioneered by Professor Sang Yup Lee.
Professor Lee’s team used metabolic engineering, a technique for manipulating microbial metabolic pathways, to construct a synthetic metabolic pathway for valerolactam production in Corynebacteriam glutamicum, a bacterium commonly used for amino acid production. With this, they successfully developed a microbial strain that utilizes biomass-derived glucose as a carbon source to produce high-value valerolactam.
In 2017, the team suggested a novel method that metabolically manipulates Escherichia coli to produce valerolactam. However, there were several limitations at the time including low producibility and the generation of harmful byproducts.
< Figure 1. Schematic graphical representation of the development of microorganisms that produce valerolactam, a nylon-5 monomer >
In this research, the team improved valerolactam producibility and incorporated an additional systems metabolic strategy to the developed microbial strain while eliminating the harmful byproducts. By removing the gene involved in the production of the main byproduct and through gene screening, the team successfully converted 5-aminovaleric acid, a byproduct and a precursor, into valerolactam.
Furthermore, by employing a strategy where the 5-aminovaleric acid-converting gene is inserted multiple times into the genome, the team strengthened the metabolic flux for valerolactam production. As a result, they reached a world-record concentration of 76.1 g/L, which is 6.17 times greater than what was previously reported.
This study was published in Metabolic Engineering on July 12, under the title, “Metabolic engineering of Corynebacterium glutamicum for the high-level production of valerolactam, a nylon-5 monomer”.
Dr. Taehee Han, the first author of the paper, said, “The significance of this research lies in our development of an environmentally friendly technology that efficiently produces monomer lactam for nylon production using microorganisms.” She added, “Through this technology, we will be able to take a step forward in replacing the petrochemical industry with a microorganism-based biopolymer industry.”
This work was supported by the “Development of Next-Generation Biofinery Platform Technologies for Leading Bio-based Chemicals Industry Project” funded by the Korean Ministry of Science and ICT.
2023.08.24 View 5645
A KAIST Research Team Produces Eco-Friendly Nylon with Engineered Bacterium
With worsening climate change and environmental issues, in recent years, there has been increased interest in the eco-friendly production of polymers like nylon.
On August 10, Dr. Taehee Han from a KAIST research team led by Distinguished Professor Sang Yup Lee in the Department of Chemical and Biomolecular Engineering revealed the successful development of a microbial strain that produces valerolactam, a monomer of nylon-5.
Valerolactam is an important monomer that constitutes nylon-5 and nylon-6,5. Nylon is the oldest synthetic polymer, and nylon-5 is one of its derivatives composed of monomers with five carbons, while nylon-5,6 is composed of two types of monomers with either five or six carbons. They not only have excellent processability, but are also light and tough, which allows them to be applied in a wide range of industrial sectors including clothing, badminton rackets, fishing nets, tents, and gear parts. Monomers are materials that can be built into polymers, and synthetic processes are what connects them into a polymer.
The chemical production of valerolactam, however, is based on petrochemistry, where extreme reaction conditions are required and toxic waste is produced. To solve these problems, efforts are being made to develop environmentally friendly and highly efficient microbial cell factories for lactam production. Systems metabolic engineering, a key strategy for effective microbial strain development, is a research field pioneered by Professor Sang Yup Lee.
Professor Lee’s team used metabolic engineering, a technique for manipulating microbial metabolic pathways, to construct a synthetic metabolic pathway for valerolactam production in Corynebacteriam glutamicum, a bacterium commonly used for amino acid production. With this, they successfully developed a microbial strain that utilizes biomass-derived glucose as a carbon source to produce high-value valerolactam.
In 2017, the team suggested a novel method that metabolically manipulates Escherichia coli to produce valerolactam. However, there were several limitations at the time including low producibility and the generation of harmful byproducts.
< Figure 1. Schematic graphical representation of the development of microorganisms that produce valerolactam, a nylon-5 monomer >
In this research, the team improved valerolactam producibility and incorporated an additional systems metabolic strategy to the developed microbial strain while eliminating the harmful byproducts. By removing the gene involved in the production of the main byproduct and through gene screening, the team successfully converted 5-aminovaleric acid, a byproduct and a precursor, into valerolactam.
Furthermore, by employing a strategy where the 5-aminovaleric acid-converting gene is inserted multiple times into the genome, the team strengthened the metabolic flux for valerolactam production. As a result, they reached a world-record concentration of 76.1 g/L, which is 6.17 times greater than what was previously reported.
This study was published in Metabolic Engineering on July 12, under the title, “Metabolic engineering of Corynebacterium glutamicum for the high-level production of valerolactam, a nylon-5 monomer”.
Dr. Taehee Han, the first author of the paper, said, “The significance of this research lies in our development of an environmentally friendly technology that efficiently produces monomer lactam for nylon production using microorganisms.” She added, “Through this technology, we will be able to take a step forward in replacing the petrochemical industry with a microorganism-based biopolymer industry.”
This work was supported by the “Development of Next-Generation Biofinery Platform Technologies for Leading Bio-based Chemicals Industry Project” funded by the Korean Ministry of Science and ICT.
2023.08.24 View 5645 -
 KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 8041
KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 8041 -
 2023 Global Entrepreneurship Summer School in Silicon Valley Successfully Concluded
< 2023 Silicon Valley Global Entrepreneurship Summer School Participants >
The 2023 KAIST Global Entrepreneurship Summer School (GESS) was successfully held. Co-hosted by the Center for Global Strategies and Planning (GSP) (Director Man-Sung Yim) and the Startup KAIST (Director Hyeonmin Bae), the 2023 KAIST GESS was the second one of the summer programs, repeating the Silicon Valley global entrepreneurship bootcamp of 2022 (2022 GESC), based on industry-academia collaboration. This program was designed to provide students with the opportunity to visit Silicon Valley, the global hub of entrepreneurship, and personally experience the Silicon Valley culture while developing human networks that would serve as a foundation for their overseas startup development.
A total of 20 participants were selected earlier this year, including potential KAISTian entrepreneurs and early-stage entrepreneurs from KAIST within one year of incorporation. In particular, a number of foreign students of various nationalities such as Vietnam, Azerbaijan, Honduras, Indonesia, Philippines, and Kazakhstan, increased significantly, demonstrating the enthusiasm for entrepreneurship across national boundaries along with the program's growing international status. This year's event was also open to 20 Impact MBA and Social Entrepreneur (SE) students from KAIST's College of Business for the Silicon Valley program.
