research
-
 KAIST-UCSD researchers build an enzyme discovering AI
- A joint research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Bernhard Palsson of UCSD developed ‘DeepECtransformer’, an artificial intelligence that can predict Enzyme Commission (EC) number of proteins.
- The AI is tasked to discover new enzymes that have not been discovered yet, which would allow prediction for a total of 5,360 types of Enzyme Commission (EC) numbers
- It is expected to be used in the development of microbial cell factories that produce environmentally friendly chemicals as a core technology for analyzing the metabolic network of a genome.
While E. coli is one of the most studied organisms, the function of 30% of proteins that make up E. coli has not yet been clearly revealed. For this, an artificial intelligence was used to discover 464 types of enzymes from the proteins that were unknown, and the researchers went on to verify the predictions of 3 types of proteins were successfully identified through in vitro enzyme assay.
KAIST (President Kwang-Hyung Lee) announced on the 24th that a joint research team comprised of Gi Bae Kim, Ji Yeon Kim, Dr. Jong An Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Dr. Charles J. Norsigian and Professor Bernhard O. Palsson of the Department of Bioengineering at UCSD has developed DeepECtransformer, an artificial intelligence that can predict the enzyme functions from the protein sequence, and has established a prediction system by utilizing the AI to quickly and accurately identify the enzyme function.
Enzymes are proteins that catalyze biological reactions, and identifying the function of each enzyme is essential to understanding the various chemical reactions that exist in living organisms and the metabolic characteristics of those organisms. Enzyme Commission (EC) number is an enzyme function classification system designed by the International Union of Biochemistry and Molecular Biology, and in order to understand the metabolic characteristics of various organisms, it is necessary to develop a technology that can quickly analyze enzymes and EC numbers of the enzymes present in the genome.
Various methodologies based on deep learning have been developed to analyze the features of biological sequences, including protein function prediction, but most of them have a problem of a black box, where the inference process of AI cannot be interpreted. Various prediction systems that utilize AI for enzyme function prediction have also been reported, but they do not solve this black box problem, or cannot interpret the reasoning process in fine-grained level (e.g., the level of amino acid residues in the enzyme sequence).
The joint team developed DeepECtransformer, an AI that utilizes deep learning and a protein homology analysis module to predict the enzyme function of a given protein sequence. To better understand the features of protein sequences, the transformer architecture, which is commonly used in natural language processing, was additionally used to extract important features about enzyme functions in the context of the entire protein sequence, which enabled the team to accurately predict the EC number of the enzyme. The developed DeepECtransformer can predict a total of 5360 EC numbers.
The joint team further analyzed the transformer architecture to understand the inference process of DeepECtransformer, and found that in the inference process, the AI utilizes information on catalytic active sites and/or the cofactor binding sites which are important for enzyme function. By analyzing the black box of DeepECtransformer, it was confirmed that the AI was able to identify the features that are important for enzyme function on its own during the learning process.
"By utilizing the prediction system we developed, we were able to predict the functions of enzymes that had not yet been identified and verify them experimentally," said Gi Bae Kim, the first author of the paper. "By using DeepECtransformer to identify previously unknown enzymes in living organisms, we will be able to more accurately analyze various facets involved in the metabolic processes of organisms, such as the enzymes needed to biosynthesize various useful compounds or the enzymes needed to biodegrade plastics." he added.
"DeepECtransformer, which quickly and accurately predicts enzyme functions, is a key technology in functional genomics, enabling us to analyze the function of entire enzymes at the systems level," said Professor Sang Yup Lee. He added, “We will be able to use it to develop eco-friendly microbial factories based on comprehensive genome-scale metabolic models, potentially minimizing missing information of metabolism.”
The joint team’s work on DeepECtransformer is described in the paper titled "Functional annotation of enzyme-encoding genes using deep learning with transformer layers" written by Gi Bae Kim, Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST and their colleagues. The paper was published via peer-review on the 14th of November on “Nature Communications”.
This research was conducted with the support by “the Development of next-generation biorefinery platform technologies for leading bio-based chemicals industry project (2022M3J5A1056072)” and by “Development of platform technologies of microbial cell factories for the next-generation biorefineries project (2022M3J5A1056117)” from National Research Foundation supported by the Korean Ministry of Science and ICT (Project Leader: Distinguished Professor Sang Yup Lee, KAIST).
< Figure 1. The structure of DeepECtransformer's artificial neural network >
2023.11.24 View 6459
KAIST-UCSD researchers build an enzyme discovering AI
- A joint research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Bernhard Palsson of UCSD developed ‘DeepECtransformer’, an artificial intelligence that can predict Enzyme Commission (EC) number of proteins.
- The AI is tasked to discover new enzymes that have not been discovered yet, which would allow prediction for a total of 5,360 types of Enzyme Commission (EC) numbers
- It is expected to be used in the development of microbial cell factories that produce environmentally friendly chemicals as a core technology for analyzing the metabolic network of a genome.
While E. coli is one of the most studied organisms, the function of 30% of proteins that make up E. coli has not yet been clearly revealed. For this, an artificial intelligence was used to discover 464 types of enzymes from the proteins that were unknown, and the researchers went on to verify the predictions of 3 types of proteins were successfully identified through in vitro enzyme assay.
KAIST (President Kwang-Hyung Lee) announced on the 24th that a joint research team comprised of Gi Bae Kim, Ji Yeon Kim, Dr. Jong An Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Dr. Charles J. Norsigian and Professor Bernhard O. Palsson of the Department of Bioengineering at UCSD has developed DeepECtransformer, an artificial intelligence that can predict the enzyme functions from the protein sequence, and has established a prediction system by utilizing the AI to quickly and accurately identify the enzyme function.
Enzymes are proteins that catalyze biological reactions, and identifying the function of each enzyme is essential to understanding the various chemical reactions that exist in living organisms and the metabolic characteristics of those organisms. Enzyme Commission (EC) number is an enzyme function classification system designed by the International Union of Biochemistry and Molecular Biology, and in order to understand the metabolic characteristics of various organisms, it is necessary to develop a technology that can quickly analyze enzymes and EC numbers of the enzymes present in the genome.
Various methodologies based on deep learning have been developed to analyze the features of biological sequences, including protein function prediction, but most of them have a problem of a black box, where the inference process of AI cannot be interpreted. Various prediction systems that utilize AI for enzyme function prediction have also been reported, but they do not solve this black box problem, or cannot interpret the reasoning process in fine-grained level (e.g., the level of amino acid residues in the enzyme sequence).
The joint team developed DeepECtransformer, an AI that utilizes deep learning and a protein homology analysis module to predict the enzyme function of a given protein sequence. To better understand the features of protein sequences, the transformer architecture, which is commonly used in natural language processing, was additionally used to extract important features about enzyme functions in the context of the entire protein sequence, which enabled the team to accurately predict the EC number of the enzyme. The developed DeepECtransformer can predict a total of 5360 EC numbers.
The joint team further analyzed the transformer architecture to understand the inference process of DeepECtransformer, and found that in the inference process, the AI utilizes information on catalytic active sites and/or the cofactor binding sites which are important for enzyme function. By analyzing the black box of DeepECtransformer, it was confirmed that the AI was able to identify the features that are important for enzyme function on its own during the learning process.
"By utilizing the prediction system we developed, we were able to predict the functions of enzymes that had not yet been identified and verify them experimentally," said Gi Bae Kim, the first author of the paper. "By using DeepECtransformer to identify previously unknown enzymes in living organisms, we will be able to more accurately analyze various facets involved in the metabolic processes of organisms, such as the enzymes needed to biosynthesize various useful compounds or the enzymes needed to biodegrade plastics." he added.
"DeepECtransformer, which quickly and accurately predicts enzyme functions, is a key technology in functional genomics, enabling us to analyze the function of entire enzymes at the systems level," said Professor Sang Yup Lee. He added, “We will be able to use it to develop eco-friendly microbial factories based on comprehensive genome-scale metabolic models, potentially minimizing missing information of metabolism.”
The joint team’s work on DeepECtransformer is described in the paper titled "Functional annotation of enzyme-encoding genes using deep learning with transformer layers" written by Gi Bae Kim, Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST and their colleagues. The paper was published via peer-review on the 14th of November on “Nature Communications”.
This research was conducted with the support by “the Development of next-generation biorefinery platform technologies for leading bio-based chemicals industry project (2022M3J5A1056072)” and by “Development of platform technologies of microbial cell factories for the next-generation biorefineries project (2022M3J5A1056117)” from National Research Foundation supported by the Korean Ministry of Science and ICT (Project Leader: Distinguished Professor Sang Yup Lee, KAIST).
< Figure 1. The structure of DeepECtransformer's artificial neural network >
2023.11.24 View 6459 -
 An intravenous needle that irreversibly softens via body temperature on insertion
- A joint research team at KAIST developed an intravenous (IV) needle that softens upon insertion, minimizing risk of damage to blood vessels and tissues.
- Once used, it remains soft even at room temperature, preventing accidental needle stick injuries and unethical multiple use of needle.
- A thin-film temperature sensor can be embedded with this needle, enabling real-time monitoring of the patient's core body temperature, or detection of unintended fluid leakage, during IV medication.
Intravenous (IV) injection is a method commonly used in patient’s treatment worldwide as it induces rapid effects and allows treatment through continuous administration of medication by directly injecting drugs into the blood vessel. However, medical IV needles, made of hard materials such as stainless steel or plastic which do not mechanically match the soft biological tissues of the body, can cause critical problems in healthcare settings, starting from minor tissue damages in the injection sites to serious inflammations.
The structure and dexterity of rigid medical IV devices also enable unethical reuse of needles for reduction of injection costs, leading to transmission of deadly blood-borne disease infections such as human immunodeficiency virus (HIV) and hepatitis B/C viruses. Furthermore, unintended needlestick injuries are frequently occurring in medical settings worldwide, that are viable sources of such infections, with IV needles having the greatest susceptibility of being the medium of transmissible diseases. For these reasons, the World Health Organization (WHO) in 2015 launched a policy on safe injection practices to encourage the development and use of “smart” syringes that have features to prevent re-use, after a tremendous increase in the number of deadly infectious disease worldwide due to medical-sharps related issues.
KAIST announced on the 13th that Professor Jae-Woong Jeong and his research team of its School of Electrical Engineering succeeded in developing the Phase-Convertible, Adapting and non-REusable (P-CARE) needle with variable stiffness that can improve patient health and ensure the safety of medical staff through convergent joint research with another team led by Professor Won-Il Jeong of the Graduate School of Medical Sciences.
The new technology is expected to allow patients to move without worrying about pain at the injection site as it reduces the risk of damage to the wall of the blood vessel as patients receive IV medication. This is possible with the needle’s stiffness-tunable characteristics which will make it soft and flexible upon insertion into the body due to increased temperature, adapting to the movement of thin-walled vein. It is also expected to prevent blood-borne disease infections caused by accidental needlestick injuries or unethical re-using of syringes as the deformed needle remains perpetually soft even after it is retracted from the injection site.
