research
-
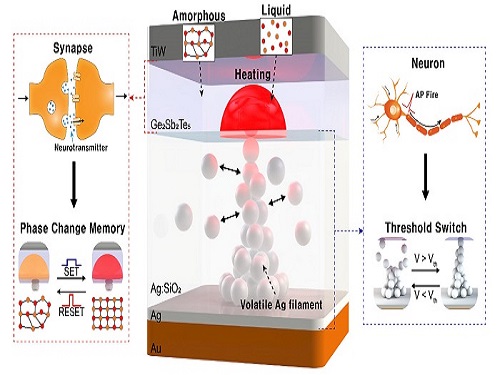 Neuromorphic Memory Device Simulates Neurons and Synapses
Simultaneous emulation of neuronal and synaptic properties promotes the development of brain-like artificial intelligence
Researchers have reported a nano-sized neuromorphic memory device that emulates neurons and synapses simultaneously in a unit cell, another step toward completing the goal of neuromorphic computing designed to rigorously mimic the human brain with semiconductor devices.
Neuromorphic computing aims to realize artificial intelligence (AI) by mimicking the mechanisms of neurons and synapses that make up the human brain. Inspired by the cognitive functions of the human brain that current computers cannot provide, neuromorphic devices have been widely investigated. However, current Complementary Metal-Oxide Semiconductor (CMOS)-based neuromorphic circuits simply connect artificial neurons and synapses without synergistic interactions, and the concomitant implementation of neurons and synapses still remains a challenge. To address these issues, a research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering implemented the biological working mechanisms of humans by introducing the neuron-synapse interactions in a single memory cell, rather than the conventional approach of electrically connecting artificial neuronal and synaptic devices.
Similar to commercial graphics cards, the artificial synaptic devices previously studied often used to accelerate parallel computations, which shows clear differences from the operational mechanisms of the human brain. The research team implemented the synergistic interactions between neurons and synapses in the neuromorphic memory device, emulating the mechanisms of the biological neural network. In addition, the developed neuromorphic device can replace complex CMOS neuron circuits with a single device, providing high scalability and cost efficiency.
The human brain consists of a complex network of 100 billion neurons and 100 trillion synapses. The functions and structures of neurons and synapses can flexibly change according to the external stimuli, adapting to the surrounding environment. The research team developed a neuromorphic device in which short-term and long-term memories coexist using volatile and non-volatile memory devices that mimic the characteristics of neurons and synapses, respectively. A threshold switch device is used as volatile memory and phase-change memory is used as a non-volatile device. Two thin-film devices are integrated without intermediate electrodes, implementing the functional adaptability of neurons and synapses in the neuromorphic memory.
Professor Keon Jae Lee explained, "Neurons and synapses interact with each other to establish cognitive functions such as memory and learning, so simulating both is an essential element for brain-inspired artificial intelligence. The developed neuromorphic memory device also mimics the retraining effect that allows quick learning of the forgotten information by implementing a positive feedback effect between neurons and synapses.”
This result entitled “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse” was published in the May 19, 2022 issue of Nature Communications.
-Publication:Sang Hyun Sung, Tae Jin Kim, Hyera Shin, Tae Hong Im, and Keon Jae Lee (2022) “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse,” Nature Communications May 19, 2022 (DOI: 10.1038/s41467-022-30432-2)
-Profile:Professor Keon Jae Leehttp://fand.kaist.ac.kr
Department of Materials Science and EngineeringKAIST
2022.05.20 View 13467
Neuromorphic Memory Device Simulates Neurons and Synapses
Simultaneous emulation of neuronal and synaptic properties promotes the development of brain-like artificial intelligence
Researchers have reported a nano-sized neuromorphic memory device that emulates neurons and synapses simultaneously in a unit cell, another step toward completing the goal of neuromorphic computing designed to rigorously mimic the human brain with semiconductor devices.
Neuromorphic computing aims to realize artificial intelligence (AI) by mimicking the mechanisms of neurons and synapses that make up the human brain. Inspired by the cognitive functions of the human brain that current computers cannot provide, neuromorphic devices have been widely investigated. However, current Complementary Metal-Oxide Semiconductor (CMOS)-based neuromorphic circuits simply connect artificial neurons and synapses without synergistic interactions, and the concomitant implementation of neurons and synapses still remains a challenge. To address these issues, a research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering implemented the biological working mechanisms of humans by introducing the neuron-synapse interactions in a single memory cell, rather than the conventional approach of electrically connecting artificial neuronal and synaptic devices.
Similar to commercial graphics cards, the artificial synaptic devices previously studied often used to accelerate parallel computations, which shows clear differences from the operational mechanisms of the human brain. The research team implemented the synergistic interactions between neurons and synapses in the neuromorphic memory device, emulating the mechanisms of the biological neural network. In addition, the developed neuromorphic device can replace complex CMOS neuron circuits with a single device, providing high scalability and cost efficiency.
The human brain consists of a complex network of 100 billion neurons and 100 trillion synapses. The functions and structures of neurons and synapses can flexibly change according to the external stimuli, adapting to the surrounding environment. The research team developed a neuromorphic device in which short-term and long-term memories coexist using volatile and non-volatile memory devices that mimic the characteristics of neurons and synapses, respectively. A threshold switch device is used as volatile memory and phase-change memory is used as a non-volatile device. Two thin-film devices are integrated without intermediate electrodes, implementing the functional adaptability of neurons and synapses in the neuromorphic memory.
Professor Keon Jae Lee explained, "Neurons and synapses interact with each other to establish cognitive functions such as memory and learning, so simulating both is an essential element for brain-inspired artificial intelligence. The developed neuromorphic memory device also mimics the retraining effect that allows quick learning of the forgotten information by implementing a positive feedback effect between neurons and synapses.”
This result entitled “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse” was published in the May 19, 2022 issue of Nature Communications.
-Publication:Sang Hyun Sung, Tae Jin Kim, Hyera Shin, Tae Hong Im, and Keon Jae Lee (2022) “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse,” Nature Communications May 19, 2022 (DOI: 10.1038/s41467-022-30432-2)
-Profile:Professor Keon Jae Leehttp://fand.kaist.ac.kr
Department of Materials Science and EngineeringKAIST
2022.05.20 View 13467 -
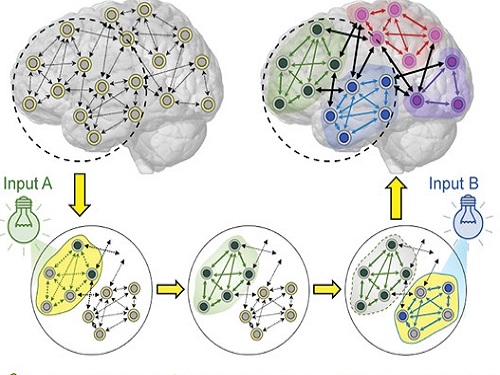 Energy-Efficient AI Hardware Technology Via a Brain-Inspired Stashing System
Researchers demonstrate neuromodulation-inspired stashing system for the energy-efficient learning of a spiking neural network using a self-rectifying memristor array
Researchers have proposed a novel system inspired by the neuromodulation of the brain, referred to as a ‘stashing system,’ that requires less energy consumption. The research group led by Professor Kyung Min Kim from the Department of Materials Science and Engineering has developed a technology that can efficiently handle mathematical operations for artificial intelligence by imitating the continuous changes in the topology of the neural network according to the situation. The human brain changes its neural topology in real time, learning to store or recall memories as needed. The research group presented a new artificial intelligence learning method that directly implements these neural coordination circuit configurations.
