deep+learning
-
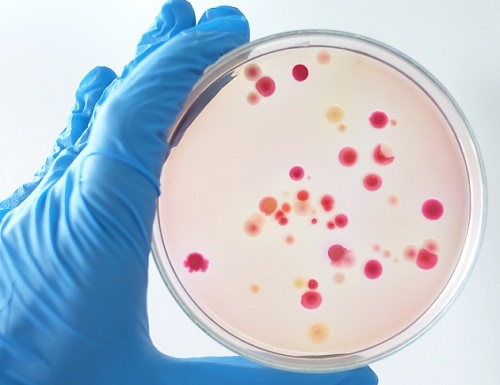 'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media
Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
Their results were made available online on Jan. 18 in Biosensors and Bioelectronics, ahead of publication in the journal’s April issue.
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. “By using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model, we demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures,” said Professor Sungho Jo from the School of Computing.
Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample — the spectral fingerprint — allowing researchers to identify its molecules. The surface-enhanced version places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. “Moreover, strong signals of surrounding media are also enhanced to overwhelm target signals, requiring time-consuming and tedious bacterial separation steps,” said Professor Yeon Sik Jung from the Department of Materials Science and Engineering.
To parse through the noisy signals, the researchers implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such an ability is critical for analyzing one-dimensional spectral data, according to Professor Jo.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that were almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” Professor Jo said. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
The National R&D Program, through a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, supported this research.
-PublicationEojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deepneural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
-ProfileProfessor Yeon Sik JungDepartment of Materials Science and EngineeringKAIST
Professor Sungho JoSchool of ComputingKAIST
2022.03.04 View 21835
'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media
Bacterial identification can take hours and often longer, precious time when diagnosing infections and selecting appropriate treatments. There may be a quicker, more accurate process according to researchers at KAIST. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
Their results were made available online on Jan. 18 in Biosensors and Bioelectronics, ahead of publication in the journal’s April issue.
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. “By using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model, we demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures,” said Professor Sungho Jo from the School of Computing.
Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample — the spectral fingerprint — allowing researchers to identify its molecules. The surface-enhanced version places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. “Moreover, strong signals of surrounding media are also enhanced to overwhelm target signals, requiring time-consuming and tedious bacterial separation steps,” said Professor Yeon Sik Jung from the Department of Materials Science and Engineering.
To parse through the noisy signals, the researchers implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such an ability is critical for analyzing one-dimensional spectral data, according to Professor Jo.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that were almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” Professor Jo said. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
The National R&D Program, through a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, supported this research.
-PublicationEojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deepneural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
-ProfileProfessor Yeon Sik JungDepartment of Materials Science and EngineeringKAIST
Professor Sungho JoSchool of ComputingKAIST
2022.03.04 View 21835 -
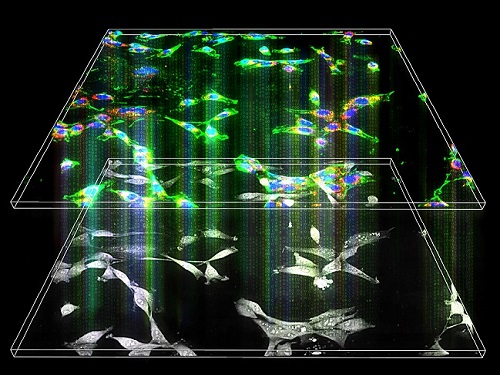 Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 10894
Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 10894 -
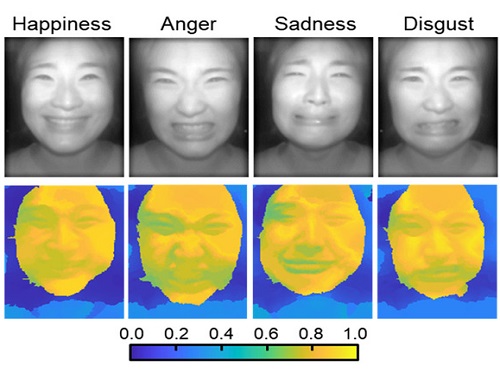 AI Light-Field Camera Reads 3D Facial Expressions
Machine-learned, light-field camera reads facial expressions from high-contrast illumination invariant 3D facial images
A joint research team led by Professors Ki-Hun Jeong and Doheon Lee from the KAIST Department of Bio and Brain Engineering reported the development of a technique for facial expression detection by merging near-infrared light-field camera techniques with artificial intelligence (AI) technology.