The results of this research, in which Karen-Christian Agno, a doctoral researcher of the School of Electrical Engineering at and Dr. Keungmo Yang of the Graduate School of Medical Sciences participated as co-first authors, was published in Nature Biomedical Engineering on October 30. (Paper title: A temperature-responsive intravenous needle that irreversibly softens on insertion)
< Figure 1. Disposable variable stiffness intravenous needle. (a) Conceptual illustration of the key features of the P-CARE needle whose mechanical properties can be changed by body temperature, (b) Photograph of commonly used IV access devices and the P-CARE needle, (c) Performance of common IV access devices and the P-CARE needle >
“We’ve developed this special needle using advanced materials and micro/nano engineering techniques, and it can solve many global problems related to conventional medical needles used in healthcare worldwide”, said Jae-Woong Jeong, Ph.D., an associate professor of Electrical Engineering at KAIST and a lead senior author of the study.
The softening IV needle created by the research team is made up of liquid metal gallium that forms the hollow, mechanical needle frame encapsulated within an ultra-soft silicone material. In its solid state, gallium has sufficient hardness that enables puncturing of soft biological tissues. However, gallium melts when it is exposed to body temperature upon insertion, and changes it into a soft state like the surrounding tissue, enabling stable delivery of the drug without damaging blood vessels. Once used, a needle remains soft even at room temperature due to the supercooling phenomenon of gallium, fundamentally preventing needlestick accidents and reuse problems.
Biocompatibility of the softening IV needle was validated through in vivo studies in mice. The studies showed that implanted needles caused significantly less inflammation relative to the standard IV access devices of similar size made of metal needles or plastic catheters. The study also confirmed the new needle was able to deliver medications as reliably as commercial injection needles.
< Photo 1. Photo of the P-CARE needle that softens with body temperature. >
Researchers also showed possibility of integrating a customized ultra-thin temperature sensor with the softening IV needle to measure the on-site temperature which can further enhance patient’s well-being. The single assembly of sensor-needle device can be used to monitor the core body temperature, or even detect if there is a fluid leakage on-site during indwelling use, eliminating the need for additional medical tools or procedures to provide the patients with better health care services.
The researchers believe that this transformative IV needle can open new opportunities for wide range of applications particularly in clinical setups, in terms of redesigning other medical needles and sharp medical tools to reduce muscle tissue injury during indwelling use. The softening IV needle may become even more valuable in the present times as there is an estimated 16 billion medical injections administered annually in a global scale, yet not all needles are disposed of properly, based on a 2018 WHO report.
< Figure 2. Biocompatibility test for P-CARE needle: Images of H&E stained histology (the area inside the dashed box on the left is provided in an expanded view in the right), TUNEL staining (green), DAPI staining of nuclei (blue) and co-staining (TUNEL and DAPI) of muscle tissue from different organs. >
< Figure 3. Conceptual images of potential utilization for temperature monitoring function of P-CARE needle integrated with a temperature sensor. >
(a) Schematic diagram of injecting a drug through intravenous injection into the abdomen of a laboratory mouse (b) Change of body temperature upon injection of drug (c) Conceptual illustration of normal intravenous drug injection (top) and fluid leakage (bottom) (d) Comparison of body temperature during normal drug injection and fluid leakage: when the fluid leak occur due to incorrect insertion, a sudden drop of temperature is detected.
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT.
2023.11.13 View 9625
An intravenous needle that irreversibly softens via body temperature on insertion
- A joint research team at KAIST developed an intravenous (IV) needle that softens upon insertion, minimizing risk of damage to blood vessels and tissues.
- Once used, it remains soft even at room temperature, preventing accidental needle stick injuries and unethical multiple use of needle.
- A thin-film temperature sensor can be embedded with this needle, enabling real-time monitoring of the patient's core body temperature, or detection of unintended fluid leakage, during IV medication.
Intravenous (IV) injection is a method commonly used in patient’s treatment worldwide as it induces rapid effects and allows treatment through continuous administration of medication by directly injecting drugs into the blood vessel. However, medical IV needles, made of hard materials such as stainless steel or plastic which do not mechanically match the soft biological tissues of the body, can cause critical problems in healthcare settings, starting from minor tissue damages in the injection sites to serious inflammations.
The structure and dexterity of rigid medical IV devices also enable unethical reuse of needles for reduction of injection costs, leading to transmission of deadly blood-borne disease infections such as human immunodeficiency virus (HIV) and hepatitis B/C viruses. Furthermore, unintended needlestick injuries are frequently occurring in medical settings worldwide, that are viable sources of such infections, with IV needles having the greatest susceptibility of being the medium of transmissible diseases. For these reasons, the World Health Organization (WHO) in 2015 launched a policy on safe injection practices to encourage the development and use of “smart” syringes that have features to prevent re-use, after a tremendous increase in the number of deadly infectious disease worldwide due to medical-sharps related issues.
KAIST announced on the 13th that Professor Jae-Woong Jeong and his research team of its School of Electrical Engineering succeeded in developing the Phase-Convertible, Adapting and non-REusable (P-CARE) needle with variable stiffness that can improve patient health and ensure the safety of medical staff through convergent joint research with another team led by Professor Won-Il Jeong of the Graduate School of Medical Sciences.
The new technology is expected to allow patients to move without worrying about pain at the injection site as it reduces the risk of damage to the wall of the blood vessel as patients receive IV medication. This is possible with the needle’s stiffness-tunable characteristics which will make it soft and flexible upon insertion into the body due to increased temperature, adapting to the movement of thin-walled vein. It is also expected to prevent blood-borne disease infections caused by accidental needlestick injuries or unethical re-using of syringes as the deformed needle remains perpetually soft even after it is retracted from the injection site.
The results of this research, in which Karen-Christian Agno, a doctoral researcher of the School of Electrical Engineering at and Dr. Keungmo Yang of the Graduate School of Medical Sciences participated as co-first authors, was published in Nature Biomedical Engineering on October 30. (Paper title: A temperature-responsive intravenous needle that irreversibly softens on insertion)
< Figure 1. Disposable variable stiffness intravenous needle. (a) Conceptual illustration of the key features of the P-CARE needle whose mechanical properties can be changed by body temperature, (b) Photograph of commonly used IV access devices and the P-CARE needle, (c) Performance of common IV access devices and the P-CARE needle >
“We’ve developed this special needle using advanced materials and micro/nano engineering techniques, and it can solve many global problems related to conventional medical needles used in healthcare worldwide”, said Jae-Woong Jeong, Ph.D., an associate professor of Electrical Engineering at KAIST and a lead senior author of the study.
The softening IV needle created by the research team is made up of liquid metal gallium that forms the hollow, mechanical needle frame encapsulated within an ultra-soft silicone material. In its solid state, gallium has sufficient hardness that enables puncturing of soft biological tissues. However, gallium melts when it is exposed to body temperature upon insertion, and changes it into a soft state like the surrounding tissue, enabling stable delivery of the drug without damaging blood vessels. Once used, a needle remains soft even at room temperature due to the supercooling phenomenon of gallium, fundamentally preventing needlestick accidents and reuse problems.
Biocompatibility of the softening IV needle was validated through in vivo studies in mice. The studies showed that implanted needles caused significantly less inflammation relative to the standard IV access devices of similar size made of metal needles or plastic catheters. The study also confirmed the new needle was able to deliver medications as reliably as commercial injection needles.
< Photo 1. Photo of the P-CARE needle that softens with body temperature. >
Researchers also showed possibility of integrating a customized ultra-thin temperature sensor with the softening IV needle to measure the on-site temperature which can further enhance patient’s well-being. The single assembly of sensor-needle device can be used to monitor the core body temperature, or even detect if there is a fluid leakage on-site during indwelling use, eliminating the need for additional medical tools or procedures to provide the patients with better health care services.
The researchers believe that this transformative IV needle can open new opportunities for wide range of applications particularly in clinical setups, in terms of redesigning other medical needles and sharp medical tools to reduce muscle tissue injury during indwelling use. The softening IV needle may become even more valuable in the present times as there is an estimated 16 billion medical injections administered annually in a global scale, yet not all needles are disposed of properly, based on a 2018 WHO report.
< Figure 2. Biocompatibility test for P-CARE needle: Images of H&E stained histology (the area inside the dashed box on the left is provided in an expanded view in the right), TUNEL staining (green), DAPI staining of nuclei (blue) and co-staining (TUNEL and DAPI) of muscle tissue from different organs. >
< Figure 3. Conceptual images of potential utilization for temperature monitoring function of P-CARE needle integrated with a temperature sensor. >
(a) Schematic diagram of injecting a drug through intravenous injection into the abdomen of a laboratory mouse (b) Change of body temperature upon injection of drug (c) Conceptual illustration of normal intravenous drug injection (top) and fluid leakage (bottom) (d) Comparison of body temperature during normal drug injection and fluid leakage: when the fluid leak occur due to incorrect insertion, a sudden drop of temperature is detected.
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT.
2023.11.13 View 9625 -
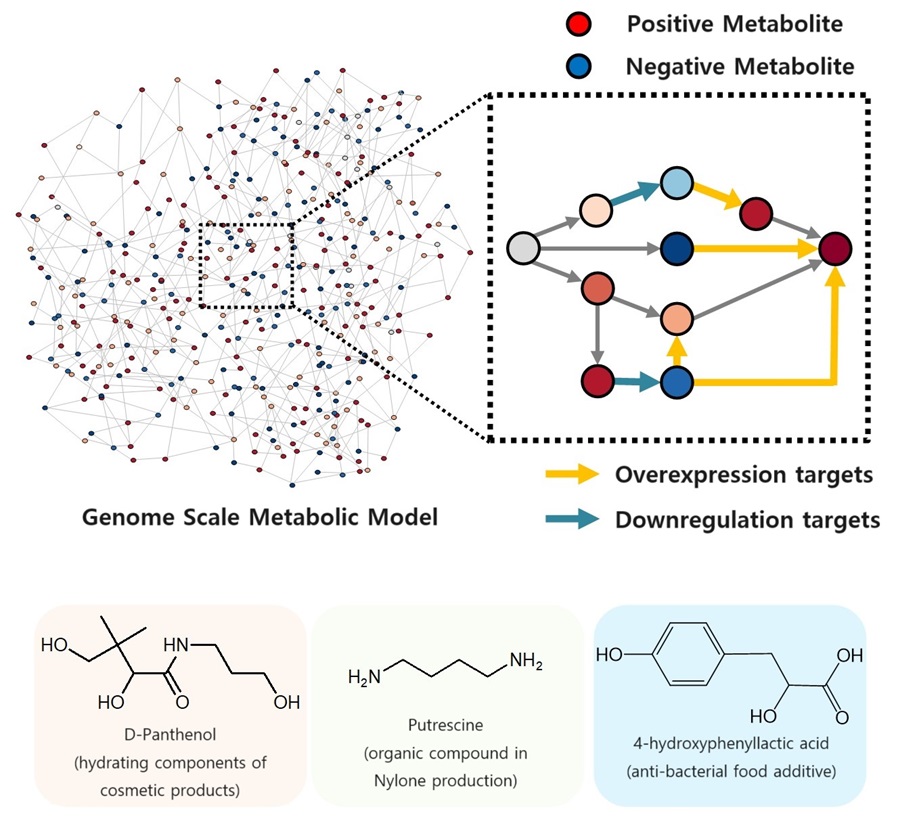 KAIST proposes alternatives to chemical factories through “iBridge”
- A computer simulation program “iBridge” was developed at KAIST that can put together microbial cell factories quickly and efficiently to produce cosmetics and food additives, and raw materials for nylons
- Eco-friendly and sustainable fermentation process to establish an alternative to chemical plants
As climate change and environmental concerns intensify, sustainable microbial cell factories garner significant attention as candidates to replace chemical plants. To develop microorganisms to be used in the microbial cell factories, it is crucial to modify their metabolic processes to induce efficient target chemical production by modulating its gene expressions. Yet, the challenge persists in determining which gene expressions to amplify and suppress, and the experimental verification of these modification targets is a time- and resource-intensive process even for experts. The challenges were addressed by a team of researchers at KAIST (President Kwang-Hyung Lee) led by Distinguished Professor Sang Yup Lee.