Research on artificial intelligence is becoming very active, and the development of artificial intelligence-based electronic devices and product releases are accelerating, especially in the Fourth Industrial Revolution age. To implement artificial intelligence in electronic devices, customized hardware development should also be supported. However most electronic devices for artificial intelligence require high power consumption and highly integrated memory arrays for large-scale tasks. It has been challenging to solve these power consumption and integration limitations, and efforts have been made to find out how the human brain solves problems.
To prove the efficiency of the developed technology, the research group created artificial neural network hardware equipped with a self-rectifying synaptic array and algorithm called a ‘stashing system’ that was developed to conduct artificial intelligence learning. As a result, it was able to reduce energy by 37% within the stashing system without any accuracy degradation. This result proves that emulating the neuromodulation in humans is possible.
Professor Kim said, "In this study, we implemented the learning method of the human brain with only a simple circuit composition and through this we were able to reduce the energy needed by nearly 40 percent.”
This neuromodulation-inspired stashing system that mimics the brain’s neural activity is compatible with existing electronic devices and commercialized semiconductor hardware. It is expected to be used in the design of next-generation semiconductor chips for artificial intelligence.
This study was published in Advanced Functional Materials in March 2022 and supported by KAIST, the National Research Foundation of Korea, the National NanoFab Center, and SK Hynix.
-Publication:
Woon Hyung Cheong, Jae Bum Jeon†, Jae Hyun In, Geunyoung Kim, Hanchan Song, Janho An, Juseong Park, Young Seok Kim, Cheol Seong Hwang, and Kyung Min Kim (2022)
“Demonstration of Neuromodulation-inspired Stashing System for Energy-efficient Learning of Spiking Neural Network using a Self-Rectifying Memristor Array,” Advanced FunctionalMaterials March 31, 2022 (DOI: 10.1002/adfm.202200337)
-Profile:
Professor Kyung Min Kimhttp://semi.kaist.ac.kr https://scholar.google.com/citations?user=BGw8yDYAAAAJ&hl=ko
Department of Materials Science and EngineeringKAIST
2022.05.18 View 11072
Energy-Efficient AI Hardware Technology Via a Brain-Inspired Stashing System
Researchers demonstrate neuromodulation-inspired stashing system for the energy-efficient learning of a spiking neural network using a self-rectifying memristor array
Researchers have proposed a novel system inspired by the neuromodulation of the brain, referred to as a ‘stashing system,’ that requires less energy consumption. The research group led by Professor Kyung Min Kim from the Department of Materials Science and Engineering has developed a technology that can efficiently handle mathematical operations for artificial intelligence by imitating the continuous changes in the topology of the neural network according to the situation. The human brain changes its neural topology in real time, learning to store or recall memories as needed. The research group presented a new artificial intelligence learning method that directly implements these neural coordination circuit configurations.
Research on artificial intelligence is becoming very active, and the development of artificial intelligence-based electronic devices and product releases are accelerating, especially in the Fourth Industrial Revolution age. To implement artificial intelligence in electronic devices, customized hardware development should also be supported. However most electronic devices for artificial intelligence require high power consumption and highly integrated memory arrays for large-scale tasks. It has been challenging to solve these power consumption and integration limitations, and efforts have been made to find out how the human brain solves problems.
To prove the efficiency of the developed technology, the research group created artificial neural network hardware equipped with a self-rectifying synaptic array and algorithm called a ‘stashing system’ that was developed to conduct artificial intelligence learning. As a result, it was able to reduce energy by 37% within the stashing system without any accuracy degradation. This result proves that emulating the neuromodulation in humans is possible.
Professor Kim said, "In this study, we implemented the learning method of the human brain with only a simple circuit composition and through this we were able to reduce the energy needed by nearly 40 percent.”
This neuromodulation-inspired stashing system that mimics the brain’s neural activity is compatible with existing electronic devices and commercialized semiconductor hardware. It is expected to be used in the design of next-generation semiconductor chips for artificial intelligence.
This study was published in Advanced Functional Materials in March 2022 and supported by KAIST, the National Research Foundation of Korea, the National NanoFab Center, and SK Hynix.
-Publication:
Woon Hyung Cheong, Jae Bum Jeon†, Jae Hyun In, Geunyoung Kim, Hanchan Song, Janho An, Juseong Park, Young Seok Kim, Cheol Seong Hwang, and Kyung Min Kim (2022)
“Demonstration of Neuromodulation-inspired Stashing System for Energy-efficient Learning of Spiking Neural Network using a Self-Rectifying Memristor Array,” Advanced FunctionalMaterials March 31, 2022 (DOI: 10.1002/adfm.202200337)
-Profile:
Professor Kyung Min Kimhttp://semi.kaist.ac.kr https://scholar.google.com/citations?user=BGw8yDYAAAAJ&hl=ko
Department of Materials Science and EngineeringKAIST
2022.05.18 View 11072 -
 Machine Learning-Based Algorithm to Speed up DNA Sequencing
The algorithm presents the first full-fledged, short-read alignment software that leverages learned indices for solving the exact match search problem for efficient seeding
The human genome consists of a complete set of DNA, which is about 6.4 billion letters long. Because of its size, reading the whole genome sequence at once is challenging. So scientists use DNA sequencers to produce hundreds of millions of DNA sequence fragments, or short reads, up to 300 letters long. Then the DNA sequencer assembles all the short reads like a giant jigsaw puzzle to reconstruct the entire genome sequence. Even with very fast computers, this job can take hours to complete.
A research team at KAIST has achieved up to 3.45x faster speeds by developing the first short-read alignment software that uses a recent advance in machine-learning called a learned index.
The research team reported their findings on March 7, 2022 in the journal Bioinformatics. The software has been released as open source and can be found on github (https://github.com/kaist-ina/BWA-MEME).
Next-generation sequencing (NGS) is a state-of-the-art DNA sequencing method. Projects are underway with the goal of producing genome sequencing at population scale. Modern NGS hardware is capable of generating billions of short reads in a single run. Then the short reads have to be aligned with the reference DNA sequence. With large-scale DNA sequencing operations running hundreds of next-generation sequences, the need for an efficient short read alignment tool has become even more critical. Accelerating the DNA sequence alignment would be a step toward achieving the goal of population-scale sequencing. However, existing algorithms are limited in their performance because of their frequent memory accesses.
BWA-MEM2 is a popular short-read alignment software package currently used to sequence the DNA. However, it has its limitations. The state-of-the-art alignment has two phases – seeding and extending. During the seeding phase, searches find exact matches of short reads in the reference DNA sequence. During the extending phase, the short reads from the seeding phase are extended. In the current process, bottlenecks occur in the seeding phase. Finding the exact matches slows the process.
The researchers set out to solve the problem of accelerating the DNA sequence alignment. To speed the process, they applied machine learning techniques to create an algorithmic improvement. Their algorithm, BWA-MEME (BWA-MEM emulated) leverages learned indices to solve the exact match search problem. The original software compared one character at a time for an exact match search. The team’s new algorithm achieves up to 3.45x faster speeds in seeding throughput over BWA-MEM2 by reducing the number of instructions by 4.60x and memory accesses by 8.77x. “Through this study, it has been shown that full genome big data analysis can be performed faster and less costly than conventional methods by applying machine learning technology,” said Professor Dongsu Han from the School of Electrical Engineering at KAIST.