Unlike a conventional camera, the light-field camera contains micro-lens arrays in front of the image sensor, which makes the camera small enough to fit into a smart phone, while allowing it to acquire the spatial and directional information of the light with a single shot. The technique has received attention as it can reconstruct images in a variety of ways including multi-views, refocusing, and 3D image acquisition, giving rise to many potential applications.
However, the optical crosstalk between shadows caused by external light sources in the environment and the micro-lens has limited existing light-field cameras from being able to provide accurate image contrast and 3D reconstruction.
The joint research team applied a vertical-cavity surface-emitting laser (VCSEL) in the near-IR range to stabilize the accuracy of 3D image reconstruction that previously depended on environmental light. When an external light source is shone on a face at 0-, 30-, and 60-degree angles, the light field camera reduces 54% of image reconstruction errors. Additionally, by inserting a light-absorbing layer for visible and near-IR wavelengths between the micro-lens arrays, the team could minimize optical crosstalk while increasing the image contrast by 2.1 times.
Through this technique, the team could overcome the limitations of existing light-field cameras and was able to develop their NIR-based light-field camera (NIR-LFC), optimized for the 3D image reconstruction of facial expressions. Using the NIR-LFC, the team acquired high-quality 3D reconstruction images of facial expressions expressing various emotions regardless of the lighting conditions of the surrounding environment.
The facial expressions in the acquired 3D images were distinguished through machine learning with an average of 85% accuracy – a statistically significant figure compared to when 2D images were used. Furthermore, by calculating the interdependency of distance information that varies with facial expression in 3D images, the team could identify the information a light-field camera utilizes to distinguish human expressions.
Professor Ki-Hun Jeong said, “The sub-miniature light-field camera developed by the research team has the potential to become the new platform to quantitatively analyze the facial expressions and emotions of humans.” To highlight the significance of this research, he added, “It could be applied in various fields including mobile healthcare, field diagnosis, social cognition, and human-machine interactions.”
This research was published in Advanced Intelligent Systems online on December 16, under the title, “Machine-Learned Light-field Camera that Reads Facial Expression from High-Contrast and Illumination Invariant 3D Facial Images.” This research was funded by the Ministry of Science and ICT and the Ministry of Trade, Industry and Energy.
-Publication“Machine-learned light-field camera that reads fascial expression from high-contrast and illumination invariant 3D facial images,” Sang-In Bae, Sangyeon Lee, Jae-Myeong Kwon, Hyun-Kyung Kim. Kyung-Won Jang, Doheon Lee, Ki-Hun Jeong, Advanced Intelligent Systems, December 16, 2021 (doi.org/10.1002/aisy.202100182)
ProfileProfessor Ki-Hun JeongBiophotonic LaboratoryDepartment of Bio and Brain EngineeringKAIST
Professor Doheon LeeDepartment of Bio and Brain EngineeringKAIST
2022.01.21 View 12475
AI Light-Field Camera Reads 3D Facial Expressions
Machine-learned, light-field camera reads facial expressions from high-contrast illumination invariant 3D facial images
A joint research team led by Professors Ki-Hun Jeong and Doheon Lee from the KAIST Department of Bio and Brain Engineering reported the development of a technique for facial expression detection by merging near-infrared light-field camera techniques with artificial intelligence (AI) technology.
Unlike a conventional camera, the light-field camera contains micro-lens arrays in front of the image sensor, which makes the camera small enough to fit into a smart phone, while allowing it to acquire the spatial and directional information of the light with a single shot. The technique has received attention as it can reconstruct images in a variety of ways including multi-views, refocusing, and 3D image acquisition, giving rise to many potential applications.
However, the optical crosstalk between shadows caused by external light sources in the environment and the micro-lens has limited existing light-field cameras from being able to provide accurate image contrast and 3D reconstruction.
The joint research team applied a vertical-cavity surface-emitting laser (VCSEL) in the near-IR range to stabilize the accuracy of 3D image reconstruction that previously depended on environmental light. When an external light source is shone on a face at 0-, 30-, and 60-degree angles, the light field camera reduces 54% of image reconstruction errors. Additionally, by inserting a light-absorbing layer for visible and near-IR wavelengths between the micro-lens arrays, the team could minimize optical crosstalk while increasing the image contrast by 2.1 times.