It was announced on the 9th by the school that a method for building a microbial factory at low cost, quickly and efficiently, was presented by a novel computer simulation program developed by the team under Professor Lee’s guidance, which is named “iBridge”. This innovative system is designed to predict gene targets to either overexpress or downregulate in the goal of producing a desired compound to enable the cost-effective and efficient construction of microbial cell factories specifically tailored for producing the chemical compound in demand from renewable biomass.
Systems metabolic engineering is a field of research and engineering pioneered by KAIST’s Distinguished Professor Sang Yup Lee that seeks to produce valuable compounds in industrial demands using microorganisms that are re-configured by a combination of methods including, but not limited to, metabolic engineering, synthetic biology, systems biology, and fermentation engineering.
In order to improve microorganisms’ capability to produce useful compounds, it is essential to delete, suppress, or overexpress microbial genes. However, it is difficult even for the experts to identify the gene targets to modify without experimental confirmations for each of them, which can take up immeasurable amount of time and resources.
The newly developed iBridge identifies positive and negative metabolites within cells, which exert positive and/or negative impact on formation of the products, by calculating the sum of covariances of their outgoing (consuming) reaction fluxes for a target chemical. Subsequently, it pinpoints "bridge" reactions responsible for converting negative metabolites into positive ones as candidates for overexpression, while identifying the opposites as targets for downregulation.
The research team successfully utilized the iBridge simulation to establish E. coli microbial cell factories each capable of producing three of the compounds that are in high demands at a production capacity that has not been reported around the world. They developed E. coli strains that can each produce panthenol, a moisturizing agent found in many cosmetics, putrescine, which is one of the key components in nylon production, and 4-hydroxyphenyllactic acid, an anti-bacterial food additive. In addition to these three compounds, the study presents predictions for overexpression and suppression genes to construct microbial factories for 298 other industrially valuable compounds.
Dr. Youngjoon Lee, the co-first author of this paper from KAIST, emphasized the accelerated construction of various microbial factories the newly developed simulation enabled. He stated, "With the use of this simulation, multiple microbial cell factories have been established significantly faster than it would have been using the conventional methods. Microbial cell factories producing a wider range of valuable compounds can now be constructed quickly using this technology."
Professor Sang Yup Lee said, "Systems metabolic engineering is a crucial technology for addressing the current climate change issues." He added, "This simulation could significantly expedite the transition from resorting to conventional chemical factories to utilizing environmentally friendly microbial factories."
< Figure. Conceptual diagram of the flow of iBridge simulation >
The team’s work on iBridge is described in a paper titled "Genome-Wide Identification of Overexpression and Downregulation Gene Targets Based on the Sum of Covariances of the Outgoing Reaction Fluxes" written by Dr. Won Jun Kim, and Dr. Youngjoon Lee of the Bioprocess Research Center and Professors Hyun Uk Kim and Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST. The paper was published via peer-review on the 6th of November on “Cell Systems” by Cell Press.
This research was conducted with the support from the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and Development of Platform Technology for the Production of Novel Aromatic Bioplastic using Microbial Cell Factories Project (Project Leader: Research Professor So Young Choi, KAIST) of the Korean Ministry of Science and ICT.
2023.11.09 View 8653
KAIST proposes alternatives to chemical factories through “iBridge”
- A computer simulation program “iBridge” was developed at KAIST that can put together microbial cell factories quickly and efficiently to produce cosmetics and food additives, and raw materials for nylons
- Eco-friendly and sustainable fermentation process to establish an alternative to chemical plants
As climate change and environmental concerns intensify, sustainable microbial cell factories garner significant attention as candidates to replace chemical plants. To develop microorganisms to be used in the microbial cell factories, it is crucial to modify their metabolic processes to induce efficient target chemical production by modulating its gene expressions. Yet, the challenge persists in determining which gene expressions to amplify and suppress, and the experimental verification of these modification targets is a time- and resource-intensive process even for experts. The challenges were addressed by a team of researchers at KAIST (President Kwang-Hyung Lee) led by Distinguished Professor Sang Yup Lee.
It was announced on the 9th by the school that a method for building a microbial factory at low cost, quickly and efficiently, was presented by a novel computer simulation program developed by the team under Professor Lee’s guidance, which is named “iBridge”. This innovative system is designed to predict gene targets to either overexpress or downregulate in the goal of producing a desired compound to enable the cost-effective and efficient construction of microbial cell factories specifically tailored for producing the chemical compound in demand from renewable biomass.
Systems metabolic engineering is a field of research and engineering pioneered by KAIST’s Distinguished Professor Sang Yup Lee that seeks to produce valuable compounds in industrial demands using microorganisms that are re-configured by a combination of methods including, but not limited to, metabolic engineering, synthetic biology, systems biology, and fermentation engineering.
In order to improve microorganisms’ capability to produce useful compounds, it is essential to delete, suppress, or overexpress microbial genes. However, it is difficult even for the experts to identify the gene targets to modify without experimental confirmations for each of them, which can take up immeasurable amount of time and resources.
The newly developed iBridge identifies positive and negative metabolites within cells, which exert positive and/or negative impact on formation of the products, by calculating the sum of covariances of their outgoing (consuming) reaction fluxes for a target chemical. Subsequently, it pinpoints "bridge" reactions responsible for converting negative metabolites into positive ones as candidates for overexpression, while identifying the opposites as targets for downregulation.
The research team successfully utilized the iBridge simulation to establish E. coli microbial cell factories each capable of producing three of the compounds that are in high demands at a production capacity that has not been reported around the world. They developed E. coli strains that can each produce panthenol, a moisturizing agent found in many cosmetics, putrescine, which is one of the key components in nylon production, and 4-hydroxyphenyllactic acid, an anti-bacterial food additive. In addition to these three compounds, the study presents predictions for overexpression and suppression genes to construct microbial factories for 298 other industrially valuable compounds.
Dr. Youngjoon Lee, the co-first author of this paper from KAIST, emphasized the accelerated construction of various microbial factories the newly developed simulation enabled. He stated, "With the use of this simulation, multiple microbial cell factories have been established significantly faster than it would have been using the conventional methods. Microbial cell factories producing a wider range of valuable compounds can now be constructed quickly using this technology."
Professor Sang Yup Lee said, "Systems metabolic engineering is a crucial technology for addressing the current climate change issues." He added, "This simulation could significantly expedite the transition from resorting to conventional chemical factories to utilizing environmentally friendly microbial factories."
< Figure. Conceptual diagram of the flow of iBridge simulation >
The team’s work on iBridge is described in a paper titled "Genome-Wide Identification of Overexpression and Downregulation Gene Targets Based on the Sum of Covariances of the Outgoing Reaction Fluxes" written by Dr. Won Jun Kim, and Dr. Youngjoon Lee of the Bioprocess Research Center and Professors Hyun Uk Kim and Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST. The paper was published via peer-review on the 6th of November on “Cell Systems” by Cell Press.
This research was conducted with the support from the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and Development of Platform Technology for the Production of Novel Aromatic Bioplastic using Microbial Cell Factories Project (Project Leader: Research Professor So Young Choi, KAIST) of the Korean Ministry of Science and ICT.
2023.11.09 View 8653 -
 KAIST builds a high-resolution 3D holographic sensor using a single mask
Holographic cameras can provide more realistic images than ordinary cameras thanks to their ability to acquire 3D information about objects. However, existing holographic cameras use interferometers that measure the wavelength and refraction of light through the interference of light waves, which makes them complex and sensitive to their surrounding environment.
On August 23, a KAIST research team led by Professor YongKeun Park from the Department of Physics announced a new leap forward in 3D holographic imaging sensor technology.
The team proposed an innovative holographic camera technology that does not use complex interferometry. Instead, it uses a mask to precisely measure the phase information of light and reconstruct the 3D information of an object with higher accuracy.
< Figure 1. Structure and principle of the proposed holographic camera. The amplitude and phase information of light scattered from a holographic camera can be measured. >
The team used a mask that fulfills certain mathematical conditions and incorporated it into an ordinary camera, and the light scattered from a laser is measured through the mask and analyzed using a computer. This does not require a complex interferometer and allows the phase information of light to be collected through a simplified optical system. With this technique, the mask that is placed between the two lenses and behind an object plays an important role. The mask selectively filters specific parts of light,, and the intensity of the light passing through the lens can be measured using an ordinary commercial camera. This technique combines the image data received from the camera with the unique pattern received from the mask and reconstructs an object’s precise 3D information using an algorithm.
This method allows a high-resolution 3D image of an object to be captured in any position. In practical situations, one can construct a laser-based holographic 3D image sensor by adding a mask with a simple design to a general image sensor. This makes the design and construction of the optical system much easier. In particular, this novel technology can capture high-resolution holographic images of objects moving at high speeds, which widens its potential field of application.
< Figure 2. A moving doll captured by a conventional camera and the proposed holographic camera. When taking a picture without focusing on the object, only a blurred image of the doll can be obtained from a general camera, but the proposed holographic camera can restore the blurred image of the doll into a clear image. >
The results of this study, conducted by Dr. Jeonghun Oh from the KAIST Department of Physics as the first author, were published in Nature Communications on August 12 under the title, "Non-interferometric stand-alone single-shot holographic camera using reciprocal diffractive imaging".
Dr. Oh said, “The holographic camera module we are suggesting can be built by adding a filter to an ordinary camera, which would allow even non-experts to handle it easily in everyday life if it were to be commercialized.” He added, “In particular, it is a promising candidate with the potential to replace existing remote sensing technologies.”
This research was supported by the National Research Foundation’s Leader Research Project, the Korean Ministry of Science and ICT’s Core Hologram Technology Support Project, and the Nano and Material Technology Development Project.
2023.09.05 View 8885
KAIST builds a high-resolution 3D holographic sensor using a single mask
Holographic cameras can provide more realistic images than ordinary cameras thanks to their ability to acquire 3D information about objects. However, existing holographic cameras use interferometers that measure the wavelength and refraction of light through the interference of light waves, which makes them complex and sensitive to their surrounding environment.
On August 23, a KAIST research team led by Professor YongKeun Park from the Department of Physics announced a new leap forward in 3D holographic imaging sensor technology.
The team proposed an innovative holographic camera technology that does not use complex interferometry. Instead, it uses a mask to precisely measure the phase information of light and reconstruct the 3D information of an object with higher accuracy.