The researchers’ ultimate goal was to develop efficient software that scientists from academia and industry could use on a daily basis for analyzing big data in genomics. “With the recent advances in artificial intelligence and machine learning, we see so many opportunities for designing better software for genomic data analysis. The potential is there for accelerating existing analysis as well as enabling new types of analysis, and our goal is to develop such software,” added Han.
Whole genome sequencing has traditionally been used for discovering genomic mutations and identifying the root causes of diseases, which leads to the discovery and development of new drugs and cures. There could be many potential applications. Whole genome sequencing is used not only for research, but also for clinical purposes. “The science and technology for analyzing genomic data is making rapid progress to make it more accessible for scientists and patients. This will enhance our understanding about diseases and develop a better cure for patients of various diseases.”
The research was funded by the National Research Foundation of the Korean government’s Ministry of Science and ICT.
-PublicationYoungmok Jung, Dongsu Han, “BWA-MEME:BWA-MEM emulated with a machine learning approach,” Bioinformatics, Volume 38, Issue 9, May 2022
(https://doi.org/10.1093/bioinformatics/btac137)
-ProfileProfessor Dongsu HanSchool of Electrical EngineeringKAIST
2022.05.10 View 9265
Machine Learning-Based Algorithm to Speed up DNA Sequencing
The algorithm presents the first full-fledged, short-read alignment software that leverages learned indices for solving the exact match search problem for efficient seeding
The human genome consists of a complete set of DNA, which is about 6.4 billion letters long. Because of its size, reading the whole genome sequence at once is challenging. So scientists use DNA sequencers to produce hundreds of millions of DNA sequence fragments, or short reads, up to 300 letters long. Then the DNA sequencer assembles all the short reads like a giant jigsaw puzzle to reconstruct the entire genome sequence. Even with very fast computers, this job can take hours to complete.
A research team at KAIST has achieved up to 3.45x faster speeds by developing the first short-read alignment software that uses a recent advance in machine-learning called a learned index.
The research team reported their findings on March 7, 2022 in the journal Bioinformatics. The software has been released as open source and can be found on github (https://github.com/kaist-ina/BWA-MEME).
Next-generation sequencing (NGS) is a state-of-the-art DNA sequencing method. Projects are underway with the goal of producing genome sequencing at population scale. Modern NGS hardware is capable of generating billions of short reads in a single run. Then the short reads have to be aligned with the reference DNA sequence. With large-scale DNA sequencing operations running hundreds of next-generation sequences, the need for an efficient short read alignment tool has become even more critical. Accelerating the DNA sequence alignment would be a step toward achieving the goal of population-scale sequencing. However, existing algorithms are limited in their performance because of their frequent memory accesses.
BWA-MEM2 is a popular short-read alignment software package currently used to sequence the DNA. However, it has its limitations. The state-of-the-art alignment has two phases – seeding and extending. During the seeding phase, searches find exact matches of short reads in the reference DNA sequence. During the extending phase, the short reads from the seeding phase are extended. In the current process, bottlenecks occur in the seeding phase. Finding the exact matches slows the process.
The researchers set out to solve the problem of accelerating the DNA sequence alignment. To speed the process, they applied machine learning techniques to create an algorithmic improvement. Their algorithm, BWA-MEME (BWA-MEM emulated) leverages learned indices to solve the exact match search problem. The original software compared one character at a time for an exact match search. The team’s new algorithm achieves up to 3.45x faster speeds in seeding throughput over BWA-MEM2 by reducing the number of instructions by 4.60x and memory accesses by 8.77x. “Through this study, it has been shown that full genome big data analysis can be performed faster and less costly than conventional methods by applying machine learning technology,” said Professor Dongsu Han from the School of Electrical Engineering at KAIST.
The researchers’ ultimate goal was to develop efficient software that scientists from academia and industry could use on a daily basis for analyzing big data in genomics. “With the recent advances in artificial intelligence and machine learning, we see so many opportunities for designing better software for genomic data analysis. The potential is there for accelerating existing analysis as well as enabling new types of analysis, and our goal is to develop such software,” added Han.
Whole genome sequencing has traditionally been used for discovering genomic mutations and identifying the root causes of diseases, which leads to the discovery and development of new drugs and cures. There could be many potential applications. Whole genome sequencing is used not only for research, but also for clinical purposes. “The science and technology for analyzing genomic data is making rapid progress to make it more accessible for scientists and patients. This will enhance our understanding about diseases and develop a better cure for patients of various diseases.”
The research was funded by the National Research Foundation of the Korean government’s Ministry of Science and ICT.
-PublicationYoungmok Jung, Dongsu Han, “BWA-MEME:BWA-MEM emulated with a machine learning approach,” Bioinformatics, Volume 38, Issue 9, May 2022
(https://doi.org/10.1093/bioinformatics/btac137)
-ProfileProfessor Dongsu HanSchool of Electrical EngineeringKAIST
2022.05.10 View 9265 -
 A New Strategy for Active Metasurface Design Provides a Full 360° Phase Tunable Metasurface
The new strategy displays an unprecedented upper limit of dynamic phase modulation with no significant variations in optical amplitude
An international team of researchers led by Professor Min Seok Jang of KAIST and Professor Victor W. Brar of the University of Wisconsin-Madison has demonstrated a widely applicable methodology enabling a full 360° active phase modulation for metasurfaces while maintaining significant levels of uniform light amplitude. This strategy can be fundamentally applied to any spectral region with any structures and resonances that fit the bill.
Metasurfaces are optical components with specialized functionalities indispensable for real-life applications ranging from LIDAR and spectroscopy to futuristic technologies such as invisibility cloaks and holograms. They are known for their compact and micro/nano-sized nature, which enables them to be integrated into electronic computerized systems with sizes that are ever decreasing as predicted by Moore’s law.
In order to allow for such innovations, metasurfaces must be capable of manipulating the impinging light, doing so by manipulating either the light’s amplitude or phase (or both) and emitting it back out. However, dynamically modulating the phase with the full circle range has been a notoriously difficult task, with very few works managing to do so by sacrificing a substantial amount of amplitude control.
Challenged by these limitations, the team proposed a general methodology that enables metasurfaces to implement a dynamic phase modulation with the complete 360° phase range, all the while uniformly maintaining significant levels of amplitude.
The underlying reason for the difficulty achieving such a feat is that there is a fundamental trade-off regarding dynamically controlling the optical phase of light. Metasurfaces generally perform such a function through optical resonances, an excitation of electrons inside the metasurface structure that harmonically oscillate together with the incident light. In order to be able to modulate through the entire range of 0-360°, the optical resonance frequency (the center of the spectrum) must be tuned by a large amount while the linewidth (the width of the spectrum) is kept to a minimum. However, to electrically tune the optical resonance frequency of the metasurface on demand, there needs to be a controllable influx and outflux of electrons into the metasurface and this inevitably leads to a larger linewidth of the aforementioned optical resonance.
The problem is further compounded by the fact that the phase and the amplitude of optical resonances are closely correlated in a complex, non-linear fashion, making it very difficult to hold substantial control over the amplitude while changing the phase.
The team’s work circumvented both problems by using two optical resonances, each with specifically designated properties. One resonance provides the decoupling between the phase and amplitude so that the phase is able to be tuned while significant and uniform levels of amplitude are maintained, as well as providing a narrow linewidth.
The other resonance provides the capability of being sufficiently tuned to a large degree so that the complete full circle range of phase modulation is achievable. The quintessence of the work is then to combine the different properties of the two resonances through a phenomenon called avoided crossing, so that the interactions between the two resonances lead to an amalgamation of the desired traits that achieves and even surpasses the full 360° phase modulation with uniform amplitude.