For the past two months, the participants have trained on business model development and pitching at KAIST's main campus in Daejeon. From June 21st to the 30th, they visited the campuses of leading universities, such as, Stanford University, UC Santa Cruz, and UC Berkeley, as well as KOTRA Silicon Valley Trade Center (Manager Hyoung il Kim), and local alumni companies and Apple company to experience the global technology startups. The start-ups by KAIST alums including B Garage (CEO Aiden Kim), ImpriMed (CEO Sungwon Lim), Medic Life Sciences (CEO Kyuho Han), and VESSL AI (CEO Jaeman Ahn) participated in the program and gave lectures and company tours to inspire the participants to have passion to take on the entrepreneurial endeavors and challenges.
On the last day, the participants gave presentations on their team’s business items in front of local venture capitalists in Silicon Valley. After receiving continuous coaching from Silicon Valley's professional accelerators through remote video conferencing and face-to-face mentoring for the last two months, the participants developed their business models and presented their creative and innovative ideas, revealing their potential as future global entrepreneurs. At the final competition, Team Sparky that developed “Snoove” won the first prize. Snoove is a scientifically-proven mattress accessory that applies mild vibration to the mattress to aid users in achieving better sleep, a method previously used to soothe infants.
< GESS Pitching Day Presentation >
Kevin Choi from the Team Sparky said, "Seeing and experiencing the realities of entrepreneurship in Silicon Valley, a global startup scene, made me think about the importance of unlearning, challenging, and failing to be a global entrepreneur who contributes to our society." Man-Sung Yim, the Associate Vice President of the International Office, who organized the event added, "Through this experience, we expect KAIST students to grow to become global leaders who would create global values and enhance the international reputation of our university."
Meanwhile, the GSP and Startup KAIST commented that they will to continue to develop the KAIST GESS program to foster prospective entrepreneurs who can compete in the global market based on the success of this program.
2023.07.05 View 10150
2023 Global Entrepreneurship Summer School in Silicon Valley Successfully Concluded
< 2023 Silicon Valley Global Entrepreneurship Summer School Participants >
The 2023 KAIST Global Entrepreneurship Summer School (GESS) was successfully held. Co-hosted by the Center for Global Strategies and Planning (GSP) (Director Man-Sung Yim) and the Startup KAIST (Director Hyeonmin Bae), the 2023 KAIST GESS was the second one of the summer programs, repeating the Silicon Valley global entrepreneurship bootcamp of 2022 (2022 GESC), based on industry-academia collaboration. This program was designed to provide students with the opportunity to visit Silicon Valley, the global hub of entrepreneurship, and personally experience the Silicon Valley culture while developing human networks that would serve as a foundation for their overseas startup development.
A total of 20 participants were selected earlier this year, including potential KAISTian entrepreneurs and early-stage entrepreneurs from KAIST within one year of incorporation. In particular, a number of foreign students of various nationalities such as Vietnam, Azerbaijan, Honduras, Indonesia, Philippines, and Kazakhstan, increased significantly, demonstrating the enthusiasm for entrepreneurship across national boundaries along with the program's growing international status. This year's event was also open to 20 Impact MBA and Social Entrepreneur (SE) students from KAIST's College of Business for the Silicon Valley program.
For the past two months, the participants have trained on business model development and pitching at KAIST's main campus in Daejeon. From June 21st to the 30th, they visited the campuses of leading universities, such as, Stanford University, UC Santa Cruz, and UC Berkeley, as well as KOTRA Silicon Valley Trade Center (Manager Hyoung il Kim), and local alumni companies and Apple company to experience the global technology startups. The start-ups by KAIST alums including B Garage (CEO Aiden Kim), ImpriMed (CEO Sungwon Lim), Medic Life Sciences (CEO Kyuho Han), and VESSL AI (CEO Jaeman Ahn) participated in the program and gave lectures and company tours to inspire the participants to have passion to take on the entrepreneurial endeavors and challenges.
On the last day, the participants gave presentations on their team’s business items in front of local venture capitalists in Silicon Valley. After receiving continuous coaching from Silicon Valley's professional accelerators through remote video conferencing and face-to-face mentoring for the last two months, the participants developed their business models and presented their creative and innovative ideas, revealing their potential as future global entrepreneurs. At the final competition, Team Sparky that developed “Snoove” won the first prize. Snoove is a scientifically-proven mattress accessory that applies mild vibration to the mattress to aid users in achieving better sleep, a method previously used to soothe infants.
< GESS Pitching Day Presentation >
Kevin Choi from the Team Sparky said, "Seeing and experiencing the realities of entrepreneurship in Silicon Valley, a global startup scene, made me think about the importance of unlearning, challenging, and failing to be a global entrepreneur who contributes to our society." Man-Sung Yim, the Associate Vice President of the International Office, who organized the event added, "Through this experience, we expect KAIST students to grow to become global leaders who would create global values and enhance the international reputation of our university."
Meanwhile, the GSP and Startup KAIST commented that they will to continue to develop the KAIST GESS program to foster prospective entrepreneurs who can compete in the global market based on the success of this program.
2023.07.05 View 10150 -
 Synthetic sRNAs to knockdown genes in medical and industrial bacteria
Bacteria are intimately involved in our daily lives. These microorganisms have been used in human history for food such as cheese, yogurt, and wine, In more recent years, through metabolic engineering, microorganisms been used extensively as microbial cell factories to manufacture plastics, feed for livestock, dietary supplements, and drugs. However, in addition to these bacteria that are beneficial to human lives, pathogens such as Pneumonia, Salmonella, and Staphylococcus that cause various infectious diseases are also ubiquitously present. It is important to be able to metabolically control these beneficial industrial bacteria for high value-added chemicals production and to manipulate harmful pathogens to suppress its pathogenic traits.
KAIST (President Kwang Hyung Lee) announced on the 10th that a research team led by Distinguished Professor Sang Yup Lee of the Department of Biochemical Engineering has developed a new sRNA tool that can effectively inhibit target genes in various bacteria, including both Gram-negative and Gram-positive bacteria. The research results were published online on April 24 in Nature Communications.