< Figure 1. Structure and principle of the proposed holographic camera. The amplitude and phase information of light scattered from a holographic camera can be measured. >
The team used a mask that fulfills certain mathematical conditions and incorporated it into an ordinary camera, and the light scattered from a laser is measured through the mask and analyzed using a computer. This does not require a complex interferometer and allows the phase information of light to be collected through a simplified optical system. With this technique, the mask that is placed between the two lenses and behind an object plays an important role. The mask selectively filters specific parts of light,, and the intensity of the light passing through the lens can be measured using an ordinary commercial camera. This technique combines the image data received from the camera with the unique pattern received from the mask and reconstructs an object’s precise 3D information using an algorithm.
This method allows a high-resolution 3D image of an object to be captured in any position. In practical situations, one can construct a laser-based holographic 3D image sensor by adding a mask with a simple design to a general image sensor. This makes the design and construction of the optical system much easier. In particular, this novel technology can capture high-resolution holographic images of objects moving at high speeds, which widens its potential field of application.
< Figure 2. A moving doll captured by a conventional camera and the proposed holographic camera. When taking a picture without focusing on the object, only a blurred image of the doll can be obtained from a general camera, but the proposed holographic camera can restore the blurred image of the doll into a clear image. >
The results of this study, conducted by Dr. Jeonghun Oh from the KAIST Department of Physics as the first author, were published in Nature Communications on August 12 under the title, "Non-interferometric stand-alone single-shot holographic camera using reciprocal diffractive imaging".
Dr. Oh said, “The holographic camera module we are suggesting can be built by adding a filter to an ordinary camera, which would allow even non-experts to handle it easily in everyday life if it were to be commercialized.” He added, “In particular, it is a promising candidate with the potential to replace existing remote sensing technologies.”
This research was supported by the National Research Foundation’s Leader Research Project, the Korean Ministry of Science and ICT’s Core Hologram Technology Support Project, and the Nano and Material Technology Development Project.
2023.09.05 View 8885 -
 A KAIST Research Team Develops a Smart Color-Changing Flexible Battery with Ultra-high Efficiency
With the rapid growth of the smart and wearable electronic devices market, smart next-generation energy storage systems that have energy storage functions as well as additional color-changing properties are receiving a great deal of attention. However, existing electrochromic devices have low electrical conductivity, leading to low efficiency in electron and ion mobility, and low storage capacities. Such batteries have therefore been limited to use in flexible and wearable devices.
On August 21, a joint research team led by Professor Il-Doo Kim from the KAIST Department of Materials Science and Engineering (DMSE) and Professor Tae Gwang Yun from the Myongji University Department of Materials Science and Engineering announced the development of a smart electrochromic Zn-ion battery that can visually represent its charging and discharging processes using an electrochromic polymer anode incorporated with a “π-bridge spacer”, which increases electron and ion mobility efficiency.
Batteries topped with electrochromic properties are groundbreaking inventions that can visually represent their charged and discharged states using colors, and can be used as display devices that cut down energy consumption for indoor cooling by controlling solar absorbance. The research team successfully built a flexible and electrochromic smart Zn-ion battery that can maintain its excellent electrochromic and electrochemical properties, even under long-term exposure to the atmosphere and mechanical deformations.
< Figure 1. Electrochromic zinc ion battery whose anode is made of a polymer that turns dark blue when charged and transparent when discharged. >
To maximize the efficiency of electron and ion mobility, the team modelled and synthesized the first π-bridge spacer-incorporated polymer anode in the world. π-bonds can improve the mobility of electrons within a structure to speed up ion movement and maximize ion adsorption efficiency, which improves its energy storage capacity.
In anode-based batteries with a π-bridge spacer, the spacer provides room for quicker ion movement. This allows fast charging, an improved zinc-ion discharging capacity of 110 mAh/g, which is 40% greater than previously reported, and a 30% increase in electrochromic function that switches from dark blue to transparent when the device is charged/discharged. In addition, should the transparent flexible battery technology be applied to smart windows, they would display darker colors during the day while they absorb solar energy, and function as a futuristic energy storage technique that can block out UV radiation and replace curtains.
< Figure 2. A schematic diagram of the structure of the electrochromic polymer with π-π spacer and the operation of a smart flexible battery using this cathode material. >
< Figure 3. (A) Density Functional Theory (DFT) theory-based atomic and electronic structure analysis. (B) Comparison of rate characteristics for polymers with and without π-bridge spacers. (C) Electrochemical performance comparison graph with previously reported zinc ion batteries. The anode material, which has an electron donor-acceptor structure with a built-in π-bridge spacer, shows better electrochemical performance and electrochromic properties than existing zinc ion batteries and electrochromic devices. >
Professor Il-Doo Kim said, “We have developed a polymer incorporated with a π-bridge spacer and successfully built a smart Zn-ion battery with excellent electrochromic efficiency and high energy storage capacity.” He added, “This technique goes beyond the existing concept of batteries that are used simply as energy storage devices, and we expect this technology to be used as a futuristic energy storage system that accelerates innovation in smart batteries and wearable technologies.”
This research, co-first authored by the alums of KAIST Departments of Material Sciences of Engineering, Professor Tae Gwang Yun of Myongji University, Dr. Jiyoung Lee, a post-doctoral associate at Northwestern University, and Professor Han Seul Kim at Chungbuk National University, was published as an inside cover article for Advanced Materials on August 3 under the title, “A π-Bridge Spacer Embedded Electron Donor-Acceptor Polymer for Flexible Electrochromic Zn-Ion Batteries”.
< Figure 4. Advanced Materials Inside Cover (August Issue) >
This research was supported by the Nanomaterial Technology Development Project under the Korean Ministry of Science and ICT, the Nano and Material Technology Development Project under the National Research Foundation of Korea, the Successive Academic Generation Development Project under the Korean Ministry of Education, and the Alchemist Project under the Korean Ministry of Trade, Industry & Energy.
2023.09.01 View 8881
A KAIST Research Team Develops a Smart Color-Changing Flexible Battery with Ultra-high Efficiency
With the rapid growth of the smart and wearable electronic devices market, smart next-generation energy storage systems that have energy storage functions as well as additional color-changing properties are receiving a great deal of attention. However, existing electrochromic devices have low electrical conductivity, leading to low efficiency in electron and ion mobility, and low storage capacities. Such batteries have therefore been limited to use in flexible and wearable devices.
On August 21, a joint research team led by Professor Il-Doo Kim from the KAIST Department of Materials Science and Engineering (DMSE) and Professor Tae Gwang Yun from the Myongji University Department of Materials Science and Engineering announced the development of a smart electrochromic Zn-ion battery that can visually represent its charging and discharging processes using an electrochromic polymer anode incorporated with a “π-bridge spacer”, which increases electron and ion mobility efficiency.
Batteries topped with electrochromic properties are groundbreaking inventions that can visually represent their charged and discharged states using colors, and can be used as display devices that cut down energy consumption for indoor cooling by controlling solar absorbance. The research team successfully built a flexible and electrochromic smart Zn-ion battery that can maintain its excellent electrochromic and electrochemical properties, even under long-term exposure to the atmosphere and mechanical deformations.
< Figure 1. Electrochromic zinc ion battery whose anode is made of a polymer that turns dark blue when charged and transparent when discharged. >
To maximize the efficiency of electron and ion mobility, the team modelled and synthesized the first π-bridge spacer-incorporated polymer anode in the world. π-bonds can improve the mobility of electrons within a structure to speed up ion movement and maximize ion adsorption efficiency, which improves its energy storage capacity.
In anode-based batteries with a π-bridge spacer, the spacer provides room for quicker ion movement. This allows fast charging, an improved zinc-ion discharging capacity of 110 mAh/g, which is 40% greater than previously reported, and a 30% increase in electrochromic function that switches from dark blue to transparent when the device is charged/discharged. In addition, should the transparent flexible battery technology be applied to smart windows, they would display darker colors during the day while they absorb solar energy, and function as a futuristic energy storage technique that can block out UV radiation and replace curtains.
< Figure 2. A schematic diagram of the structure of the electrochromic polymer with π-π spacer and the operation of a smart flexible battery using this cathode material. >
< Figure 3. (A) Density Functional Theory (DFT) theory-based atomic and electronic structure analysis. (B) Comparison of rate characteristics for polymers with and without π-bridge spacers. (C) Electrochemical performance comparison graph with previously reported zinc ion batteries. The anode material, which has an electron donor-acceptor structure with a built-in π-bridge spacer, shows better electrochemical performance and electrochromic properties than existing zinc ion batteries and electrochromic devices. >
Professor Il-Doo Kim said, “We have developed a polymer incorporated with a π-bridge spacer and successfully built a smart Zn-ion battery with excellent electrochromic efficiency and high energy storage capacity.” He added, “This technique goes beyond the existing concept of batteries that are used simply as energy storage devices, and we expect this technology to be used as a futuristic energy storage system that accelerates innovation in smart batteries and wearable technologies.”
This research, co-first authored by the alums of KAIST Departments of Material Sciences of Engineering, Professor Tae Gwang Yun of Myongji University, Dr. Jiyoung Lee, a post-doctoral associate at Northwestern University, and Professor Han Seul Kim at Chungbuk National University, was published as an inside cover article for Advanced Materials on August 3 under the title, “A π-Bridge Spacer Embedded Electron Donor-Acceptor Polymer for Flexible Electrochromic Zn-Ion Batteries”.
< Figure 4. Advanced Materials Inside Cover (August Issue) >
This research was supported by the Nanomaterial Technology Development Project under the Korean Ministry of Science and ICT, the Nano and Material Technology Development Project under the National Research Foundation of Korea, the Successive Academic Generation Development Project under the Korean Ministry of Education, and the Alchemist Project under the Korean Ministry of Trade, Industry & Energy.
2023.09.01 View 8881 -
 A KAIST Research Team Produces Eco-Friendly Nylon with Engineered Bacterium
With worsening climate change and environmental issues, in recent years, there has been increased interest in the eco-friendly production of polymers like nylon.
On August 10, Dr. Taehee Han from a KAIST research team led by Distinguished Professor Sang Yup Lee in the Department of Chemical and Biomolecular Engineering revealed the successful development of a microbial strain that produces valerolactam, a monomer of nylon-5.
Valerolactam is an important monomer that constitutes nylon-5 and nylon-6,5. Nylon is the oldest synthetic polymer, and nylon-5 is one of its derivatives composed of monomers with five carbons, while nylon-5,6 is composed of two types of monomers with either five or six carbons. They not only have excellent processability, but are also light and tough, which allows them to be applied in a wide range of industrial sectors including clothing, badminton rackets, fishing nets, tents, and gear parts. Monomers are materials that can be built into polymers, and synthetic processes are what connects them into a polymer.
The chemical production of valerolactam, however, is based on petrochemistry, where extreme reaction conditions are required and toxic waste is produced. To solve these problems, efforts are being made to develop environmentally friendly and highly efficient microbial cell factories for lactam production. Systems metabolic engineering, a key strategy for effective microbial strain development, is a research field pioneered by Professor Sang Yup Lee.