Professor Jang said, “Our research proposes a new methodology in dynamic phase modulation that breaks through the conventional limits and trade-offs, while being broadly applicable in diverse types of metasurfaces. We hope that this idea helps researchers implement and realize many key applications of metasurfaces, such as LIDAR and holograms, so that the nanophotonics industry keeps growing and provides a brighter technological future.”
The research paper authored by Ju Young Kim and Juho Park, et al., and titled "Full 2π Tunable Phase Modulation Using Avoided Crossing of Resonances" was published in Nature Communications on April 19. The research was funded by the Samsung Research Funding & Incubation Center of Samsung Electronics.
-Publication:Ju Young Kim, Juho Park, Gregory R. Holdman, Jacob T. Heiden, Shinho Kim, Victor W. Brar, and Min Seok Jang, “Full 2π Tunable Phase Modulation Using Avoided Crossing ofResonances” Nature Communications on April 19 (2022). doi.org/10.1038/s41467-022-29721-7
-ProfileProfessor Min Seok JangSchool of Electrical EngineeringKAIST
2022.05.02 View 7866
A New Strategy for Active Metasurface Design Provides a Full 360° Phase Tunable Metasurface
The new strategy displays an unprecedented upper limit of dynamic phase modulation with no significant variations in optical amplitude
An international team of researchers led by Professor Min Seok Jang of KAIST and Professor Victor W. Brar of the University of Wisconsin-Madison has demonstrated a widely applicable methodology enabling a full 360° active phase modulation for metasurfaces while maintaining significant levels of uniform light amplitude. This strategy can be fundamentally applied to any spectral region with any structures and resonances that fit the bill.
Metasurfaces are optical components with specialized functionalities indispensable for real-life applications ranging from LIDAR and spectroscopy to futuristic technologies such as invisibility cloaks and holograms. They are known for their compact and micro/nano-sized nature, which enables them to be integrated into electronic computerized systems with sizes that are ever decreasing as predicted by Moore’s law.
In order to allow for such innovations, metasurfaces must be capable of manipulating the impinging light, doing so by manipulating either the light’s amplitude or phase (or both) and emitting it back out. However, dynamically modulating the phase with the full circle range has been a notoriously difficult task, with very few works managing to do so by sacrificing a substantial amount of amplitude control.
Challenged by these limitations, the team proposed a general methodology that enables metasurfaces to implement a dynamic phase modulation with the complete 360° phase range, all the while uniformly maintaining significant levels of amplitude.
The underlying reason for the difficulty achieving such a feat is that there is a fundamental trade-off regarding dynamically controlling the optical phase of light. Metasurfaces generally perform such a function through optical resonances, an excitation of electrons inside the metasurface structure that harmonically oscillate together with the incident light. In order to be able to modulate through the entire range of 0-360°, the optical resonance frequency (the center of the spectrum) must be tuned by a large amount while the linewidth (the width of the spectrum) is kept to a minimum. However, to electrically tune the optical resonance frequency of the metasurface on demand, there needs to be a controllable influx and outflux of electrons into the metasurface and this inevitably leads to a larger linewidth of the aforementioned optical resonance.
The problem is further compounded by the fact that the phase and the amplitude of optical resonances are closely correlated in a complex, non-linear fashion, making it very difficult to hold substantial control over the amplitude while changing the phase.
The team’s work circumvented both problems by using two optical resonances, each with specifically designated properties. One resonance provides the decoupling between the phase and amplitude so that the phase is able to be tuned while significant and uniform levels of amplitude are maintained, as well as providing a narrow linewidth.
The other resonance provides the capability of being sufficiently tuned to a large degree so that the complete full circle range of phase modulation is achievable. The quintessence of the work is then to combine the different properties of the two resonances through a phenomenon called avoided crossing, so that the interactions between the two resonances lead to an amalgamation of the desired traits that achieves and even surpasses the full 360° phase modulation with uniform amplitude.
Professor Jang said, “Our research proposes a new methodology in dynamic phase modulation that breaks through the conventional limits and trade-offs, while being broadly applicable in diverse types of metasurfaces. We hope that this idea helps researchers implement and realize many key applications of metasurfaces, such as LIDAR and holograms, so that the nanophotonics industry keeps growing and provides a brighter technological future.”
The research paper authored by Ju Young Kim and Juho Park, et al., and titled "Full 2π Tunable Phase Modulation Using Avoided Crossing of Resonances" was published in Nature Communications on April 19. The research was funded by the Samsung Research Funding & Incubation Center of Samsung Electronics.
-Publication:Ju Young Kim, Juho Park, Gregory R. Holdman, Jacob T. Heiden, Shinho Kim, Victor W. Brar, and Min Seok Jang, “Full 2π Tunable Phase Modulation Using Avoided Crossing ofResonances” Nature Communications on April 19 (2022). doi.org/10.1038/s41467-022-29721-7
-ProfileProfessor Min Seok JangSchool of Electrical EngineeringKAIST
2022.05.02 View 7866 -
 LightPC Presents a Resilient System Using Only Non-Volatile Memory
Lightweight Persistence Centric System (LightPC) ensures both data and execution persistence for energy-efficient full system persistence
A KAIST research team has developed hardware and software technology that ensures both data and execution persistence. The Lightweight Persistence Centric System (LightPC) makes the systems resilient against power failures by utilizing only non-volatile memory as the main memory.
“We mounted non-volatile memory on a system board prototype and created an operating system to verify the effectiveness of LightPC,” said Professor Myoungsoo Jung. The team confirmed that LightPC validated its execution while powering up and down in the middle of execution, showing up to eight times more memory, 4.3 times faster application execution, and 73% lower power consumption compared to traditional systems.
Professor Jung said that LightPC can be utilized in a variety of fields such as data centers and high-performance computing to provide large-capacity memory, high performance, low power consumption, and service reliability.
In general, power failures on legacy systems can lead to the loss of data stored in the DRAM-based main memory. Unlike volatile memory such as DRAM, non-volatile memory can retain its data without power. Although non-volatile memory has the characteristics of lower power consumption and larger capacity than DRAM, non-volatile memory is typically used for the task of secondary storage due to its lower write performance. For this reason, nonvolatile memory is often used with DRAM. However, modern systems employing non-volatile memory-based main memory experience unexpected performance degradation due to the complicated memory microarchitecture.
To enable both data and execution persistent in legacy systems, it is necessary to transfer the data from the volatile memory to the non-volatile memory. Checkpointing is one possible solution. It periodically transfers the data in preparation for a sudden power failure. While this technology is essential for ensuring high mobility and reliability for users, checkpointing also has fatal drawbacks. It takes additional time and power to move data and requires a data recovery process as well as restarting the system.
In order to address these issues, the research team developed a processor and memory controller to raise the performance of non-volatile memory-only memory. LightPC matches the performance of DRAM by minimizing the internal volatile memory components from non-volatile memory, exposing the non-volatile memory (PRAM) media to the host, and increasing parallelism to service on-the-fly requests as soon as possible.
The team also presented operating system technology that quickly makes execution states of running processes persistent without the need for a checkpointing process. The operating system prevents all modifications to execution states and data by keeping all program executions idle before transferring data in order to support consistency within a period much shorter than the standard power hold-up time of about 16 minutes. For consistency, when the power is recovered, the computer almost immediately revives itself and re-executes all the offline processes immediately without the need for a boot process.