※ Thesis title: Targeted and high-throughput gene knockdown in diverse bacteria using synthetic sRNAs
※ Author information : Jae Sung Cho (co-1st), Dongsoo Yang (co-1st), Cindy Pricilia Surya Prabowo (co-author), Mohammad Rifqi Ghiffary (co-author), Taehee Han (co-author), Kyeong Rok Choi (co-author), Cheon Woo Moon (co-author), Hengrui Zhou (co-author), Jae Yong Ryu (co-author), Hyun Uk Kim (co-author) and Sang Yup Lee (corresponding author).
sRNA is an effective tool for synthesizing and regulating target genes in E. coli, but it has been difficult to apply to industrially useful Gram-positive bacteria such as Bacillus subtilis and Corynebacterium in addition to Gram-negative bacteria such as E. coli.
To address this issue, a research team led by Distinguished Professor Lee Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST developed a new sRNA platform that can effectively suppress target genes in various bacteria, including both Gram-negative and positive bacteria. The research team surveyed thousands of microbial-derived sRNA systems in the microbial database, and eventually designated the sRNA system derived from 'Bacillus subtilis' that showed the highest gene knockdown efficiency, and designated it as “Broad-Host-Range sRNA”, or BHR-sRNA.
A similar well-known system is the CRISPR interference (CRISPRi) system, which is a modified CRISPR system that knocks down gene expression by suppressing the gene transcription process. However, the Cas9 protein in the CRISPRi system has a very high molecular weight, and there have been reports growth inhibition in bacteria. The BHR-sRNA system developed in this study did not affect bacterial growth while showing similar gene knockdown efficiencies to CRISPRi.
< Figure 1. a) Schematic illustration demonstrating the mechanism of syntetic sRNA b) Phylogenetic tree of the 16 Gram-negative and Gram-positive bacterial species tested for gene knockdown by the BHR-sRNA system. >
To validate the versatility of the BHR-sRNA system, 16 different gram-negative and gram-positive bacteria were selected and tested, where the BHR-sRNA system worked successfully in 15 of them. In addition, it was demonstrated that the gene knockdown capability was more effective than that of the existing E. coli-based sRNA system in 10 bacteria. The BHR-sRNA system proved to be a universal tool capable of effectively inhibiting gene expression in various bacteria.
In order to address the problem of antibiotic-resistant pathogens that have recently become more serious, the BHR-sRNA was demonstrated to suppress the pathogenicity by suppressing the gene producing the virulence factor. By using BHR-sRNA, biofilm formation, one of the factors resulting in antibiotic resistance, was inhibited by 73% in Staphylococcus epidermidis a pathogen that can cause hospital-acquired infections. Antibiotic resistance was also weakened by 58% in the pneumonia causing bacteria Klebsiella pneumoniae. In addition, BHR-sRNA was applied to industrial bacteria to develop microbial cell factories to produce high value-added chemicals with better production performance. Notably, superior industrial strains were constructed with the aid of BHR-sRNA to produce the following chemicals: valerolactam, a raw material for polyamide polymers, methyl-anthranilate, a grape-flavor food additive, and indigoidine, a blue-toned natural dye.
The BHR-sRNA developed through this study will help expedite the commercialization of bioprocesses to produce high value-added compounds and materials such as artificial meat, jet fuel, health supplements, pharmaceuticals, and plastics. It is also anticipated that to help eradicating antibiotic-resistant pathogens in preparation for another upcoming pandemic. “In the past, we could only develop new tools for gene knockdown for each bacterium, but now we have developed a tool that works for a variety of bacteria” said Distinguished Professor Sang Yup Lee.
This work was supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project from NRF supported by the Korean MSIT.
2023.05.10 View 7170
Synthetic sRNAs to knockdown genes in medical and industrial bacteria
Bacteria are intimately involved in our daily lives. These microorganisms have been used in human history for food such as cheese, yogurt, and wine, In more recent years, through metabolic engineering, microorganisms been used extensively as microbial cell factories to manufacture plastics, feed for livestock, dietary supplements, and drugs. However, in addition to these bacteria that are beneficial to human lives, pathogens such as Pneumonia, Salmonella, and Staphylococcus that cause various infectious diseases are also ubiquitously present. It is important to be able to metabolically control these beneficial industrial bacteria for high value-added chemicals production and to manipulate harmful pathogens to suppress its pathogenic traits.
KAIST (President Kwang Hyung Lee) announced on the 10th that a research team led by Distinguished Professor Sang Yup Lee of the Department of Biochemical Engineering has developed a new sRNA tool that can effectively inhibit target genes in various bacteria, including both Gram-negative and Gram-positive bacteria. The research results were published online on April 24 in Nature Communications.
※ Thesis title: Targeted and high-throughput gene knockdown in diverse bacteria using synthetic sRNAs
※ Author information : Jae Sung Cho (co-1st), Dongsoo Yang (co-1st), Cindy Pricilia Surya Prabowo (co-author), Mohammad Rifqi Ghiffary (co-author), Taehee Han (co-author), Kyeong Rok Choi (co-author), Cheon Woo Moon (co-author), Hengrui Zhou (co-author), Jae Yong Ryu (co-author), Hyun Uk Kim (co-author) and Sang Yup Lee (corresponding author).
sRNA is an effective tool for synthesizing and regulating target genes in E. coli, but it has been difficult to apply to industrially useful Gram-positive bacteria such as Bacillus subtilis and Corynebacterium in addition to Gram-negative bacteria such as E. coli.
To address this issue, a research team led by Distinguished Professor Lee Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST developed a new sRNA platform that can effectively suppress target genes in various bacteria, including both Gram-negative and positive bacteria. The research team surveyed thousands of microbial-derived sRNA systems in the microbial database, and eventually designated the sRNA system derived from 'Bacillus subtilis' that showed the highest gene knockdown efficiency, and designated it as “Broad-Host-Range sRNA”, or BHR-sRNA.
A similar well-known system is the CRISPR interference (CRISPRi) system, which is a modified CRISPR system that knocks down gene expression by suppressing the gene transcription process. However, the Cas9 protein in the CRISPRi system has a very high molecular weight, and there have been reports growth inhibition in bacteria. The BHR-sRNA system developed in this study did not affect bacterial growth while showing similar gene knockdown efficiencies to CRISPRi.