Professor Lee’s team used metabolic engineering, a technique for manipulating microbial metabolic pathways, to construct a synthetic metabolic pathway for valerolactam production in Corynebacteriam glutamicum, a bacterium commonly used for amino acid production. With this, they successfully developed a microbial strain that utilizes biomass-derived glucose as a carbon source to produce high-value valerolactam.
In 2017, the team suggested a novel method that metabolically manipulates Escherichia coli to produce valerolactam. However, there were several limitations at the time including low producibility and the generation of harmful byproducts.
< Figure 1. Schematic graphical representation of the development of microorganisms that produce valerolactam, a nylon-5 monomer >
In this research, the team improved valerolactam producibility and incorporated an additional systems metabolic strategy to the developed microbial strain while eliminating the harmful byproducts. By removing the gene involved in the production of the main byproduct and through gene screening, the team successfully converted 5-aminovaleric acid, a byproduct and a precursor, into valerolactam.
Furthermore, by employing a strategy where the 5-aminovaleric acid-converting gene is inserted multiple times into the genome, the team strengthened the metabolic flux for valerolactam production. As a result, they reached a world-record concentration of 76.1 g/L, which is 6.17 times greater than what was previously reported.
This study was published in Metabolic Engineering on July 12, under the title, “Metabolic engineering of Corynebacterium glutamicum for the high-level production of valerolactam, a nylon-5 monomer”.
Dr. Taehee Han, the first author of the paper, said, “The significance of this research lies in our development of an environmentally friendly technology that efficiently produces monomer lactam for nylon production using microorganisms.” She added, “Through this technology, we will be able to take a step forward in replacing the petrochemical industry with a microorganism-based biopolymer industry.”
This work was supported by the “Development of Next-Generation Biofinery Platform Technologies for Leading Bio-based Chemicals Industry Project” funded by the Korean Ministry of Science and ICT.
2023.08.24 View 6664
A KAIST Research Team Produces Eco-Friendly Nylon with Engineered Bacterium
With worsening climate change and environmental issues, in recent years, there has been increased interest in the eco-friendly production of polymers like nylon.
On August 10, Dr. Taehee Han from a KAIST research team led by Distinguished Professor Sang Yup Lee in the Department of Chemical and Biomolecular Engineering revealed the successful development of a microbial strain that produces valerolactam, a monomer of nylon-5.
Valerolactam is an important monomer that constitutes nylon-5 and nylon-6,5. Nylon is the oldest synthetic polymer, and nylon-5 is one of its derivatives composed of monomers with five carbons, while nylon-5,6 is composed of two types of monomers with either five or six carbons. They not only have excellent processability, but are also light and tough, which allows them to be applied in a wide range of industrial sectors including clothing, badminton rackets, fishing nets, tents, and gear parts. Monomers are materials that can be built into polymers, and synthetic processes are what connects them into a polymer.
The chemical production of valerolactam, however, is based on petrochemistry, where extreme reaction conditions are required and toxic waste is produced. To solve these problems, efforts are being made to develop environmentally friendly and highly efficient microbial cell factories for lactam production. Systems metabolic engineering, a key strategy for effective microbial strain development, is a research field pioneered by Professor Sang Yup Lee.
Professor Lee’s team used metabolic engineering, a technique for manipulating microbial metabolic pathways, to construct a synthetic metabolic pathway for valerolactam production in Corynebacteriam glutamicum, a bacterium commonly used for amino acid production. With this, they successfully developed a microbial strain that utilizes biomass-derived glucose as a carbon source to produce high-value valerolactam.
In 2017, the team suggested a novel method that metabolically manipulates Escherichia coli to produce valerolactam. However, there were several limitations at the time including low producibility and the generation of harmful byproducts.
< Figure 1. Schematic graphical representation of the development of microorganisms that produce valerolactam, a nylon-5 monomer >
In this research, the team improved valerolactam producibility and incorporated an additional systems metabolic strategy to the developed microbial strain while eliminating the harmful byproducts. By removing the gene involved in the production of the main byproduct and through gene screening, the team successfully converted 5-aminovaleric acid, a byproduct and a precursor, into valerolactam.
Furthermore, by employing a strategy where the 5-aminovaleric acid-converting gene is inserted multiple times into the genome, the team strengthened the metabolic flux for valerolactam production. As a result, they reached a world-record concentration of 76.1 g/L, which is 6.17 times greater than what was previously reported.
This study was published in Metabolic Engineering on July 12, under the title, “Metabolic engineering of Corynebacterium glutamicum for the high-level production of valerolactam, a nylon-5 monomer”.
Dr. Taehee Han, the first author of the paper, said, “The significance of this research lies in our development of an environmentally friendly technology that efficiently produces monomer lactam for nylon production using microorganisms.” She added, “Through this technology, we will be able to take a step forward in replacing the petrochemical industry with a microorganism-based biopolymer industry.”
This work was supported by the “Development of Next-Generation Biofinery Platform Technologies for Leading Bio-based Chemicals Industry Project” funded by the Korean Ministry of Science and ICT.
2023.08.24 View 6664 -
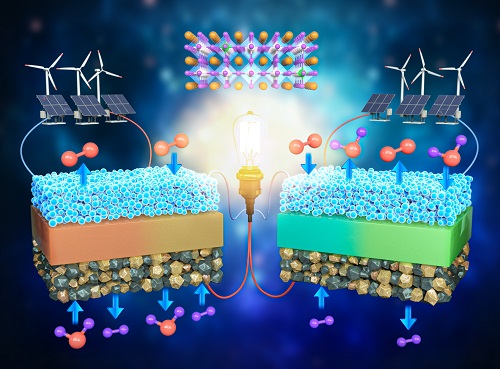 A KAIST Research Team Develops an Ultra-High Performing “Universal Electrode” for Next-Generation Fuel Cells
Fuel cells are devices that generate electricity with high efficiency using hydrogen, a clean energy source, and are expected to play an important part in the upcoming hydrogen society. The recent development of an excellent universal electrode material that is applicable to all next-generation fuel cells and can withstand 700 hours of operation has therefore garnered a great deal of attention.
On August 9, a joint research team led by Prof. WooChul Jung from the KAIST Department of Materials Science and Engineering, Prof. Kang Taek Lee from the KAIST Department of Mechanical Engineering, and Prof. Jun Hyuk Kim from the Department of Chemical Engineering at Hongik University announced the development of an electrode material that is applicable to both oxygen- and proton-conducting solid oxide cells.
Depending on the type of ion conducted by the electrolyte, ceramic fuel cells are categorized into either solid oxide fuel cells (SOFC) or protonic ceramic fuel cells (PCFC). As they can both convert between electricity and hydrogen production, fuel cells can be categorized into a total of four device types. These devices are applicable in hydrogen fuel cell vehicles, hydrogen charging stations, and power generation systems, and are henceforth emerging as core next-generation technologies for a carbon-neutral society.
However, these devices have a chronic problem where the speed of their slowest reaction would decrease with a drop of driving temperature, which greatly reduces device efficiency. Various studies have been conducted to solve this, but most reported that electrode materials have low catalytic activity and their applications are limited to specific devices, which limits them from being used as SOFCs that require reversible power conversion and hydrogen production.
< Figure 1. Schematic diagram of high-performance oxygen ion conductive solid oxide fuel cell (SOFC) and proton conductive ceramic fuel cell (PCFC) operates with the new universal electrodes >
To solve this issue, the research team doped a perovskite oxide material with Ta5+, a high valence ion that did not receive much attention in the field. Through this, the team successfully stabilized what is usually a highly unstable crystal structure, and confirmed that catalytic activity improved by 100 times.
The electrode material developed by the team was applied to all four of the mentioned device types. Furthermore, their efficiencies were greater than any of the devices reported thus far, and showed excellent performance by stably running for much longer (700 hours) compared to existing materials that deteriorated within the first 100 hours of operation.
< Figure 2. (a) Power conversion and hydrogen production performance chart for the protonic ceramic fuel cell (PCFC) with the new universal electrodes (b) and performance comparison with other reported devices >
This research, in which KAIST’s Ph.D. candidates Dongyeon Kim and Sejong Ahn, and Professor Jun Hyuk Kim from Hongik University contributed as co-first authors, was published in the internationally renowned Energy & Environmental Science under the title, "Oxygen-Electrode for Reversible Solid Oxide Electrochemical Cells at Reduced Temperatures".
Prof. WooChul Jung said, “We broke free from the idea that we must develop a completely new material to solve an existing problem, and instead suggested a way to control the crystal structure of a lesser-known material to develop a high-efficiency fuel cell, and that’s what makes these results more significant.”
Prof. Kang Taek Lee added, “Unlike previously reported materials that could only be applied to one device type at a time, our material has the flexibility of being applicable to all four. We therefore look forward to its contribution in the commercialization of eco-friendly energy technology including fuel cells and water-splitting equipment for hydrogen production.”
This research was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean Ministry of Science and ICT.
2023.08.22 View 8654
A KAIST Research Team Develops an Ultra-High Performing “Universal Electrode” for Next-Generation Fuel Cells
Fuel cells are devices that generate electricity with high efficiency using hydrogen, a clean energy source, and are expected to play an important part in the upcoming hydrogen society. The recent development of an excellent universal electrode material that is applicable to all next-generation fuel cells and can withstand 700 hours of operation has therefore garnered a great deal of attention.
On August 9, a joint research team led by Prof. WooChul Jung from the KAIST Department of Materials Science and Engineering, Prof. Kang Taek Lee from the KAIST Department of Mechanical Engineering, and Prof. Jun Hyuk Kim from the Department of Chemical Engineering at Hongik University announced the development of an electrode material that is applicable to both oxygen- and proton-conducting solid oxide cells.
Depending on the type of ion conducted by the electrolyte, ceramic fuel cells are categorized into either solid oxide fuel cells (SOFC) or protonic ceramic fuel cells (PCFC). As they can both convert between electricity and hydrogen production, fuel cells can be categorized into a total of four device types. These devices are applicable in hydrogen fuel cell vehicles, hydrogen charging stations, and power generation systems, and are henceforth emerging as core next-generation technologies for a carbon-neutral society.
However, these devices have a chronic problem where the speed of their slowest reaction would decrease with a drop of driving temperature, which greatly reduces device efficiency. Various studies have been conducted to solve this, but most reported that electrode materials have low catalytic activity and their applications are limited to specific devices, which limits them from being used as SOFCs that require reversible power conversion and hydrogen production.
< Figure 1. Schematic diagram of high-performance oxygen ion conductive solid oxide fuel cell (SOFC) and proton conductive ceramic fuel cell (PCFC) operates with the new universal electrodes >
To solve this issue, the research team doped a perovskite oxide material with Ta5+, a high valence ion that did not receive much attention in the field. Through this, the team successfully stabilized what is usually a highly unstable crystal structure, and confirmed that catalytic activity improved by 100 times.
The electrode material developed by the team was applied to all four of the mentioned device types. Furthermore, their efficiencies were greater than any of the devices reported thus far, and showed excellent performance by stably running for much longer (700 hours) compared to existing materials that deteriorated within the first 100 hours of operation.