The researchers will present their work (LightPC: Hardware and Software Co-Design for Energy-Efficient Full System Persistence) at the International Symposium on Computer Architecture (ISCA) 2022 in New York in June. More information is available at the CAMELab website (http://camelab.org).
-Profile: Professor Myoungsoo Jung Computer Architecture and Memory Systems Laboratory (CAMEL)http://camelab.org School of Electrical EngineeringKAIST
2022.04.25 View 22948
LightPC Presents a Resilient System Using Only Non-Volatile Memory
Lightweight Persistence Centric System (LightPC) ensures both data and execution persistence for energy-efficient full system persistence
A KAIST research team has developed hardware and software technology that ensures both data and execution persistence. The Lightweight Persistence Centric System (LightPC) makes the systems resilient against power failures by utilizing only non-volatile memory as the main memory.
“We mounted non-volatile memory on a system board prototype and created an operating system to verify the effectiveness of LightPC,” said Professor Myoungsoo Jung. The team confirmed that LightPC validated its execution while powering up and down in the middle of execution, showing up to eight times more memory, 4.3 times faster application execution, and 73% lower power consumption compared to traditional systems.
Professor Jung said that LightPC can be utilized in a variety of fields such as data centers and high-performance computing to provide large-capacity memory, high performance, low power consumption, and service reliability.
In general, power failures on legacy systems can lead to the loss of data stored in the DRAM-based main memory. Unlike volatile memory such as DRAM, non-volatile memory can retain its data without power. Although non-volatile memory has the characteristics of lower power consumption and larger capacity than DRAM, non-volatile memory is typically used for the task of secondary storage due to its lower write performance. For this reason, nonvolatile memory is often used with DRAM. However, modern systems employing non-volatile memory-based main memory experience unexpected performance degradation due to the complicated memory microarchitecture.
To enable both data and execution persistent in legacy systems, it is necessary to transfer the data from the volatile memory to the non-volatile memory. Checkpointing is one possible solution. It periodically transfers the data in preparation for a sudden power failure. While this technology is essential for ensuring high mobility and reliability for users, checkpointing also has fatal drawbacks. It takes additional time and power to move data and requires a data recovery process as well as restarting the system.
In order to address these issues, the research team developed a processor and memory controller to raise the performance of non-volatile memory-only memory. LightPC matches the performance of DRAM by minimizing the internal volatile memory components from non-volatile memory, exposing the non-volatile memory (PRAM) media to the host, and increasing parallelism to service on-the-fly requests as soon as possible.
The team also presented operating system technology that quickly makes execution states of running processes persistent without the need for a checkpointing process. The operating system prevents all modifications to execution states and data by keeping all program executions idle before transferring data in order to support consistency within a period much shorter than the standard power hold-up time of about 16 minutes. For consistency, when the power is recovered, the computer almost immediately revives itself and re-executes all the offline processes immediately without the need for a boot process.
The researchers will present their work (LightPC: Hardware and Software Co-Design for Energy-Efficient Full System Persistence) at the International Symposium on Computer Architecture (ISCA) 2022 in New York in June. More information is available at the CAMELab website (http://camelab.org).
-Profile: Professor Myoungsoo Jung Computer Architecture and Memory Systems Laboratory (CAMEL)http://camelab.org School of Electrical EngineeringKAIST
2022.04.25 View 22948 -
 Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 10037
Mathematicians Identify a Key Source of Cell-to-Cell Variability in Cell Signaling
Systematic inferences identify a major source of heterogeneity in cell signaling dynamics
Why do genetically identical cells respond differently to the same external stimuli, such as antibiotics? This long-standing mystery has been solved by KAIST and IBS mathematicians who have developed a new framework for analyzing cell responses to some stimuli. The team found that the cell-to-cell variability in antibiotic stress response increases as the effective length of the cell signaling pathway (i.e., the number of rate-limiting steps) increases. This finding could identify more effective chemotherapies to overcome the fractional killing of cancer cells caused by cell-to-cell variability.
Cells in the human body contain signal transduction systems that respond to various external stimuli such as antibiotics and changes in osmotic pressure. When an external stimulus is detected, various biochemical reactions occur sequentially. This leads to the expression of relevant genes, allowing the cells to respond to the perturbed external environment. Furthermore, signal transduction leads to a drug response (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
However, even when the same external stimuli are detected, the responses of individual cells are greatly heterogeneous. This leads to the emergence of persister cells that are highly resistant to drugs. To identify potential sources of this cell-to cell variability, many studies have been conducted. However, most of the intermediate signal transduction reactions are unobservable with current experimental techniques.
A group of researchers including Dae Wook Kim and Hyukpyo Hong and led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences and IBS Biomedical Mathematics Group solved the mystery by exploiting queueing theory and Bayesian inference methodology. They proposed a queueing process that describes the signal transduction system in cells. Based on this, they developed Bayesian inference computational software using MBI (the Moment-based Bayesian Inference method). This enables the analysis of the signal transduction system without a direct observation of the intermediate steps. This study was published in Science Advances.
By analyzing experimental data from Escherichia coli using MBI, the research team found that cell-to-cell variability increases as the number of rate-limiting steps in the signaling pathway increases.
The rate-limiting steps denote the slowest steps (i.e., bottlenecks) in sequential biochemical reaction steps composing cell signaling pathways and thus dominates most of the signaling time. As the number of the rate-limiting steps increases, the intensity of the transduced signal becomes greatly heterogeneous even in a population of genetically identical cells. This finding is expected to provide a new paradigm for studying the heterogeneous antibiotic resistance of cells, which is a big challenge in cancer medicine.
Professor Kim said, “As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies.”
This work was supported by the Samsung Science and Technology Foundation, the National Research Foundation of Korea, and the Institute for Basic Science.
-Publication:Dae Wook Kim, Hyukpyo Hong, and Jae Kyoung Kim (2022) “Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number,”Science Advances March 18, 2022 (DOI: 10.1126/sciadv.abl4598)
-Profile:Professor Jae Kyoung Kimhttp://mathsci.kaist.ac.kr/~jaekkim
jaekkim@kaist.ac.kr@umichkim on TwitterDepartment of Mathematical SciencesKAIST
2022.03.29 View 10037 -
 Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 8181
Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 8181 -
 Decoding Brain Signals to Control a Robotic Arm
Advanced brain-machine interface system successfully interprets arm movement directions from neural signals in the brain
Researchers have developed a mind-reading system for decoding neural signals from the brain during arm movement. The method, described in the journal Applied Soft Computing, can be used by a person to control a robotic arm through a brain-machine interface (BMI).
A BMI is a device that translates nerve signals into commands to control a machine, such as a computer or a robotic limb. There are two main techniques for monitoring neural signals in BMIs: electroencephalography (EEG) and electrocorticography (ECoG).
The EEG exhibits signals from electrodes on the surface of the scalp and is widely employed because it is non-invasive, relatively cheap, safe and easy to use. However, the EEG has low spatial resolution and detects irrelevant neural signals, which makes it difficult to interpret the intentions of individuals from the EEG.
On the other hand, the ECoG is an invasive method that involves placing electrodes directly on the surface of the cerebral cortex below the scalp. Compared with the EEG, the ECoG can monitor neural signals with much higher spatial resolution and less background noise. However, this technique has several drawbacks.
“The ECoG is primarily used to find potential sources of epileptic seizures, meaning the electrodes are placed in different locations for different patients and may not be in the optimal regions of the brain for detecting sensory and movement signals,” explained Professor Jaeseung Jeong, a brain scientist at KAIST. “This inconsistency makes it difficult to decode brain signals to predict movements.”