< Figure 1. a) Schematic illustration demonstrating the mechanism of syntetic sRNA b) Phylogenetic tree of the 16 Gram-negative and Gram-positive bacterial species tested for gene knockdown by the BHR-sRNA system. >
To validate the versatility of the BHR-sRNA system, 16 different gram-negative and gram-positive bacteria were selected and tested, where the BHR-sRNA system worked successfully in 15 of them. In addition, it was demonstrated that the gene knockdown capability was more effective than that of the existing E. coli-based sRNA system in 10 bacteria. The BHR-sRNA system proved to be a universal tool capable of effectively inhibiting gene expression in various bacteria.
In order to address the problem of antibiotic-resistant pathogens that have recently become more serious, the BHR-sRNA was demonstrated to suppress the pathogenicity by suppressing the gene producing the virulence factor. By using BHR-sRNA, biofilm formation, one of the factors resulting in antibiotic resistance, was inhibited by 73% in Staphylococcus epidermidis a pathogen that can cause hospital-acquired infections. Antibiotic resistance was also weakened by 58% in the pneumonia causing bacteria Klebsiella pneumoniae. In addition, BHR-sRNA was applied to industrial bacteria to develop microbial cell factories to produce high value-added chemicals with better production performance. Notably, superior industrial strains were constructed with the aid of BHR-sRNA to produce the following chemicals: valerolactam, a raw material for polyamide polymers, methyl-anthranilate, a grape-flavor food additive, and indigoidine, a blue-toned natural dye.
The BHR-sRNA developed through this study will help expedite the commercialization of bioprocesses to produce high value-added compounds and materials such as artificial meat, jet fuel, health supplements, pharmaceuticals, and plastics. It is also anticipated that to help eradicating antibiotic-resistant pathogens in preparation for another upcoming pandemic. “In the past, we could only develop new tools for gene knockdown for each bacterium, but now we have developed a tool that works for a variety of bacteria” said Distinguished Professor Sang Yup Lee.
This work was supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project from NRF supported by the Korean MSIT.
2023.05.10 View 7170 -
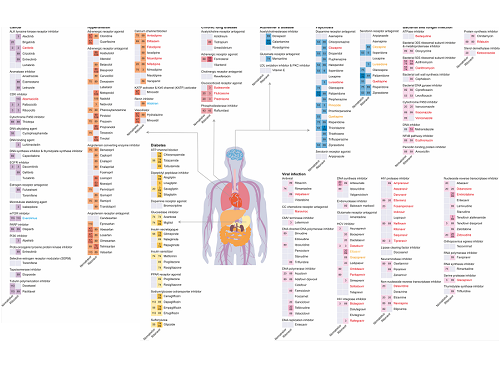 KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 7964
KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 7964 -
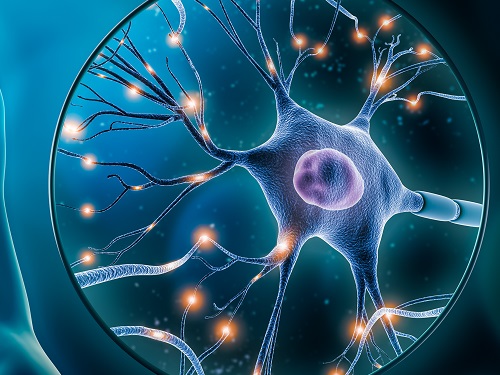 KAIST team develops smart immune system that can pin down on malignant tumors
A joint research team led by Professor Jung Kyoon Choi of the KAIST Department of Bio and Brain Engineering and Professor Jong-Eun Park of the KAIST Graduate School of Medical Science and Engineering (GSMSE) announced the development of the key technologies to treat cancers using smart immune cells designed based on AI and big data analysis. This technology is expected to be a next-generation immunotherapy that allows precision targeting of tumor cells by having the chimeric antigen receptors (CARs) operate through a logical circuit. Professor Hee Jung An of CHA Bundang Medical Center and Professor Hae-Ock Lee of the Catholic University of Korea also participated in this research to contribute joint effort.
Professor Jung Kyoon Choi’s team built a gene expression database from millions of cells, and used this to successfully develop and verify a deep-learning algorithm that could detect the differences in gene expression patterns between tumor cells and normal cells through a logical circuit. CAR immune cells that were fitted with the logic circuits discovered through this methodology could distinguish between tumorous and normal cells as a computer would, and therefore showed potentials to strike only on tumor cells accurately without causing unwanted side effects.
This research, conducted by co-first authors Dr. Joonha Kwon of the KAIST Department of Bio and Brain Engineering and Ph.D. candidate Junho Kang of KAIST GSMSE, was published by Nature Biotechnology on February 16, under the title Single-cell mapping of combinatorial target antigens for CAR switches using logic gates.
An area in cancer research where the most attempts and advances have been made in recent years is immunotherapy. This field of treatment, which utilizes the patient’s own immune system in order to overcome cancer, has several methods including immune checkpoint inhibitors, cancer vaccines and cellular treatments. Immune cells like CAR-T or CAR-NK equipped with chimera antigen receptors, in particular, can recognize cancer antigens and directly destroy cancer cells.
Starting with its success in blood cancer treatment, scientists have been trying to expand the application of CAR cell therapy to treat solid cancer. But there have been difficulties to develop CAR cells with effective killing abilities against solid cancer cells with minimized side effects. Accordingly, in recent years, the development of smarter CAR engineering technologies, i.e., computational logic gates such as AND, OR, and NOT, to effectively target cancer cells has been underway.
At this point in time, the research team built a large-scale database for cancer and normal cells to discover the exact genes that are expressed only from cancer cells at a single-cell level. The team followed this up by developing an AI algorithm that could search for a combination of genes that best distinguishes cancer cells from normal cells. This algorithm, in particular, has been used to find a logic circuit that can specifically target cancer cells through cell-level simulations of all gene combinations. CAR-T cells equipped with logic circuits discovered through this methodology are expected to distinguish cancerous cells from normal cells like computers, thereby minimizing side effects and maximizing the effects of chemotherapy.