< Figure 2. (a) Power conversion and hydrogen production performance chart for the protonic ceramic fuel cell (PCFC) with the new universal electrodes (b) and performance comparison with other reported devices >
This research, in which KAIST’s Ph.D. candidates Dongyeon Kim and Sejong Ahn, and Professor Jun Hyuk Kim from Hongik University contributed as co-first authors, was published in the internationally renowned Energy & Environmental Science under the title, "Oxygen-Electrode for Reversible Solid Oxide Electrochemical Cells at Reduced Temperatures".
Prof. WooChul Jung said, “We broke free from the idea that we must develop a completely new material to solve an existing problem, and instead suggested a way to control the crystal structure of a lesser-known material to develop a high-efficiency fuel cell, and that’s what makes these results more significant.”
Prof. Kang Taek Lee added, “Unlike previously reported materials that could only be applied to one device type at a time, our material has the flexibility of being applicable to all four. We therefore look forward to its contribution in the commercialization of eco-friendly energy technology including fuel cells and water-splitting equipment for hydrogen production.”
This research was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean Ministry of Science and ICT.
2023.08.22 View 8654 -
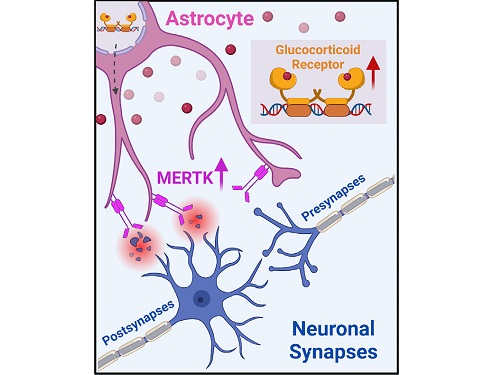 A KAIST research team identifies a cause of mental diseases induced by childhood abuse
Childhood neglect and/or abuse can induce extreme stress that significantly changes neural networks and functions during growth. This can lead to mental illnesses, including depression and schizophrenia, but the exact mechanism and means to control it were yet to be discovered.
On August 1, a KAIST research team led by Professor Won-Suk Chung from the Department of Biological Sciences announced the identification of excessive synapse removal mediated by astrocytes as the cause of mental diseases induced by childhood abuse trauma. Their research was published in Immunity, a top international journal in the field of immunology.
The research team discovered that the excessive astrocyte-mediated removal of excitatory synapses in the brain in response to stress hormones is a cause of mental diseases induced by childhood neglect and abuse. Clinical data have previously shown that high levels of stress can lead to various mental diseases, but the exact mechanism has been unknown. The results of this research therefore are expected to be widely applied to the prevention and treatment of such diseases.
The research team clinically screened an FDA-approved drug to uncover the mechanism that regulates the phagocytotic role of astrocytes, in which they capture external substances and eliminate them. As a result, the team found that synthetic glucocorticoids, namely stress hormones, enhanced astrocyte-mediated phagocytosis to an abnormal level. Glucocorticoids play essential roles in processes that maintain life, such as carbohydrate metabolism and anti-inflammation, but are also secreted in response to external stimuli such as stress, allowing the body to respond appropriately. However, excessive and long-term exposure to glucocorticoids caused by chronic stress can lead to various mental diseases including depression, cognitive disorders, and anxiety.
< Figure 1. Results of screening for compounds that increase astrocyte phagocytosis
(A) Discovered that synthetic glucocorticoid (stress hormone) increases the phagocytosis of astrocytes through screening of FDA-approved clinical compounds. (B-C) When treated with stress hormones, the phagocytosis of astrocytes is greatly increased, but this phenomenon is strongly suppressed by the GR antagonist (Mifepristone). CORT: corticosterone (stress hormone), Eplerenone: mineralocorticoid receptor (MR) antagonist, Mifepristone: glucocorticoid receptor (GR) antagonist >
To understand the changes in astrocyte functions caused by childhood stress, the research team used mice models with early social deprivation, and discovered that stress hormones bind to the glucocorticoid receptors (GRs) of astrocytes. This significantly increased the expression of Mer tyrosine kinase (MERK), which plays an essential role in astrocyte phagocytosis. Surprisingly, out of the various neurons in the cerebral cortex, astrocytes would eliminate only the excitatory synapses of specific neurons. The team found that this builds abnormal neural networks, which can lead to complex behavioral abnormalities such as social deficiencies and depression in adulthood.
The team also observed that microglia, which also play an important role in cerebral immunity, did not contribute to synapse removal in the mice models with early social deprivation. This confirms that the response to stress hormones during childhood is specifically astrocyte-mediated.
To find out whether these results are also applicable in humans, the research team used a brain organoid grown from human-induced pluripotent stem cells to observe human responses to stress hormones. The team observed that the stress hormones induced astrocyte GRs and phagocyte activation in the human brain organoid as well, and confirmed that the astrocytes subsequently eliminated excessive amounts of excitatory synapses. By showing that mice and humans both showed the same synapse control mechanism in response to stress, the team suggested that this discovery is applicable to mental disorders in humans.
< Figure 2. A schematic diagram of the study published in Immunity. Excessive stress hormone secretion in childhood increases the expression of the MERTK phagocytic receptor through the glucocorticoid receptor (GR) of astrocytes, resulting in excessive elimination of excitatory synapses. Excessive synaptic elimination by astrocytes during brain development causes permanent damage to brain circuits, resulting in abnormal neural activity in the adult brain and psychiatric behaviors such as depression and anti-social tendencies. >
Prof. Won-Suk Chung said, “Until now, we did not know the exact mechanism for how childhood stress caused brain diseases. This research was the first to show that the excessive phagocytosis of astrocytes could be an important cause of such diseases.” He added, “In the future, controlling the immune response of astrocytes will be used as a fundamental target for understanding and treating brain diseases.”
This research, written by co-first authors Youkyeong Byun (Ph.D. candidate) and Nam-Shik Kim (post-doctoral associate) from the KAIST Department of Biological Sciences, was published in the internationally renowned journal Immunity, a sister magazine of Cell and one of the best journal in the field of immunology, on July 31 under the title "Stress induces behavioral abnormalities by increasing expression of phagocytic receptor MERTK in astrocytes to promote synapse phagocytosis."
This work was supported by a National Research Foundation of Korea grant, the Korea Health Industry Development Institute (KHIDI), and the Korea Dementia Research Center (KDRC).
2023.08.04 View 8180
A KAIST research team identifies a cause of mental diseases induced by childhood abuse
Childhood neglect and/or abuse can induce extreme stress that significantly changes neural networks and functions during growth. This can lead to mental illnesses, including depression and schizophrenia, but the exact mechanism and means to control it were yet to be discovered.
On August 1, a KAIST research team led by Professor Won-Suk Chung from the Department of Biological Sciences announced the identification of excessive synapse removal mediated by astrocytes as the cause of mental diseases induced by childhood abuse trauma. Their research was published in Immunity, a top international journal in the field of immunology.
The research team discovered that the excessive astrocyte-mediated removal of excitatory synapses in the brain in response to stress hormones is a cause of mental diseases induced by childhood neglect and abuse. Clinical data have previously shown that high levels of stress can lead to various mental diseases, but the exact mechanism has been unknown. The results of this research therefore are expected to be widely applied to the prevention and treatment of such diseases.
The research team clinically screened an FDA-approved drug to uncover the mechanism that regulates the phagocytotic role of astrocytes, in which they capture external substances and eliminate them. As a result, the team found that synthetic glucocorticoids, namely stress hormones, enhanced astrocyte-mediated phagocytosis to an abnormal level. Glucocorticoids play essential roles in processes that maintain life, such as carbohydrate metabolism and anti-inflammation, but are also secreted in response to external stimuli such as stress, allowing the body to respond appropriately. However, excessive and long-term exposure to glucocorticoids caused by chronic stress can lead to various mental diseases including depression, cognitive disorders, and anxiety.
< Figure 1. Results of screening for compounds that increase astrocyte phagocytosis
(A) Discovered that synthetic glucocorticoid (stress hormone) increases the phagocytosis of astrocytes through screening of FDA-approved clinical compounds. (B-C) When treated with stress hormones, the phagocytosis of astrocytes is greatly increased, but this phenomenon is strongly suppressed by the GR antagonist (Mifepristone). CORT: corticosterone (stress hormone), Eplerenone: mineralocorticoid receptor (MR) antagonist, Mifepristone: glucocorticoid receptor (GR) antagonist >
To understand the changes in astrocyte functions caused by childhood stress, the research team used mice models with early social deprivation, and discovered that stress hormones bind to the glucocorticoid receptors (GRs) of astrocytes. This significantly increased the expression of Mer tyrosine kinase (MERK), which plays an essential role in astrocyte phagocytosis. Surprisingly, out of the various neurons in the cerebral cortex, astrocytes would eliminate only the excitatory synapses of specific neurons. The team found that this builds abnormal neural networks, which can lead to complex behavioral abnormalities such as social deficiencies and depression in adulthood.
The team also observed that microglia, which also play an important role in cerebral immunity, did not contribute to synapse removal in the mice models with early social deprivation. This confirms that the response to stress hormones during childhood is specifically astrocyte-mediated.
To find out whether these results are also applicable in humans, the research team used a brain organoid grown from human-induced pluripotent stem cells to observe human responses to stress hormones. The team observed that the stress hormones induced astrocyte GRs and phagocyte activation in the human brain organoid as well, and confirmed that the astrocytes subsequently eliminated excessive amounts of excitatory synapses. By showing that mice and humans both showed the same synapse control mechanism in response to stress, the team suggested that this discovery is applicable to mental disorders in humans.
< Figure 2. A schematic diagram of the study published in Immunity. Excessive stress hormone secretion in childhood increases the expression of the MERTK phagocytic receptor through the glucocorticoid receptor (GR) of astrocytes, resulting in excessive elimination of excitatory synapses. Excessive synaptic elimination by astrocytes during brain development causes permanent damage to brain circuits, resulting in abnormal neural activity in the adult brain and psychiatric behaviors such as depression and anti-social tendencies. >
Prof. Won-Suk Chung said, “Until now, we did not know the exact mechanism for how childhood stress caused brain diseases. This research was the first to show that the excessive phagocytosis of astrocytes could be an important cause of such diseases.” He added, “In the future, controlling the immune response of astrocytes will be used as a fundamental target for understanding and treating brain diseases.”
This research, written by co-first authors Youkyeong Byun (Ph.D. candidate) and Nam-Shik Kim (post-doctoral associate) from the KAIST Department of Biological Sciences, was published in the internationally renowned journal Immunity, a sister magazine of Cell and one of the best journal in the field of immunology, on July 31 under the title "Stress induces behavioral abnormalities by increasing expression of phagocytic receptor MERTK in astrocytes to promote synapse phagocytosis."
This work was supported by a National Research Foundation of Korea grant, the Korea Health Industry Development Institute (KHIDI), and the Korea Dementia Research Center (KDRC).
2023.08.04 View 8180 -
 KAIST Research Team Develops World’s First Humanoid Pilot, PIBOT
In the Spring of last year, the legendary, fictional pilot “Maverick” flew his plane in the film “Top Gun: Maverick” that drew crowds to theatres around the world. This year, the appearance of a humanoid pilot, PIBOT, has stolen the spotlight at KAIST.