To overcome these problems, Professor Jeong’s team developed a new method for decoding ECoG neural signals during arm movement. The system is based on a machine-learning system for analysing and predicting neural signals called an ‘echo-state network’ and a mathematical probability model called the Gaussian distribution.
In the study, the researchers recorded ECoG signals from four individuals with epilepsy while they were performing a reach-and-grasp task. Because the ECoG electrodes were placed according to the potential sources of each patient’s epileptic seizures, only 22% to 44% of the electrodes were located in the regions of the brain responsible for controlling movement.
During the movement task, the participants were given visual cues, either by placing a real tennis ball in front of them, or via a virtual reality headset showing a clip of a human arm reaching forward in first-person view. They were asked to reach forward, grasp an object, then return their hand and release the object, while wearing motion sensors on their wrists and fingers. In a second task, they were instructed to imagine reaching forward without moving their arms.
The researchers monitored the signals from the ECoG electrodes during real and imaginary arm movements, and tested whether the new system could predict the direction of this movement from the neural signals. They found that the novel decoder successfully classified arm movements in 24 directions in three-dimensional space, both in the real and virtual tasks, and that the results were at least five times more accurate than chance. They also used a computer simulation to show that the novel ECoG decoder could control the movements of a robotic arm.
Overall, the results suggest that the new machine learning-based BCI system successfully used ECoG signals to interpret the direction of the intended movements. The next steps will be to improve the accuracy and efficiency of the decoder. In the future, it could be used in a real-time BMI device to help people with movement or sensory impairments.
This research was supported by the KAIST Global Singularity Research Program of 2021, Brain Research Program of the National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning, and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education.
-PublicationHoon-Hee Kim, Jaeseung Jeong, “An electrocorticographic decoder for arm movement for brain-machine interface using an echo state network and Gaussian readout,” Applied SoftComputing online December 31, 2021 (doi.org/10.1016/j.asoc.2021.108393)
-ProfileProfessor Jaeseung JeongDepartment of Bio and Brain EngineeringCollege of EngineeringKAIST
2022.03.18 View 12058
Decoding Brain Signals to Control a Robotic Arm
Advanced brain-machine interface system successfully interprets arm movement directions from neural signals in the brain
Researchers have developed a mind-reading system for decoding neural signals from the brain during arm movement. The method, described in the journal Applied Soft Computing, can be used by a person to control a robotic arm through a brain-machine interface (BMI).
A BMI is a device that translates nerve signals into commands to control a machine, such as a computer or a robotic limb. There are two main techniques for monitoring neural signals in BMIs: electroencephalography (EEG) and electrocorticography (ECoG).
The EEG exhibits signals from electrodes on the surface of the scalp and is widely employed because it is non-invasive, relatively cheap, safe and easy to use. However, the EEG has low spatial resolution and detects irrelevant neural signals, which makes it difficult to interpret the intentions of individuals from the EEG.
On the other hand, the ECoG is an invasive method that involves placing electrodes directly on the surface of the cerebral cortex below the scalp. Compared with the EEG, the ECoG can monitor neural signals with much higher spatial resolution and less background noise. However, this technique has several drawbacks.
“The ECoG is primarily used to find potential sources of epileptic seizures, meaning the electrodes are placed in different locations for different patients and may not be in the optimal regions of the brain for detecting sensory and movement signals,” explained Professor Jaeseung Jeong, a brain scientist at KAIST. “This inconsistency makes it difficult to decode brain signals to predict movements.”
To overcome these problems, Professor Jeong’s team developed a new method for decoding ECoG neural signals during arm movement. The system is based on a machine-learning system for analysing and predicting neural signals called an ‘echo-state network’ and a mathematical probability model called the Gaussian distribution.
In the study, the researchers recorded ECoG signals from four individuals with epilepsy while they were performing a reach-and-grasp task. Because the ECoG electrodes were placed according to the potential sources of each patient’s epileptic seizures, only 22% to 44% of the electrodes were located in the regions of the brain responsible for controlling movement.
During the movement task, the participants were given visual cues, either by placing a real tennis ball in front of them, or via a virtual reality headset showing a clip of a human arm reaching forward in first-person view. They were asked to reach forward, grasp an object, then return their hand and release the object, while wearing motion sensors on their wrists and fingers. In a second task, they were instructed to imagine reaching forward without moving their arms.
The researchers monitored the signals from the ECoG electrodes during real and imaginary arm movements, and tested whether the new system could predict the direction of this movement from the neural signals. They found that the novel decoder successfully classified arm movements in 24 directions in three-dimensional space, both in the real and virtual tasks, and that the results were at least five times more accurate than chance. They also used a computer simulation to show that the novel ECoG decoder could control the movements of a robotic arm.
Overall, the results suggest that the new machine learning-based BCI system successfully used ECoG signals to interpret the direction of the intended movements. The next steps will be to improve the accuracy and efficiency of the decoder. In the future, it could be used in a real-time BMI device to help people with movement or sensory impairments.
This research was supported by the KAIST Global Singularity Research Program of 2021, Brain Research Program of the National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning, and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education.
-PublicationHoon-Hee Kim, Jaeseung Jeong, “An electrocorticographic decoder for arm movement for brain-machine interface using an echo state network and Gaussian readout,” Applied SoftComputing online December 31, 2021 (doi.org/10.1016/j.asoc.2021.108393)
-ProfileProfessor Jaeseung JeongDepartment of Bio and Brain EngineeringCollege of EngineeringKAIST
2022.03.18 View 12058 -
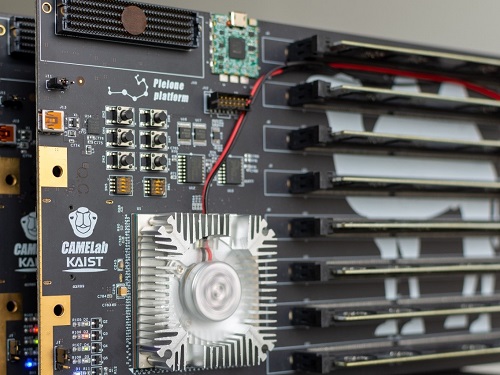 CXL-Based Memory Disaggregation Technology Opens Up a New Direction for Big Data Solution Frameworks
A KAIST team’s compute express link (CXL) provides new insights on memory disaggregation and ensures direct access and high-performance capabilities
A team from the Computer Architecture and Memory Systems Laboratory (CAMEL) at KAIST presented a new compute express link (CXL) solution whose directly accessible, and high-performance memory disaggregation opens new directions for big data memory processing. Professor Myoungsoo Jung said the team’s technology significantly improves performance compared to existing remote direct memory access (RDMA)-based memory disaggregation.
CXL is a peripheral component interconnect-express (PCIe)-based new dynamic multi-protocol made for efficiently utilizing memory devices and accelerators. Many enterprise data centers and memory vendors are paying attention to it as the next-generation multi-protocol for the era of big data.
Emerging big data applications such as machine learning, graph analytics, and in-memory databases require large memory capacities. However, scaling out the memory capacity via a prior memory interface like double data rate (DDR) is limited by the number of the central processing units (CPUs) and memory controllers. Therefore, memory disaggregation, which allows connecting a host to another host’s memory or memory nodes, has appeared.