Through this technique, the team could overcome the limitations of existing light-field cameras and was able to develop their NIR-based light-field camera (NIR-LFC), optimized for the 3D image reconstruction of facial expressions. Using the NIR-LFC, the team acquired high-quality 3D reconstruction images of facial expressions expressing various emotions regardless of the lighting conditions of the surrounding environment.
The facial expressions in the acquired 3D images were distinguished through machine learning with an average of 85% accuracy – a statistically significant figure compared to when 2D images were used. Furthermore, by calculating the interdependency of distance information that varies with facial expression in 3D images, the team could identify the information a light-field camera utilizes to distinguish human expressions.
Professor Ki-Hun Jeong said, “The sub-miniature light-field camera developed by the research team has the potential to become the new platform to quantitatively analyze the facial expressions and emotions of humans.” To highlight the significance of this research, he added, “It could be applied in various fields including mobile healthcare, field diagnosis, social cognition, and human-machine interactions.”
This research was published in Advanced Intelligent Systems online on December 16, under the title, “Machine-Learned Light-field Camera that Reads Facial Expression from High-Contrast and Illumination Invariant 3D Facial Images.” This research was funded by the Ministry of Science and ICT and the Ministry of Trade, Industry and Energy.
-Publication“Machine-learned light-field camera that reads fascial expression from high-contrast and illumination invariant 3D facial images,” Sang-In Bae, Sangyeon Lee, Jae-Myeong Kwon, Hyun-Kyung Kim. Kyung-Won Jang, Doheon Lee, Ki-Hun Jeong, Advanced Intelligent Systems, December 16, 2021 (doi.org/10.1002/aisy.202100182)
ProfileProfessor Ki-Hun JeongBiophotonic LaboratoryDepartment of Bio and Brain EngineeringKAIST
Professor Doheon LeeDepartment of Bio and Brain EngineeringKAIST
2022.01.21 View 12475 -
 Deep Learning Framework to Enable Material Design in Unseen Domain
Researchers propose a deep neural network-based forward design space exploration using active transfer learning and data augmentation
A new study proposed a deep neural network-based forward design approach that enables an efficient search for superior materials far beyond the domain of the initial training set. This approach compensates for the weak predictive power of neural networks on an unseen domain through gradual updates of the neural network with active transfer learning and data augmentation methods.
Professor Seungwha Ryu believes that this study will help address a variety of optimization problems that have an astronomical number of possible design configurations. For the grid composite optimization problem, the proposed framework was able to provide excellent designs close to the global optima, even with the addition of a very small dataset corresponding to less than 0.5% of the initial training data-set size. This study was reported in npj Computational Materials last month.
“We wanted to mitigate the limitation of the neural network, weak predictive power beyond the training set domain for the material or structure design,” said Professor Ryu from the Department of Mechanical Engineering.
Neural network-based generative models have been actively investigated as an inverse design method for finding novel materials in a vast design space. However, the applicability of conventional generative models is limited because they cannot access data outside the range of training sets. Advanced generative models that were devised to overcome this limitation also suffer from weak predictive power for the unseen domain.
Professor Ryu’s team, in collaboration with researchers from Professor Grace Gu’s group at UC Berkeley, devised a design method that simultaneously expands the domain using the strong predictive power of a deep neural network and searches for the optimal design by repetitively performing three key steps.
First, it searches for few candidates with improved properties located close to the training set via genetic algorithms, by mixing superior designs within the training set. Then, it checks to see if the candidates really have improved properties, and expands the training set by duplicating the validated designs via a data augmentation method. Finally, they can expand the reliable prediction domain by updating the neural network with the new superior designs via transfer learning. Because the expansion proceeds along relatively narrow but correct routes toward the optimal design (depicted in the schematic of Fig. 1), the framework enables an efficient search.
As a data-hungry method, a deep neural network model tends to have reliable predictive power only within and near the domain of the training set. When the optimal configuration of materials and structures lies far beyond the initial training set, which frequently is the case, neural network-based design methods suffer from weak predictive power and become inefficient.
Researchers expect that the framework will be applicable for a wide range of optimization problems in other science and engineering disciplines with astronomically large design space, because it provides an efficient way of gradually expanding the reliable prediction domain toward the target design while avoiding the risk of being stuck in local minima. Especially, being a less-data-hungry method, design problems in which data generation is time-consuming and expensive will benefit most from this new framework.