< Photo 1. Humanoid pilot robot, PIBOT >
A KAIST research team has developed a humanoid robot that can understand manuals written in natural language and fly a plane on its own. The team also announced their plans to commercialize the humanoid pilot.
< Photo 2. PIBOT on flight simulator (view from above) >
The project was led by KAIST Professor David Hyunchul Shim, and was conducted as a joint research project with Professors Jaegul Choo, Kuk-Jin Yoon, and Min Jun Kim. The study was supported by Future Challenge Funding under the project title, “Development of Human-like Pilot Robot based on Natural Language Processing”. The team utilized AI and robotics technologies, and demonstrated that the humanoid could sit itself in a real cockpit and operate the various pieces of equipment without modifying any part of the aircraft. This is a fundamental difference that distinguishes this technology from existing autopilot functions or unmanned aircrafts.
< Photo 3. PIBOT operating a flight simulator (side) >
The KAIST team’s humanoid pilot is still under development but it can already remember Jeppeson charts from all around the world, which is impossible for human pilots to do, and fly without error. In particular, it can make use of recent ChatGPT technology to remember the full Quick Reference Handbook (QRF) and respond immediately to various situations, as well as calculate safe routes in real time based on the flight status of the aircraft, with emergency response times quicker than human pilots.
Furthermore, while existing robots usually carry out repeated motions in a fixed position, PIBOT can analyze the state of the cockpit as well as the situation outside the aircraft using an embedded camera. PIBOT can accurately control the various switches in the cockpit and, using high-precision control technology, it can accurately control its robotic arms and hands even during harsh turbulence.
< Photo 4. PIBOT on-board KLA-100, Korea’s first light aircraft >
The humanoid pilot is currently capable of carrying out all operations from starting the aircraft to taxiing, takeoff and landing, cruising, and cycling using a flight control simulator. The research team plans to use the humanoid pilot to fly a real-life light aircraft to verify its abilities. Prof. Shim explained, “Humanoid pilot robots do not require the modification of existing aircrafts and can be applied immediately to automated flights. They are therefore highly applicable and practical. We expect them to be applied into various other vehicles like cars and military trucks since they can control a wide range of equipment. They will particularly be particularly helpful in situations where military resources are severely depleted.”
This research was supported by Future Challenge Funding (total: 5.7 bn KRW) from the Agency for Defense Development. The project started in 2022 as a joint research project by Prof. David Hyunchul Shim (chief of research) from the KAIST School of Electrical Engineering (EE), Prof. Jaegul Choo from the Kim Jaechul Graduate School of AI at KAIST, Prof. Kuk-Jin Yoon from the KAIST Department of Mechanical Engineering, and Prof. Min Jun Kim from the KAIST School of EE. The project is to be completed by 2026 and the involved researchers are also considering commercialization strategies for both military and civil use.
2023.08.03 View 15565
KAIST Research Team Develops World’s First Humanoid Pilot, PIBOT
In the Spring of last year, the legendary, fictional pilot “Maverick” flew his plane in the film “Top Gun: Maverick” that drew crowds to theatres around the world. This year, the appearance of a humanoid pilot, PIBOT, has stolen the spotlight at KAIST.
< Photo 1. Humanoid pilot robot, PIBOT >
A KAIST research team has developed a humanoid robot that can understand manuals written in natural language and fly a plane on its own. The team also announced their plans to commercialize the humanoid pilot.
< Photo 2. PIBOT on flight simulator (view from above) >
The project was led by KAIST Professor David Hyunchul Shim, and was conducted as a joint research project with Professors Jaegul Choo, Kuk-Jin Yoon, and Min Jun Kim. The study was supported by Future Challenge Funding under the project title, “Development of Human-like Pilot Robot based on Natural Language Processing”. The team utilized AI and robotics technologies, and demonstrated that the humanoid could sit itself in a real cockpit and operate the various pieces of equipment without modifying any part of the aircraft. This is a fundamental difference that distinguishes this technology from existing autopilot functions or unmanned aircrafts.
< Photo 3. PIBOT operating a flight simulator (side) >
The KAIST team’s humanoid pilot is still under development but it can already remember Jeppeson charts from all around the world, which is impossible for human pilots to do, and fly without error. In particular, it can make use of recent ChatGPT technology to remember the full Quick Reference Handbook (QRF) and respond immediately to various situations, as well as calculate safe routes in real time based on the flight status of the aircraft, with emergency response times quicker than human pilots.
Furthermore, while existing robots usually carry out repeated motions in a fixed position, PIBOT can analyze the state of the cockpit as well as the situation outside the aircraft using an embedded camera. PIBOT can accurately control the various switches in the cockpit and, using high-precision control technology, it can accurately control its robotic arms and hands even during harsh turbulence.
< Photo 4. PIBOT on-board KLA-100, Korea’s first light aircraft >
The humanoid pilot is currently capable of carrying out all operations from starting the aircraft to taxiing, takeoff and landing, cruising, and cycling using a flight control simulator. The research team plans to use the humanoid pilot to fly a real-life light aircraft to verify its abilities. Prof. Shim explained, “Humanoid pilot robots do not require the modification of existing aircrafts and can be applied immediately to automated flights. They are therefore highly applicable and practical. We expect them to be applied into various other vehicles like cars and military trucks since they can control a wide range of equipment. They will particularly be particularly helpful in situations where military resources are severely depleted.”
This research was supported by Future Challenge Funding (total: 5.7 bn KRW) from the Agency for Defense Development. The project started in 2022 as a joint research project by Prof. David Hyunchul Shim (chief of research) from the KAIST School of Electrical Engineering (EE), Prof. Jaegul Choo from the Kim Jaechul Graduate School of AI at KAIST, Prof. Kuk-Jin Yoon from the KAIST Department of Mechanical Engineering, and Prof. Min Jun Kim from the KAIST School of EE. The project is to be completed by 2026 and the involved researchers are also considering commercialization strategies for both military and civil use.
2023.08.03 View 15565 -
 KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 9524
KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 9524 -
 A KAIST research team develops a washable, transparent, and flexible OLED with MXene nanotechnology
Transparent and flexible displays, which have received a lot of attention in various fields including automobile displays, bio-healthcare, military, and fashion, are in fact known to break easily when experiencing small deformations. To solve this problem, active research is being conducted on many transparent and flexible conductive materials such as carbon nanotubes, graphene, silver nanowires, and conductive polymers.
On June 13, a joint research team led by Professor Kyung Cheol Choi from the KAIST School of Electrical Engineering and Dr. Yonghee Lee from the National Nano Fab Center (NNFC) announced the successful development of a water-resistant, transparent, and flexible OLED using MXene nanotechnology. The material can emit and transmit light even when exposed to water.
MXene is a 2D material with high electrical conductivity and optical transmittance, and it can be produced on a large scale through solution processes. However, despite these attractive properties, MXene’s applications were limited as a long-term electrical device due to its electrical properties being degraded easily by atmospheric moisture and water. The material was therefore unable to be systemized into the form of a matrix that can display information.
Professor Choi’s research team used an encapsulation tactic that can protect materials from oxidation caused by moisture and oxygen to develop a MXene-based OLED with a long lifespan and high stability against external environmental factors. The research team first focused on analyzing the degradation mechanism of MXene’s electrical conductivity, and then concentrated on designing an encapsulation membrane. The team blocked moisture and provided flexibility through residual stress offset, ultimately producing a double-layered encapsulation membrane. In addition, a thin plastic film with a thickness of a few micrometers was attached to the top layer to allow washing in water without degradation.
< Figure 1. (a) Transparent passive-matrix display made of MXene-based OLED, (b) Cross-sectional image of MXene-based OLED observed by transmission electron microscope (TEM), (c) Electro-optical characteristic graph of red, green, and blue MXene-based OLED >
Through this study, the research team developed a MXene-based red(R)/green(G)/blue(B) OLED that emits a brightness of over 1,000 cd/m2 that is detectable by the naked eye even under sunlight, thereby meeting the conditions for outdoor displays. As for the red MXene-based OLED, the researchers confirmed a standby storage life of 2,000 hours (under 70% luminescence), a standby operation life of 1,500 hours (under 60% luminescence), and a flexibility withstanding 1,000 cycles under a low curvature of under 1.5mm. In addition, they showed that its performance was maintained even after six hours of immersion under water (under 80% luminescence). Furthermore, a patterning technique was used to produce the MXene-based OLED in the form of a passive matrix, and the team demonstrated its use as a transparent display by displaying letters and shapes.
Ph.D. candidate So Yeong Jeong, who led this study, said, “To improve the reliability of MXene OLED, we focused on producing an adequate encapsulation structure and a suitable process design.” She added, “By producing a matrix-type MXene OLED and displaying simple letters and shapes, we have laid the foundations for MXene’s application in the field of transparent displays.”
< Image 1. Cover of ACS Nano Front Cover (Conceptual diagram of MXene-based OLED display) >
Professor Choi said, “This research will become the guideline for applying MXene in electrical devices, but we expect for it to also be applied in other fields that require flexible and transparent displays like automobiles, fashion, and functional clothing. And to widen the gap with China’s OLED technology, these new OLED convergence technologies must continue to be developed.”
This research was supported by the National Research Foundation of Korea and funded by the Ministry of Science and ICT, Korea. It was published as a front cover story of ACS Nano under the title, “Highly Air-Stable, Flexible, and Water-Resistive 2D Titanium Carbide MXene-Based RGB Organic Light-Emitting Diode Displays for Transparent Free-Form Electronics” on June 13.
2023.07.10 View 7691
A KAIST research team develops a washable, transparent, and flexible OLED with MXene nanotechnology
Transparent and flexible displays, which have received a lot of attention in various fields including automobile displays, bio-healthcare, military, and fashion, are in fact known to break easily when experiencing small deformations. To solve this problem, active research is being conducted on many transparent and flexible conductive materials such as carbon nanotubes, graphene, silver nanowires, and conductive polymers.
On June 13, a joint research team led by Professor Kyung Cheol Choi from the KAIST School of Electrical Engineering and Dr. Yonghee Lee from the National Nano Fab Center (NNFC) announced the successful development of a water-resistant, transparent, and flexible OLED using MXene nanotechnology. The material can emit and transmit light even when exposed to water.
MXene is a 2D material with high electrical conductivity and optical transmittance, and it can be produced on a large scale through solution processes. However, despite these attractive properties, MXene’s applications were limited as a long-term electrical device due to its electrical properties being degraded easily by atmospheric moisture and water. The material was therefore unable to be systemized into the form of a matrix that can display information.
Professor Choi’s research team used an encapsulation tactic that can protect materials from oxidation caused by moisture and oxygen to develop a MXene-based OLED with a long lifespan and high stability against external environmental factors. The research team first focused on analyzing the degradation mechanism of MXene’s electrical conductivity, and then concentrated on designing an encapsulation membrane. The team blocked moisture and provided flexibility through residual stress offset, ultimately producing a double-layered encapsulation membrane. In addition, a thin plastic film with a thickness of a few micrometers was attached to the top layer to allow washing in water without degradation.