RDMA is a way that a host can directly access another host’s memory via InfiniBand, the commonly used network protocol in data centers. Nowadays, most existing memory disaggregation technologies employ RDMA to get a large memory capacity. As a result, a host can share another host’s memory by transferring the data between local and remote memory.
Although RDMA-based memory disaggregation provides a large memory capacity to a host, two critical problems exist. First, scaling out the memory still needs an extra CPU to be added. Since passive memory such as dynamic random-access memory (DRAM), cannot operate by itself, it should be controlled by the CPU. Second, redundant data copies and software fabric interventions for RDMA-based memory disaggregation cause longer access latency. For example, remote memory access latency in RDMA-based memory disaggregation is multiple orders of magnitude longer than local memory access.
To address these issues, Professor Jung’s team developed the CXL-based memory disaggregation framework, including CXL-enabled customized CPUs, CXL devices, CXL switches, and CXL-aware operating system modules. The team’s CXL device is a pure passive and directly accessible memory node that contains multiple DRAM dual inline memory modules (DIMMs) and a CXL memory controller. Since the CXL memory controller supports the memory in the CXL device, a host can utilize the memory node without processor or software intervention. The team’s CXL switch enables scaling out a host’s memory capacity by hierarchically connecting multiple CXL devices to the CXL switch allowing more than hundreds of devices. Atop the switches and devices, the team’s CXL-enabled operating system removes redundant data copy and protocol conversion exhibited by conventional RDMA, which can significantly decrease access latency to the memory nodes.
In a test comparing loading 64B (cacheline) data from memory pooling devices, CXL-based memory disaggregation showed 8.2 times higher data load performance than RDMA-based memory disaggregation and even similar performance to local DRAM memory. In the team’s evaluations for a big data benchmark such as a machine learning-based test, CXL-based memory disaggregation technology also showed a maximum of 3.7 times higher performance than prior RDMA-based memory disaggregation technologies.
“Escaping from the conventional RDMA-based memory disaggregation, our CXL-based memory disaggregation framework can provide high scalability and performance for diverse datacenters and cloud service infrastructures,” said Professor Jung. He went on to stress, “Our CXL-based memory disaggregation research will bring about a new paradigm for memory solutions that will lead the era of big data.”
-Profile: Professor Myoungsoo Jung Computer Architecture and Memory Systems Laboratory (CAMEL)http://camelab.org School of Electrical EngineeringKAIST
2022.03.16 View 22604
CXL-Based Memory Disaggregation Technology Opens Up a New Direction for Big Data Solution Frameworks
A KAIST team’s compute express link (CXL) provides new insights on memory disaggregation and ensures direct access and high-performance capabilities
A team from the Computer Architecture and Memory Systems Laboratory (CAMEL) at KAIST presented a new compute express link (CXL) solution whose directly accessible, and high-performance memory disaggregation opens new directions for big data memory processing. Professor Myoungsoo Jung said the team’s technology significantly improves performance compared to existing remote direct memory access (RDMA)-based memory disaggregation.
CXL is a peripheral component interconnect-express (PCIe)-based new dynamic multi-protocol made for efficiently utilizing memory devices and accelerators. Many enterprise data centers and memory vendors are paying attention to it as the next-generation multi-protocol for the era of big data.
Emerging big data applications such as machine learning, graph analytics, and in-memory databases require large memory capacities. However, scaling out the memory capacity via a prior memory interface like double data rate (DDR) is limited by the number of the central processing units (CPUs) and memory controllers. Therefore, memory disaggregation, which allows connecting a host to another host’s memory or memory nodes, has appeared.
RDMA is a way that a host can directly access another host’s memory via InfiniBand, the commonly used network protocol in data centers. Nowadays, most existing memory disaggregation technologies employ RDMA to get a large memory capacity. As a result, a host can share another host’s memory by transferring the data between local and remote memory.
Although RDMA-based memory disaggregation provides a large memory capacity to a host, two critical problems exist. First, scaling out the memory still needs an extra CPU to be added. Since passive memory such as dynamic random-access memory (DRAM), cannot operate by itself, it should be controlled by the CPU. Second, redundant data copies and software fabric interventions for RDMA-based memory disaggregation cause longer access latency. For example, remote memory access latency in RDMA-based memory disaggregation is multiple orders of magnitude longer than local memory access.
To address these issues, Professor Jung’s team developed the CXL-based memory disaggregation framework, including CXL-enabled customized CPUs, CXL devices, CXL switches, and CXL-aware operating system modules. The team’s CXL device is a pure passive and directly accessible memory node that contains multiple DRAM dual inline memory modules (DIMMs) and a CXL memory controller. Since the CXL memory controller supports the memory in the CXL device, a host can utilize the memory node without processor or software intervention. The team’s CXL switch enables scaling out a host’s memory capacity by hierarchically connecting multiple CXL devices to the CXL switch allowing more than hundreds of devices. Atop the switches and devices, the team’s CXL-enabled operating system removes redundant data copy and protocol conversion exhibited by conventional RDMA, which can significantly decrease access latency to the memory nodes.
In a test comparing loading 64B (cacheline) data from memory pooling devices, CXL-based memory disaggregation showed 8.2 times higher data load performance than RDMA-based memory disaggregation and even similar performance to local DRAM memory. In the team’s evaluations for a big data benchmark such as a machine learning-based test, CXL-based memory disaggregation technology also showed a maximum of 3.7 times higher performance than prior RDMA-based memory disaggregation technologies.
“Escaping from the conventional RDMA-based memory disaggregation, our CXL-based memory disaggregation framework can provide high scalability and performance for diverse datacenters and cloud service infrastructures,” said Professor Jung. He went on to stress, “Our CXL-based memory disaggregation research will bring about a new paradigm for memory solutions that will lead the era of big data.”
-Profile: Professor Myoungsoo Jung Computer Architecture and Memory Systems Laboratory (CAMEL)http://camelab.org School of Electrical EngineeringKAIST
2022.03.16 View 22604 -
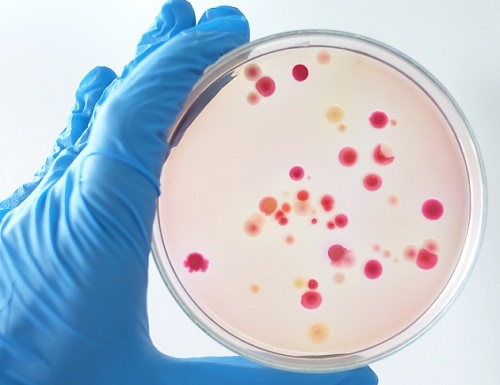 'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media
Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
Their results were made available online on Jan. 18 in Biosensors and Bioelectronics, ahead of publication in the journal’s April issue.
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. “By using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model, we demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures,” said Professor Sungho Jo from the School of Computing.
Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample — the spectral fingerprint — allowing researchers to identify its molecules. The surface-enhanced version places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. “Moreover, strong signals of surrounding media are also enhanced to overwhelm target signals, requiring time-consuming and tedious bacterial separation steps,” said Professor Yeon Sik Jung from the Department of Materials Science and Engineering.
To parse through the noisy signals, the researchers implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such an ability is critical for analyzing one-dimensional spectral data, according to Professor Jo.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that were almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” Professor Jo said. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
The National R&D Program, through a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, supported this research.
-PublicationEojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deepneural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
-ProfileProfessor Yeon Sik JungDepartment of Materials Science and EngineeringKAIST
Professor Sungho JoSchool of ComputingKAIST
2022.03.04 View 21825
'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media
Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
Their results were made available online on Jan. 18 in Biosensors and Bioelectronics, ahead of publication in the journal’s April issue.