The research team is currently applying the optimization framework for the design task of metamaterial structures, segmented thermoelectric generators, and optimal sensor distributions. “From these sets of on-going studies, we expect to better recognize the pros and cons, and the potential of the suggested algorithm. Ultimately, we want to devise more efficient machine learning-based design approaches,” explained Professor Ryu.This study was funded by the National Research Foundation of Korea and the KAIST Global Singularity Research Project.
-Publication
Yongtae Kim, Youngsoo, Charles Yang, Kundo Park, Grace X. Gu, and Seunghwa Ryu, “Deep learning framework for material design space exploration using active transfer learning and data augmentation,” npj Computational Materials (https://doi.org/10.1038/s41524-021-00609-2)
-Profile
Professor Seunghwa Ryu
Mechanics & Materials Modeling Lab
Department of Mechanical Engineering
KAIST
2021.09.29 View 12077
Deep Learning Framework to Enable Material Design in Unseen Domain
Researchers propose a deep neural network-based forward design space exploration using active transfer learning and data augmentation
A new study proposed a deep neural network-based forward design approach that enables an efficient search for superior materials far beyond the domain of the initial training set. This approach compensates for the weak predictive power of neural networks on an unseen domain through gradual updates of the neural network with active transfer learning and data augmentation methods.
Professor Seungwha Ryu believes that this study will help address a variety of optimization problems that have an astronomical number of possible design configurations. For the grid composite optimization problem, the proposed framework was able to provide excellent designs close to the global optima, even with the addition of a very small dataset corresponding to less than 0.5% of the initial training data-set size. This study was reported in npj Computational Materials last month.
“We wanted to mitigate the limitation of the neural network, weak predictive power beyond the training set domain for the material or structure design,” said Professor Ryu from the Department of Mechanical Engineering.
Neural network-based generative models have been actively investigated as an inverse design method for finding novel materials in a vast design space. However, the applicability of conventional generative models is limited because they cannot access data outside the range of training sets. Advanced generative models that were devised to overcome this limitation also suffer from weak predictive power for the unseen domain.
Professor Ryu’s team, in collaboration with researchers from Professor Grace Gu’s group at UC Berkeley, devised a design method that simultaneously expands the domain using the strong predictive power of a deep neural network and searches for the optimal design by repetitively performing three key steps.
First, it searches for few candidates with improved properties located close to the training set via genetic algorithms, by mixing superior designs within the training set. Then, it checks to see if the candidates really have improved properties, and expands the training set by duplicating the validated designs via a data augmentation method. Finally, they can expand the reliable prediction domain by updating the neural network with the new superior designs via transfer learning. Because the expansion proceeds along relatively narrow but correct routes toward the optimal design (depicted in the schematic of Fig. 1), the framework enables an efficient search.
As a data-hungry method, a deep neural network model tends to have reliable predictive power only within and near the domain of the training set. When the optimal configuration of materials and structures lies far beyond the initial training set, which frequently is the case, neural network-based design methods suffer from weak predictive power and become inefficient.
Researchers expect that the framework will be applicable for a wide range of optimization problems in other science and engineering disciplines with astronomically large design space, because it provides an efficient way of gradually expanding the reliable prediction domain toward the target design while avoiding the risk of being stuck in local minima. Especially, being a less-data-hungry method, design problems in which data generation is time-consuming and expensive will benefit most from this new framework.
The research team is currently applying the optimization framework for the design task of metamaterial structures, segmented thermoelectric generators, and optimal sensor distributions. “From these sets of on-going studies, we expect to better recognize the pros and cons, and the potential of the suggested algorithm. Ultimately, we want to devise more efficient machine learning-based design approaches,” explained Professor Ryu.This study was funded by the National Research Foundation of Korea and the KAIST Global Singularity Research Project.