Dr. Joonha Kwon, who is the first author of this paper, said, “this research suggests a new method that hasn’t been tried before. What’s particularly noteworthy is the process in which we found the optimal CAR cell circuit through simulations of millions of individual tumors and normal cells.” He added, “This is an innovative technology that can apply AI and computer logic circuits to immune cell engineering. It would contribute greatly to expanding CAR therapy, which is being successfully used for blood cancer, to solid cancers as well.”
This research was funded by the Original Technology Development Project and Research Program for Next Generation Applied Omic of the Korea Research Foundation.
Figure 1. A schematic diagram of manufacturing and administration process of CAR therapy and of cancer cell-specific dual targeting using CAR.
Figure 2. Deep learning (convolutional neural networks, CNNs) algorithm for selection of dual targets based on gene combination (left) and algorithm for calculating expressing cell fractions by gene combination according to logical circuit (right).
2023.03.09 View 9447
KAIST team develops smart immune system that can pin down on malignant tumors
A joint research team led by Professor Jung Kyoon Choi of the KAIST Department of Bio and Brain Engineering and Professor Jong-Eun Park of the KAIST Graduate School of Medical Science and Engineering (GSMSE) announced the development of the key technologies to treat cancers using smart immune cells designed based on AI and big data analysis. This technology is expected to be a next-generation immunotherapy that allows precision targeting of tumor cells by having the chimeric antigen receptors (CARs) operate through a logical circuit. Professor Hee Jung An of CHA Bundang Medical Center and Professor Hae-Ock Lee of the Catholic University of Korea also participated in this research to contribute joint effort.
Professor Jung Kyoon Choi’s team built a gene expression database from millions of cells, and used this to successfully develop and verify a deep-learning algorithm that could detect the differences in gene expression patterns between tumor cells and normal cells through a logical circuit. CAR immune cells that were fitted with the logic circuits discovered through this methodology could distinguish between tumorous and normal cells as a computer would, and therefore showed potentials to strike only on tumor cells accurately without causing unwanted side effects.
This research, conducted by co-first authors Dr. Joonha Kwon of the KAIST Department of Bio and Brain Engineering and Ph.D. candidate Junho Kang of KAIST GSMSE, was published by Nature Biotechnology on February 16, under the title Single-cell mapping of combinatorial target antigens for CAR switches using logic gates.
An area in cancer research where the most attempts and advances have been made in recent years is immunotherapy. This field of treatment, which utilizes the patient’s own immune system in order to overcome cancer, has several methods including immune checkpoint inhibitors, cancer vaccines and cellular treatments. Immune cells like CAR-T or CAR-NK equipped with chimera antigen receptors, in particular, can recognize cancer antigens and directly destroy cancer cells.
Starting with its success in blood cancer treatment, scientists have been trying to expand the application of CAR cell therapy to treat solid cancer. But there have been difficulties to develop CAR cells with effective killing abilities against solid cancer cells with minimized side effects. Accordingly, in recent years, the development of smarter CAR engineering technologies, i.e., computational logic gates such as AND, OR, and NOT, to effectively target cancer cells has been underway.
At this point in time, the research team built a large-scale database for cancer and normal cells to discover the exact genes that are expressed only from cancer cells at a single-cell level. The team followed this up by developing an AI algorithm that could search for a combination of genes that best distinguishes cancer cells from normal cells. This algorithm, in particular, has been used to find a logic circuit that can specifically target cancer cells through cell-level simulations of all gene combinations. CAR-T cells equipped with logic circuits discovered through this methodology are expected to distinguish cancerous cells from normal cells like computers, thereby minimizing side effects and maximizing the effects of chemotherapy.
Dr. Joonha Kwon, who is the first author of this paper, said, “this research suggests a new method that hasn’t been tried before. What’s particularly noteworthy is the process in which we found the optimal CAR cell circuit through simulations of millions of individual tumors and normal cells.” He added, “This is an innovative technology that can apply AI and computer logic circuits to immune cell engineering. It would contribute greatly to expanding CAR therapy, which is being successfully used for blood cancer, to solid cancers as well.”
This research was funded by the Original Technology Development Project and Research Program for Next Generation Applied Omic of the Korea Research Foundation.
Figure 1. A schematic diagram of manufacturing and administration process of CAR therapy and of cancer cell-specific dual targeting using CAR.
Figure 2. Deep learning (convolutional neural networks, CNNs) algorithm for selection of dual targets based on gene combination (left) and algorithm for calculating expressing cell fractions by gene combination according to logical circuit (right).
2023.03.09 View 9447 -
 KAIST presents a fundamental technology to remove metastatic traits from lung cancer cells
KAIST (President Kwang Hyung Lee) announced on January 30th that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering succeeded in using systems biology research to change the properties of carcinogenic cells in the lungs and eliminate both drug resistance and their ability to proliferate out to other areas of the body.
As the incidences of cancer increase within aging populations, cancer has become the most lethal disease threatening healthy life. Fatality rates are especially high when early detection does not happen in time and metastasis has occurred in various organs. In order to resolve this problem, a series of attempts were made to remove or lower the ability of cancer cells to spread, but they resulted in cancer cells in the intermediate state becoming more unstable and even more malignant, which created serious treatment challenges.
Professor Kwang-Hyun Cho's research team simulated various cancer cell states in the Epithelial-to-Mesenchymal Transition (EMT) of lung cancer cells, between epithelial cells without metastatic ability and mesenchymal cells with metastatic ability. A mathematical model of molecular network was established, and key regulators that could reverse the state of invasive and drug resistant mesenchymal cells back to the epithelial state were discovered through computer simulation analysis and molecular cell experiments. In particular, this process succeeded in properly reverting the mesenchymal lung cancer cells to a state where they were sensitive to chemotherapy treatment while avoiding the unstable EMT hybrid cell state in the middle process, which had remained a difficult problem.
The results of this research, in which KAIST Ph.D. student Namhee Kim, Dr. Chae Young Hwang, Researcher Taeyoung Kim, and Ph.D. student Hyunjin Kim participated, were published as an online paper in the international journal “Cancer Research” published by the American Association for Cancer Research (AACR) on January 30th. (Paper title: A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states)
Cells in an EMT hybrid state, which are caused by incomplete transitions during the EMT process in cancer cells, have the characteristics of both epithelial cells and mesenchymal cells, and are known to have high drug resistance and metastatic potential by acquiring high stem cell capacity. In particular, EMT is further enhanced through factors such as transforming growth factor-beta (TGF-β) secreted from the tumor microenvironment (TME) and, as a result, various cell states with high plasticity appear. Due to the complexity of EMT, it has been very difficult to completely reverse the transitional process of the mesenchymal cancer cells to an epithelial cell state in which metastatic ability and drug resistance are eliminated while avoiding the EMT hybrid cell state with high metastatic ability and drug resistance.