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. “By using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model, we demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures,” said Professor Sungho Jo from the School of Computing.
Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample — the spectral fingerprint — allowing researchers to identify its molecules. The surface-enhanced version places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. “Moreover, strong signals of surrounding media are also enhanced to overwhelm target signals, requiring time-consuming and tedious bacterial separation steps,” said Professor Yeon Sik Jung from the Department of Materials Science and Engineering.
To parse through the noisy signals, the researchers implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such an ability is critical for analyzing one-dimensional spectral data, according to Professor Jo.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that were almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” Professor Jo said. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
The National R&D Program, through a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, supported this research.
-PublicationEojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deepneural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
-ProfileProfessor Yeon Sik JungDepartment of Materials Science and EngineeringKAIST
Professor Sungho JoSchool of ComputingKAIST
2022.03.04 View 21825 -
 Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 9819
Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 9819 -
 A Mathematical Model Shows High Viral Transmissions Reduce the Progression Rates for Severe Covid-19
The model suggests a clue as to when a pandemic will turn into an endemic
A mathematical model demonstrated that high transmission rates among highly vaccinated populations of COVID-19 ultimately reduce the numbers of severe cases. This model suggests a clue as to when this pandemic will turn into an endemic.
With the future of the pandemic remaining uncertain, a research team of mathematicians and medical scientists analyzed a mathematical model that may predict how the changing transmission rate of COVID-19 would affect the settlement process of the virus as a mild respiratory virus.
The team led by Professor Jae Kyoung Kim from the Department of Mathematical Science and Professor Eui-Cheol Shin from the Graduate School of Medical Science and Engineering used a new approach by dividing the human immune responses to SARS-CoV-2 into a shorter-term neutralizing antibody response and a longer-term T-cell immune response, and applying them each to a mathematical model. Additionally, the analysis was based on the fact that although breakthrough infection may occur frequently, the immune response of the patient will be boosted after recovery from each breakthrough infection.
The results showed that in an environment with a high vaccination rate, although COVID-19 cases may rise temporarily when the transmission rate increases, the ratio of critical cases would ultimately decline, thereby decreasing the total number of critical cases and in fact settling COVID-19 as a mild respiratory disease more quickly.
Conditions in which the number of cases may spike include relaxing social distancing measures or the rise of variants with higher transmission rates like the Omicron variant. This research did not take the less virulent characteristic of the Omicron variant into account but focused on the results of its high transmission rate, thereby predicting what may happen in the process of the endemic transition of COVID-19.
The research team pointed out the limitations of their mathematical model, such as the lack of consideration for age or patients with underlying diseases, and explained that the results of this study must be applied with care when compared against high-risk groups. Additionally, as medical systems may collapse when the number of cases rises sharply, this study must be interpreted with prudence and applied accordingly. The research team therefore emphasized that for policies that encourage a step-wise return to normality to succeed, the sustainable maintenance of public health systems is indispensable.
Professor Kim said, “We have drawn a counter-intuitive conclusion amid the unpredictable pandemic through an adequate mathematical model,” asserting the importance of applying mathematical models to medical research.
Professor Shin said, “Although the Omicron variant has become the dominant strain and the number of cases is rising rapidly in South Korea, it is important to use scientific approaches to predict the future and apply them to policies rather than fearing the current situation.”
The results of the research were published on medRxiv.org on February 11, under the title “Increasing viral transmission paradoxically reduces progression rates to severe COVID-19 during endemic transition.”
This research was funded by the Institute of Basic Science, the Korea Health Industry Development Institute, and the National Research Foundation of Korea.
-PublicationHyukpyo Hong, Ji Yun Noh, Hyojung Lee, Sunhwa Choi, Boseung Choi, Jae Kyung Kim, Eui-Cheol Shin, “Increasing viral transmission paradoxically reduces progression rates to
severe COVID-19 during endemic transition,” medRxiv, February 9, 2022 (doi.org/10.1101/2022.02.09.22270633)
-ProfileProfessor Jae Kyung KimDepartment of Mathematical SciencesKAIST
Professor Eui-Cheol ShinGraduate School of Medical Science and EngineeringKAIST
2022.02.22 View 10277
A Mathematical Model Shows High Viral Transmissions Reduce the Progression Rates for Severe Covid-19
The model suggests a clue as to when a pandemic will turn into an endemic
A mathematical model demonstrated that high transmission rates among highly vaccinated populations of COVID-19 ultimately reduce the numbers of severe cases. This model suggests a clue as to when this pandemic will turn into an endemic.
With the future of the pandemic remaining uncertain, a research team of mathematicians and medical scientists analyzed a mathematical model that may predict how the changing transmission rate of COVID-19 would affect the settlement process of the virus as a mild respiratory virus.
The team led by Professor Jae Kyoung Kim from the Department of Mathematical Science and Professor Eui-Cheol Shin from the Graduate School of Medical Science and Engineering used a new approach by dividing the human immune responses to SARS-CoV-2 into a shorter-term neutralizing antibody response and a longer-term T-cell immune response, and applying them each to a mathematical model. Additionally, the analysis was based on the fact that although breakthrough infection may occur frequently, the immune response of the patient will be boosted after recovery from each breakthrough infection.
The results showed that in an environment with a high vaccination rate, although COVID-19 cases may rise temporarily when the transmission rate increases, the ratio of critical cases would ultimately decline, thereby decreasing the total number of critical cases and in fact settling COVID-19 as a mild respiratory disease more quickly.
Conditions in which the number of cases may spike include relaxing social distancing measures or the rise of variants with higher transmission rates like the Omicron variant. This research did not take the less virulent characteristic of the Omicron variant into account but focused on the results of its high transmission rate, thereby predicting what may happen in the process of the endemic transition of COVID-19.
The research team pointed out the limitations of their mathematical model, such as the lack of consideration for age or patients with underlying diseases, and explained that the results of this study must be applied with care when compared against high-risk groups. Additionally, as medical systems may collapse when the number of cases rises sharply, this study must be interpreted with prudence and applied accordingly. The research team therefore emphasized that for policies that encourage a step-wise return to normality to succeed, the sustainable maintenance of public health systems is indispensable.
Professor Kim said, “We have drawn a counter-intuitive conclusion amid the unpredictable pandemic through an adequate mathematical model,” asserting the importance of applying mathematical models to medical research.
Professor Shin said, “Although the Omicron variant has become the dominant strain and the number of cases is rising rapidly in South Korea, it is important to use scientific approaches to predict the future and apply them to policies rather than fearing the current situation.”
The results of the research were published on medRxiv.org on February 11, under the title “Increasing viral transmission paradoxically reduces progression rates to severe COVID-19 during endemic transition.”
This research was funded by the Institute of Basic Science, the Korea Health Industry Development Institute, and the National Research Foundation of Korea.
-PublicationHyukpyo Hong, Ji Yun Noh, Hyojung Lee, Sunhwa Choi, Boseung Choi, Jae Kyung Kim, Eui-Cheol Shin, “Increasing viral transmission paradoxically reduces progression rates to
severe COVID-19 during endemic transition,” medRxiv, February 9, 2022 (doi.org/10.1101/2022.02.09.22270633)
-ProfileProfessor Jae Kyung KimDepartment of Mathematical SciencesKAIST
Professor Eui-Cheol ShinGraduate School of Medical Science and EngineeringKAIST
2022.02.22 View 10277