-Publication
Yongtae Kim, Youngsoo, Charles Yang, Kundo Park, Grace X. Gu, and Seunghwa Ryu, “Deep learning framework for material design space exploration using active transfer learning and data augmentation,” npj Computational Materials (https://doi.org/10.1038/s41524-021-00609-2)
-Profile
Professor Seunghwa Ryu
Mechanics & Materials Modeling Lab
Department of Mechanical Engineering
KAIST
2021.09.29 View 12077 -
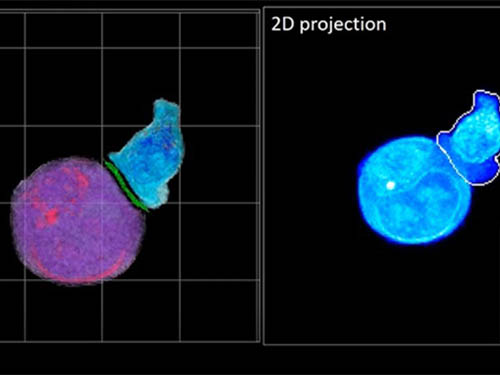 Deep-Learning and 3D Holographic Microscopy Beats Scientists at Analyzing Cancer Immunotherapy
Live tracking and analyzing of the dynamics of chimeric antigen receptor (CAR) T-cells targeting cancer cells can open new avenues for the development of cancer immunotherapy. However, imaging via conventional microscopy approaches can result in cellular damage, and assessments of cell-to-cell interactions are extremely difficult and labor-intensive. When researchers applied deep learning and 3D holographic microscopy to the task, however, they not only avoided these difficultues but found that AI was better at it than humans were.
Artificial intelligence (AI) is helping researchers decipher images from a new holographic microscopy technique needed to investigate a key process in cancer immunotherapy “live” as it takes place. The AI transformed work that, if performed manually by scientists, would otherwise be incredibly labor-intensive and time-consuming into one that is not only effortless but done better than they could have done it themselves. The research, conducted by the team of Professor YongKeun Park from the Department of Physics, appeared in the journal eLife last December.
A critical stage in the development of the human immune system’s ability to respond not just generally to any invader (such as pathogens or cancer cells) but specifically to that particular type of invader and remember it should it attempt to invade again is the formation of a junction between an immune cell called a T-cell and a cell that presents the antigen, or part of the invader that is causing the problem, to it. This process is like when a picture of a suspect is sent to a police car so that the officers can recognize the criminal they are trying to track down. The junction between the two cells, called the immunological synapse, or IS, is the key process in teaching the immune system how to recognize a specific type of invader.
Since the formation of the IS junction is such a critical step for the initiation of an antigen-specific immune response, various techniques allowing researchers to observe the process as it happens have been used to study its dynamics. Most of these live imaging techniques rely on fluorescence microscopy, where genetic tweaking causes part of a protein from a cell to fluoresce, in turn allowing the subject to be tracked via fluorescence rather than via the reflected light used in many conventional microscopy techniques.
However, fluorescence-based imaging can suffer from effects such as photo-bleaching and photo-toxicity, preventing the assessment of dynamic changes in the IS junction process over the long term. Fluorescence-based imaging still involves illumination, whereupon the fluorophores (chemical compounds that cause the fluorescence) emit light of a different color. Photo-bleaching or photo-toxicity occur when the subject is exposed to too much illumination, resulting in chemical alteration or cellular damage.
One recent option that does away with fluorescent labelling and thereby avoids such problems is 3D holographic microscopy or holotomography (HT). In this technique, the refractive index (the way that light changes direction when encountering a substance with a different density—why a straw looks like it bends in a glass of water) is recorded in 3D as a hologram.
Until now, HT has been used to study single cells, but never cell-cell interactions involved in immune responses. One of the main reasons is the difficulty of “segmentation,” or distinguishing the different parts of a cell and thus distinguishing between the interacting cells; in other words, deciphering which part belongs to which cell.
Manual segmentation, or marking out the different parts manually, is one option, but it is difficult and time-consuming, especially in three dimensions. To overcome this problem, automatic segmentation has been developed in which simple computer algorithms perform the identification.
“But these basic algorithms often make mistakes,” explained Professor YongKeun Park, “particularly with respect to adjoining segmentation, which of course is exactly what is occurring here in the immune response we’re most interested in.”
So, the researchers applied a deep learning framework to the HT segmentation problem. Deep learning is a type of machine learning in which artificial neural networks based on the human brain recognize patterns in a way that is similar to how humans do this. Regular machine learning requires data as an input that has already been labelled. The AI “learns” by understanding the labeled data and then recognizes the concept that has been labelled when it is fed novel data. For example, AI trained on a thousand images of cats labelled “cat” should be able to recognize a cat the next time it encounters an image with a cat in it. Deep learning involves multiple layers of artificial neural networks attacking much larger, but unlabeled datasets, in which the AI develops its own ‘labels’ for concepts it encounters.