Professor Kwang-Hyun Cho's research team established a mathematical model of the gene regulation network that governs the complex process of EMT, and then applied large-scale computer simulation analysis and complex system network control technology to identify and verify 'p53', 'SMAD4', and 'ERK1' and 'ERK 2' (collectively ERKs) through molecular cell experiments as the three key molecular targets that can transform lung cancer cells in the mesenchymal cell state, reversed back to an epithelial cell state that no longer demonstrates the ability to metastasize, while avoiding the EMT hybrid cell state.
In particular, by analyzing the molecular regulatory mechanism of the complex EMT process at the system level, the key pathways were identified that were linked to the positive feedback that plays an important role in completely returning cancer cells to an epithelial cell state in which metastatic ability and drug resistance are removed.
This discovery is significant in that it proved that mesenchymal cells can be reverted to the state of epithelial cells under conditions where TGF-β stimulation are present, like they are in the actual environment where cancer tissue forms in the human body.
Abnormal EMT in cancer cells leads to various malignant traits such as the migration and invasion of cancer cells, changes in responsiveness to chemotherapy treatment, enhanced stem cell function, and the dissemination of cancer. In particular, the acquisition of the metastatic ability of cancer cells is a key determinant factor for the prognosis of cancer patients. The EMT reversal technology in lung cancer cells developed in this research is a new anti-cancer treatment strategy that reprograms cancer cells to eliminate their high plasticity and metastatic potential and increase their responsiveness to chemotherapy.
Professor Kwang-Hyun Cho said, "By succeeding in reversing the state of lung cancer cells that acquired high metastatic traits and resistance to drugs and reverting them to a treatable epithelial cell state with renewed sensitivity to chemotherapy, the research findings propose a new strategy for treatments that can improve the prognosis of cancer patients.”
Professor Kwang-Hyun Cho's research team was the first to present the principle of reversal treatment to revert cancer cells to normal cells, following through with the announcement of the results of their study that reverted colon cancer cells to normal colon cells in January of 2020, and also presenting successful re-programming research where the most malignant basal type breast cancer cells turned into less-malignant luminal type breast cancer cells that were treatable with hormonal therapies in January of 2022. This latest research result is the third in the development of reversal technology where lung cancer cells that had acquired metastatic traits returned to a state in which their metastatic ability was removed and drug sensitivity was enhanced.
This research was carried out with support from the Ministry of Science and ICT and the National Research Foundation of Korea's Basic Research in Science & Engineering Program for Mid-Career Researchers.
< Figure 1. Construction of the mathematical model of the regulatory network to represent the EMT phenotype based on the interaction between various molecules related to EMT.
(A) Professor Kwang-Hyun Cho's research team investigated numerous literatures and databases related to complex EMT, and based on comparative analysis of cell line data showing epithelial and mesenchymal cell conditions, they extracted key signaling pathways related to EMT and built a mathematical model of regulatory network (B) By comparing the results of computer simulation analysis and the molecular cell experiments, it was verified how well the constructed mathematical model simulated the actual cellular phenomena. >
< Figure 2. Understanding of various EMT phenotypes through large-scale computer simulation analysis and complex system network control technology.
(A) Through computer simulation analysis and experiments, Professor Kwang-Hyun Cho's research team found that complete control of EMT is impossible with single-molecule control alone. In particular, through comparison of the relative stability of attractors, it was revealed that the cell state exhibiting EMT hybrid characteristics has unstable properties. (B), (C) Based on these results, Prof. Cho’s team identified two feedbacks (positive feedback consisting of Snail-miR-34 and ZEB1-miR-200) that play an important role in avoiding the EMT hybrid state that appeared in the TGF-β-ON state. It was found through computer simulation analysis that the two feedbacks restore relatively high stability when the excavated p53 and SMAD4 are regulated. In addition, molecular cell experiments demonstrated that the expression levels of E-cad and ZEB1, which are representative phenotypic markers of EMT, changed similarly to the expression profile in the epithelial cell state, despite the TGF-β-ON state. >
< Figure 3. Complex molecular network analysis and discovery of reprogramming molecular targets for intact elimination of EMT hybrid features.
(A) Controlling the expression of p53 and SMAD4 in lung cancer cell lines was expected to overcome drug resistance, but contrary to expectations, chemotherapy responsiveness was not restored. (B) Professor Kwang-Hyun Cho's research team additionally analyzed computer simulations, genome data, and experimental results and found that high expression levels of TWIST1 and EPCAM were related to drug resistance. (C) Prof. Cho’s team identified three key molecular targets: p53, SMAD4 and ERK1 & ERK2. (D), (E) Furthermore, they identified a key pathway that plays an important role in completely reversing into epithelial cells while avoiding EMT hybrid characteristics, and confirmed through network analysis and attractor analysis that high stability of the key pathway was restored when the proposed molecular target was controlled. >
< Figure 4. Verification through experiments with lung cancer cell lines.
When p53 was activated and SMAD4 and ERK1/2 were inhibited in lung cancer cell lines, (A), (B) E-cad protein expression increased and ZEB1 protein expression decreased, and (C) mesenchymal cell status including TWIST1 and EPCAM and gene expression of markers related to stem cell potential characteristics were completely inhibited. In addition, (D) it was confirmed that resistance to chemotherapy treatment was also overcome as the cell state was reversed by the regulated target. >
< Figure 5. A schematic representation of the research results.
Prof. Cho’s research team identified key molecular regulatory pathways to avoid high plasticity formed by abnormal EMT of cancer cells and reverse it to an epithelial cell state through systems biology research. From this analysis, a reprogramming molecular target that can reverse the state of mesenchymal cells with acquired invasiveness and drug resistance to the state of epithelial cells with restored drug responsiveness was discovered.