< Figure 1. (a) Transparent passive-matrix display made of MXene-based OLED, (b) Cross-sectional image of MXene-based OLED observed by transmission electron microscope (TEM), (c) Electro-optical characteristic graph of red, green, and blue MXene-based OLED >
Through this study, the research team developed a MXene-based red(R)/green(G)/blue(B) OLED that emits a brightness of over 1,000 cd/m2 that is detectable by the naked eye even under sunlight, thereby meeting the conditions for outdoor displays. As for the red MXene-based OLED, the researchers confirmed a standby storage life of 2,000 hours (under 70% luminescence), a standby operation life of 1,500 hours (under 60% luminescence), and a flexibility withstanding 1,000 cycles under a low curvature of under 1.5mm. In addition, they showed that its performance was maintained even after six hours of immersion under water (under 80% luminescence). Furthermore, a patterning technique was used to produce the MXene-based OLED in the form of a passive matrix, and the team demonstrated its use as a transparent display by displaying letters and shapes.
Ph.D. candidate So Yeong Jeong, who led this study, said, “To improve the reliability of MXene OLED, we focused on producing an adequate encapsulation structure and a suitable process design.” She added, “By producing a matrix-type MXene OLED and displaying simple letters and shapes, we have laid the foundations for MXene’s application in the field of transparent displays.”
< Image 1. Cover of ACS Nano Front Cover (Conceptual diagram of MXene-based OLED display) >
Professor Choi said, “This research will become the guideline for applying MXene in electrical devices, but we expect for it to also be applied in other fields that require flexible and transparent displays like automobiles, fashion, and functional clothing. And to widen the gap with China’s OLED technology, these new OLED convergence technologies must continue to be developed.”
This research was supported by the National Research Foundation of Korea and funded by the Ministry of Science and ICT, Korea. It was published as a front cover story of ACS Nano under the title, “Highly Air-Stable, Flexible, and Water-Resistive 2D Titanium Carbide MXene-Based RGB Organic Light-Emitting Diode Displays for Transparent Free-Form Electronics” on June 13.
2023.07.10 View 7691 -
 KAIST researchers find sleep delays more prevalent in countries of particular culture than others
Sleep has a huge impact on health, well-being and productivity, but how long and how well people sleep these days has not been accurately reported. Previous research on how much and how well we sleep has mostly relied on self-reports or was confined within the data from the unnatural environments of the sleep laboratories.
So, the questions remained: Is the amount and quality of sleep purely a personal choice? Could they be independent from social factors such as culture and geography?
< From left to right, Sungkyu Park of Kangwon National University, South Korea; Assem Zhunis of KAIST and IBS, South Korea; Marios Constantinides of Nokia Bell Labs, UK; Luca Maria Aiello of the IT University of Copenhagen, Denmark; Daniele Quercia of Nokia Bell Labs and King's College London, UK; and Meeyoung Cha of IBS and KAIST, South Korea >
A new study led by researchers at Korea Advanced Institute of Science and Technology (KAIST) and Nokia Bell Labs in the United Kingdom investigated the cultural and individual factors that influence sleep. In contrast to previous studies that relied on surveys or controlled experiments at labs, the team used commercially available smartwatches for extensive data collection, analyzing 52 million logs collected over a four-year period from 30,082 individuals in 11 countries. These people wore Nokia smartwatches, which allowed the team to investigate country-specific sleep patterns based on the digital logs from the devices.
< Figure comparing survey and smartwatch logs on average sleep-time, wake-time, and sleep durations. Digital logs consistently recorded delayed hours of wake- and sleep-time, resulting in shorter sleep durations. >
Digital logs collected from the smartwatches revealed discrepancies in wake-up times and sleep-times, sometimes by tens of minutes to an hour, from the data previously collected from self-report assessments. The average sleep-time overall was calculated to be around midnight, and the average wake-up time was 7:42 AM. The team discovered, however, that individuals' sleep is heavily linked to their geographical location and cultural factors. While wake-up times were similar, sleep-time varied by country. Individuals in higher GDP countries had more records of delayed bedtime. Those in collectivist culture, compared to individualist culture, also showed more records of delayed bedtime. Among the studied countries, Japan had the shortest total sleep duration, averaging a duration of under 7 hours, while Finland had the longest, averaging 8 hours.
Researchers calculated essential sleep metrics used in clinical studies, such as sleep efficiency, sleep duration, and overslept hours on weekends, to analyze the extensive sleep patterns. Using Principal Component Analysis (PCA), they further condensed these metrics into two major sleep dimensions representing sleep quality and quantity. A cross-country comparison revealed that societal factors account for 55% of the variation in sleep quality and 63% of the variation in sleep quantity.
Countries with a higher individualism index (IDV), which placed greater emphasis on individual achievements and relationships, had significantly longer sleep durations, which could be attributed to such societies having a norm of going to bed early. Spain and Japan, on the other hand, had the bedtime scheduled at the latest hours despite having the highest collectivism scores (low IDV). The study also discovered a moderate relationship between a higher uncertainty avoidance index (UAI), which measures implementation of general laws and regulation in daily lives of regular citizens, and better sleep quality.
Researchers also investigated how physical activity can affect sleep quantity and quality to see if individuals can counterbalance cultural influences through personal interventions. They discovered that increasing daily activity can improve sleep quality in terms of shortened time needed in falling asleep and waking up. Individuals who exercise more, however, did not sleep longer. The effect of exercise differed by country, with more pronounced effects observed in some countries, such as the United States and Finland. Interestingly, in Japan, no obvious effect of exercise could be observed. These findings suggest that the relationship between daily activity and sleep may differ by country and that different exercise regimens may be more effective in different cultures.
This research published on the Scientific Reports by the international journal, Nature, sheds light on the influence of social factors on sleep. (Paper Title "Social dimensions impact individual sleep quantity and quality" Article number: 9681)
One of the co-authors, Daniele Quercia, commented: “Excessive work schedules, long working hours, and late bedtime in high-income countries and social engagement due to high collectivism may cause bedtimes to be delayed.”
Commenting on the research, the first author Shaun Sungkyu Park said, "While it is intriguing to see that a society can play a role in determining the quantity and quality of an individual's sleep with large-scale data, the significance of this study is that it quantitatively shows that even within the same culture (country), individual efforts such as daily exercise can have a positive impact on sleep quantity and quality."
"Sleep not only has a great impact on one’s well-being but it is also known to be associated with health issues such as obesity and dementia," said the lead author, Meeyoung Cha. "In order to ensure adequate sleep and improve sleep quality in an aging society, not only individual efforts but also a social support must be provided to work together," she said. The research team will contribute to the development of the high-tech sleep industry by making a code that easily calculates the sleep indicators developed in this study available free of charge, as well as providing the benchmark data for various types of sleep research to follow.
2023.07.07 View 8621
KAIST researchers find sleep delays more prevalent in countries of particular culture than others
Sleep has a huge impact on health, well-being and productivity, but how long and how well people sleep these days has not been accurately reported. Previous research on how much and how well we sleep has mostly relied on self-reports or was confined within the data from the unnatural environments of the sleep laboratories.
So, the questions remained: Is the amount and quality of sleep purely a personal choice? Could they be independent from social factors such as culture and geography?
< From left to right, Sungkyu Park of Kangwon National University, South Korea; Assem Zhunis of KAIST and IBS, South Korea; Marios Constantinides of Nokia Bell Labs, UK; Luca Maria Aiello of the IT University of Copenhagen, Denmark; Daniele Quercia of Nokia Bell Labs and King's College London, UK; and Meeyoung Cha of IBS and KAIST, South Korea >
A new study led by researchers at Korea Advanced Institute of Science and Technology (KAIST) and Nokia Bell Labs in the United Kingdom investigated the cultural and individual factors that influence sleep. In contrast to previous studies that relied on surveys or controlled experiments at labs, the team used commercially available smartwatches for extensive data collection, analyzing 52 million logs collected over a four-year period from 30,082 individuals in 11 countries. These people wore Nokia smartwatches, which allowed the team to investigate country-specific sleep patterns based on the digital logs from the devices.
< Figure comparing survey and smartwatch logs on average sleep-time, wake-time, and sleep durations. Digital logs consistently recorded delayed hours of wake- and sleep-time, resulting in shorter sleep durations. >
Digital logs collected from the smartwatches revealed discrepancies in wake-up times and sleep-times, sometimes by tens of minutes to an hour, from the data previously collected from self-report assessments. The average sleep-time overall was calculated to be around midnight, and the average wake-up time was 7:42 AM. The team discovered, however, that individuals' sleep is heavily linked to their geographical location and cultural factors. While wake-up times were similar, sleep-time varied by country. Individuals in higher GDP countries had more records of delayed bedtime. Those in collectivist culture, compared to individualist culture, also showed more records of delayed bedtime. Among the studied countries, Japan had the shortest total sleep duration, averaging a duration of under 7 hours, while Finland had the longest, averaging 8 hours.
Researchers calculated essential sleep metrics used in clinical studies, such as sleep efficiency, sleep duration, and overslept hours on weekends, to analyze the extensive sleep patterns. Using Principal Component Analysis (PCA), they further condensed these metrics into two major sleep dimensions representing sleep quality and quantity. A cross-country comparison revealed that societal factors account for 55% of the variation in sleep quality and 63% of the variation in sleep quantity.
Countries with a higher individualism index (IDV), which placed greater emphasis on individual achievements and relationships, had significantly longer sleep durations, which could be attributed to such societies having a norm of going to bed early. Spain and Japan, on the other hand, had the bedtime scheduled at the latest hours despite having the highest collectivism scores (low IDV). The study also discovered a moderate relationship between a higher uncertainty avoidance index (UAI), which measures implementation of general laws and regulation in daily lives of regular citizens, and better sleep quality.
Researchers also investigated how physical activity can affect sleep quantity and quality to see if individuals can counterbalance cultural influences through personal interventions. They discovered that increasing daily activity can improve sleep quality in terms of shortened time needed in falling asleep and waking up. Individuals who exercise more, however, did not sleep longer. The effect of exercise differed by country, with more pronounced effects observed in some countries, such as the United States and Finland. Interestingly, in Japan, no obvious effect of exercise could be observed. These findings suggest that the relationship between daily activity and sleep may differ by country and that different exercise regimens may be more effective in different cultures.
This research published on the Scientific Reports by the international journal, Nature, sheds light on the influence of social factors on sleep. (Paper Title "Social dimensions impact individual sleep quantity and quality" Article number: 9681)
One of the co-authors, Daniele Quercia, commented: “Excessive work schedules, long working hours, and late bedtime in high-income countries and social engagement due to high collectivism may cause bedtimes to be delayed.”
Commenting on the research, the first author Shaun Sungkyu Park said, "While it is intriguing to see that a society can play a role in determining the quantity and quality of an individual's sleep with large-scale data, the significance of this study is that it quantitatively shows that even within the same culture (country), individual efforts such as daily exercise can have a positive impact on sleep quantity and quality."
"Sleep not only has a great impact on one’s well-being but it is also known to be associated with health issues such as obesity and dementia," said the lead author, Meeyoung Cha. "In order to ensure adequate sleep and improve sleep quality in an aging society, not only individual efforts but also a social support must be provided to work together," she said. The research team will contribute to the development of the high-tech sleep industry by making a code that easily calculates the sleep indicators developed in this study available free of charge, as well as providing the benchmark data for various types of sleep research to follow.
2023.07.07 View 8621