For lung cancer cells, when a drug that enhances the expression of p53, one of the molecular targets discovered, and inhibits the expression of SMAD4 and ERK1 & ERK2 is administered, the molecular network of genes in the state of mesenchymal cells is modified, eventually eliminating metastatic ability and it is reprogrammed to turn into epithelial cells without the resistance to chemotherapy treatments. >
2023.01.30 View 17004
KAIST presents a fundamental technology to remove metastatic traits from lung cancer cells
KAIST (President Kwang Hyung Lee) announced on January 30th that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering succeeded in using systems biology research to change the properties of carcinogenic cells in the lungs and eliminate both drug resistance and their ability to proliferate out to other areas of the body.
As the incidences of cancer increase within aging populations, cancer has become the most lethal disease threatening healthy life. Fatality rates are especially high when early detection does not happen in time and metastasis has occurred in various organs. In order to resolve this problem, a series of attempts were made to remove or lower the ability of cancer cells to spread, but they resulted in cancer cells in the intermediate state becoming more unstable and even more malignant, which created serious treatment challenges.
Professor Kwang-Hyun Cho's research team simulated various cancer cell states in the Epithelial-to-Mesenchymal Transition (EMT) of lung cancer cells, between epithelial cells without metastatic ability and mesenchymal cells with metastatic ability. A mathematical model of molecular network was established, and key regulators that could reverse the state of invasive and drug resistant mesenchymal cells back to the epithelial state were discovered through computer simulation analysis and molecular cell experiments. In particular, this process succeeded in properly reverting the mesenchymal lung cancer cells to a state where they were sensitive to chemotherapy treatment while avoiding the unstable EMT hybrid cell state in the middle process, which had remained a difficult problem.
The results of this research, in which KAIST Ph.D. student Namhee Kim, Dr. Chae Young Hwang, Researcher Taeyoung Kim, and Ph.D. student Hyunjin Kim participated, were published as an online paper in the international journal “Cancer Research” published by the American Association for Cancer Research (AACR) on January 30th. (Paper title: A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states)
Cells in an EMT hybrid state, which are caused by incomplete transitions during the EMT process in cancer cells, have the characteristics of both epithelial cells and mesenchymal cells, and are known to have high drug resistance and metastatic potential by acquiring high stem cell capacity. In particular, EMT is further enhanced through factors such as transforming growth factor-beta (TGF-β) secreted from the tumor microenvironment (TME) and, as a result, various cell states with high plasticity appear. Due to the complexity of EMT, it has been very difficult to completely reverse the transitional process of the mesenchymal cancer cells to an epithelial cell state in which metastatic ability and drug resistance are eliminated while avoiding the EMT hybrid cell state with high metastatic ability and drug resistance.
Professor Kwang-Hyun Cho's research team established a mathematical model of the gene regulation network that governs the complex process of EMT, and then applied large-scale computer simulation analysis and complex system network control technology to identify and verify 'p53', 'SMAD4', and 'ERK1' and 'ERK 2' (collectively ERKs) through molecular cell experiments as the three key molecular targets that can transform lung cancer cells in the mesenchymal cell state, reversed back to an epithelial cell state that no longer demonstrates the ability to metastasize, while avoiding the EMT hybrid cell state.
In particular, by analyzing the molecular regulatory mechanism of the complex EMT process at the system level, the key pathways were identified that were linked to the positive feedback that plays an important role in completely returning cancer cells to an epithelial cell state in which metastatic ability and drug resistance are removed.
This discovery is significant in that it proved that mesenchymal cells can be reverted to the state of epithelial cells under conditions where TGF-β stimulation are present, like they are in the actual environment where cancer tissue forms in the human body.
Abnormal EMT in cancer cells leads to various malignant traits such as the migration and invasion of cancer cells, changes in responsiveness to chemotherapy treatment, enhanced stem cell function, and the dissemination of cancer. In particular, the acquisition of the metastatic ability of cancer cells is a key determinant factor for the prognosis of cancer patients. The EMT reversal technology in lung cancer cells developed in this research is a new anti-cancer treatment strategy that reprograms cancer cells to eliminate their high plasticity and metastatic potential and increase their responsiveness to chemotherapy.
Professor Kwang-Hyun Cho said, "By succeeding in reversing the state of lung cancer cells that acquired high metastatic traits and resistance to drugs and reverting them to a treatable epithelial cell state with renewed sensitivity to chemotherapy, the research findings propose a new strategy for treatments that can improve the prognosis of cancer patients.”
Professor Kwang-Hyun Cho's research team was the first to present the principle of reversal treatment to revert cancer cells to normal cells, following through with the announcement of the results of their study that reverted colon cancer cells to normal colon cells in January of 2020, and also presenting successful re-programming research where the most malignant basal type breast cancer cells turned into less-malignant luminal type breast cancer cells that were treatable with hormonal therapies in January of 2022. This latest research result is the third in the development of reversal technology where lung cancer cells that had acquired metastatic traits returned to a state in which their metastatic ability was removed and drug sensitivity was enhanced.
This research was carried out with support from the Ministry of Science and ICT and the National Research Foundation of Korea's Basic Research in Science & Engineering Program for Mid-Career Researchers.
< Figure 1. Construction of the mathematical model of the regulatory network to represent the EMT phenotype based on the interaction between various molecules related to EMT.
(A) Professor Kwang-Hyun Cho's research team investigated numerous literatures and databases related to complex EMT, and based on comparative analysis of cell line data showing epithelial and mesenchymal cell conditions, they extracted key signaling pathways related to EMT and built a mathematical model of regulatory network (B) By comparing the results of computer simulation analysis and the molecular cell experiments, it was verified how well the constructed mathematical model simulated the actual cellular phenomena. >
< Figure 2. Understanding of various EMT phenotypes through large-scale computer simulation analysis and complex system network control technology.
(A) Through computer simulation analysis and experiments, Professor Kwang-Hyun Cho's research team found that complete control of EMT is impossible with single-molecule control alone. In particular, through comparison of the relative stability of attractors, it was revealed that the cell state exhibiting EMT hybrid characteristics has unstable properties. (B), (C) Based on these results, Prof. Cho’s team identified two feedbacks (positive feedback consisting of Snail-miR-34 and ZEB1-miR-200) that play an important role in avoiding the EMT hybrid state that appeared in the TGF-β-ON state. It was found through computer simulation analysis that the two feedbacks restore relatively high stability when the excavated p53 and SMAD4 are regulated. In addition, molecular cell experiments demonstrated that the expression levels of E-cad and ZEB1, which are representative phenotypic markers of EMT, changed similarly to the expression profile in the epithelial cell state, despite the TGF-β-ON state. >
< Figure 3. Complex molecular network analysis and discovery of reprogramming molecular targets for intact elimination of EMT hybrid features.
(A) Controlling the expression of p53 and SMAD4 in lung cancer cell lines was expected to overcome drug resistance, but contrary to expectations, chemotherapy responsiveness was not restored. (B) Professor Kwang-Hyun Cho's research team additionally analyzed computer simulations, genome data, and experimental results and found that high expression levels of TWIST1 and EPCAM were related to drug resistance. (C) Prof. Cho’s team identified three key molecular targets: p53, SMAD4 and ERK1 & ERK2. (D), (E) Furthermore, they identified a key pathway that plays an important role in completely reversing into epithelial cells while avoiding EMT hybrid characteristics, and confirmed through network analysis and attractor analysis that high stability of the key pathway was restored when the proposed molecular target was controlled. >
< Figure 4. Verification through experiments with lung cancer cell lines.
When p53 was activated and SMAD4 and ERK1/2 were inhibited in lung cancer cell lines, (A), (B) E-cad protein expression increased and ZEB1 protein expression decreased, and (C) mesenchymal cell status including TWIST1 and EPCAM and gene expression of markers related to stem cell potential characteristics were completely inhibited. In addition, (D) it was confirmed that resistance to chemotherapy treatment was also overcome as the cell state was reversed by the regulated target. >
< Figure 5. A schematic representation of the research results.
Prof. Cho’s research team identified key molecular regulatory pathways to avoid high plasticity formed by abnormal EMT of cancer cells and reverse it to an epithelial cell state through systems biology research. From this analysis, a reprogramming molecular target that can reverse the state of mesenchymal cells with acquired invasiveness and drug resistance to the state of epithelial cells with restored drug responsiveness was discovered.
For lung cancer cells, when a drug that enhances the expression of p53, one of the molecular targets discovered, and inhibits the expression of SMAD4 and ERK1 & ERK2 is administered, the molecular network of genes in the state of mesenchymal cells is modified, eventually eliminating metastatic ability and it is reprogrammed to turn into epithelial cells without the resistance to chemotherapy treatments. >
2023.01.30 View 17004 -
 Overview of the 30-year history of metabolic engineering
< Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST >
A research team comprised of Gi Bae Kim, Dr. So Young Choi, Dr. In Jin Cho, Da-Hee Ahn, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST reported the 30-year history of metabolic engineering, highlighting examples of recent progress in the field and contributions to sustainability and health. Their paper “Metabolic engineering for sustainability and health” was published online in the 40th anniversary special issue of Trends in Biotechnology on January 10, 2023.
Metabolic engineering, a discipline of engineering that modifies cell phenotypes through molecular and genetic-level manipulations to improve cellular activities, has been studied since the early 1990s, and has progressed significantly over the past 30 years. In particular, metabolic engineering has enabled the engineering of microorganisms for the development of microbial cell factories capable of efficiently producing chemicals and materials as well as degrading recalcitrant contaminants.
This review article revisited how metabolic engineering has advanced over the past 30 years, from the advent of genetic engineering techniques such as recombinant DNA technologies to recent breakthroughs in systems metabolic engineering and data science aided by artificial intelligence. The research team highlighted momentous events and achievements in metabolic engineering, providing both trends and future directions in the field. Metabolic engineering’s contributions to bio-based sustainable chemicals and clean energy, health, and bioremediation were also reviewed. Finally, the research team shared their perspectives on the future challenges impacting metabolic engineering than must be overcome in order to achieve advancements in sustainability and health.
Distinguished Professor Sang Yup Lee said, “Replacing fossil resource-based chemical processes with bio-based sustainable processes for the production of chemicals, fuels, and materials using metabolic engineering has become our essential task for the future. By looking back on the 30+ years of metabolic engineering, we aimed to highlight the contributions of metabolic engineering to achieve sustainability and good health.” He added, “Metabolic engineering will play an increasingly important role as a key solution to the climate crisis, environmental pollution, food and energy shortages, and health problems in aging societies.”
< Figure: Metabolic Engineering Timeline >
2023.01.25 View 10148
Overview of the 30-year history of metabolic engineering
< Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST >
A research team comprised of Gi Bae Kim, Dr. So Young Choi, Dr. In Jin Cho, Da-Hee Ahn, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST reported the 30-year history of metabolic engineering, highlighting examples of recent progress in the field and contributions to sustainability and health. Their paper “Metabolic engineering for sustainability and health” was published online in the 40th anniversary special issue of Trends in Biotechnology on January 10, 2023.
Metabolic engineering, a discipline of engineering that modifies cell phenotypes through molecular and genetic-level manipulations to improve cellular activities, has been studied since the early 1990s, and has progressed significantly over the past 30 years. In particular, metabolic engineering has enabled the engineering of microorganisms for the development of microbial cell factories capable of efficiently producing chemicals and materials as well as degrading recalcitrant contaminants.
This review article revisited how metabolic engineering has advanced over the past 30 years, from the advent of genetic engineering techniques such as recombinant DNA technologies to recent breakthroughs in systems metabolic engineering and data science aided by artificial intelligence. The research team highlighted momentous events and achievements in metabolic engineering, providing both trends and future directions in the field. Metabolic engineering’s contributions to bio-based sustainable chemicals and clean energy, health, and bioremediation were also reviewed. Finally, the research team shared their perspectives on the future challenges impacting metabolic engineering than must be overcome in order to achieve advancements in sustainability and health.
Distinguished Professor Sang Yup Lee said, “Replacing fossil resource-based chemical processes with bio-based sustainable processes for the production of chemicals, fuels, and materials using metabolic engineering has become our essential task for the future. By looking back on the 30+ years of metabolic engineering, we aimed to highlight the contributions of metabolic engineering to achieve sustainability and good health.” He added, “Metabolic engineering will play an increasingly important role as a key solution to the climate crisis, environmental pollution, food and energy shortages, and health problems in aging societies.”
< Figure: Metabolic Engineering Timeline >
2023.01.25 View 10148