In essence, the deep learning framework that KAIST researchers developed, called DeepIS, came up with its own concepts by which it distinguishes the different parts of the IS junction process. To validate this method, the research team applied it to the dynamics of a particular IS junction formed between chimeric antigen receptor (CAR) T-cells and target cancer cells. They then compared the results to what they would normally have done: the laborious process of performing the segmentation manually. They found not only that DeepIS was able to define areas within the IS with high accuracy, but that the technique was even able to capture information about the total distribution of proteins within the IS that may not have been easily measured using conventional techniques.
“In addition to allowing us to avoid the drudgery of manual segmentation and the problems of photo-bleaching and photo-toxicity, we found that the AI actually did a better job,” Professor Park added.
The next step will be to combine the technique with methods of measuring how much physical force is applied by different parts of the IS junction, such as holographic optical tweezers or traction force microscopy.
-Profile
Professor YongKeun Park
Department of Physics
Biomedical Optics Laboratory
http://bmol.kaist.ac.kr
KAIST
2021.02.24 View 12667
Deep-Learning and 3D Holographic Microscopy Beats Scientists at Analyzing Cancer Immunotherapy
Live tracking and analyzing of the dynamics of chimeric antigen receptor (CAR) T-cells targeting cancer cells can open new avenues for the development of cancer immunotherapy. However, imaging via conventional microscopy approaches can result in cellular damage, and assessments of cell-to-cell interactions are extremely difficult and labor-intensive. When researchers applied deep learning and 3D holographic microscopy to the task, however, they not only avoided these difficultues but found that AI was better at it than humans were.
Artificial intelligence (AI) is helping researchers decipher images from a new holographic microscopy technique needed to investigate a key process in cancer immunotherapy “live” as it takes place. The AI transformed work that, if performed manually by scientists, would otherwise be incredibly labor-intensive and time-consuming into one that is not only effortless but done better than they could have done it themselves. The research, conducted by the team of Professor YongKeun Park from the Department of Physics, appeared in the journal eLife last December.
A critical stage in the development of the human immune system’s ability to respond not just generally to any invader (such as pathogens or cancer cells) but specifically to that particular type of invader and remember it should it attempt to invade again is the formation of a junction between an immune cell called a T-cell and a cell that presents the antigen, or part of the invader that is causing the problem, to it. This process is like when a picture of a suspect is sent to a police car so that the officers can recognize the criminal they are trying to track down. The junction between the two cells, called the immunological synapse, or IS, is the key process in teaching the immune system how to recognize a specific type of invader.
Since the formation of the IS junction is such a critical step for the initiation of an antigen-specific immune response, various techniques allowing researchers to observe the process as it happens have been used to study its dynamics. Most of these live imaging techniques rely on fluorescence microscopy, where genetic tweaking causes part of a protein from a cell to fluoresce, in turn allowing the subject to be tracked via fluorescence rather than via the reflected light used in many conventional microscopy techniques.
However, fluorescence-based imaging can suffer from effects such as photo-bleaching and photo-toxicity, preventing the assessment of dynamic changes in the IS junction process over the long term. Fluorescence-based imaging still involves illumination, whereupon the fluorophores (chemical compounds that cause the fluorescence) emit light of a different color. Photo-bleaching or photo-toxicity occur when the subject is exposed to too much illumination, resulting in chemical alteration or cellular damage.
One recent option that does away with fluorescent labelling and thereby avoids such problems is 3D holographic microscopy or holotomography (HT). In this technique, the refractive index (the way that light changes direction when encountering a substance with a different density—why a straw looks like it bends in a glass of water) is recorded in 3D as a hologram.
Until now, HT has been used to study single cells, but never cell-cell interactions involved in immune responses. One of the main reasons is the difficulty of “segmentation,” or distinguishing the different parts of a cell and thus distinguishing between the interacting cells; in other words, deciphering which part belongs to which cell.
Manual segmentation, or marking out the different parts manually, is one option, but it is difficult and time-consuming, especially in three dimensions. To overcome this problem, automatic segmentation has been developed in which simple computer algorithms perform the identification.
“But these basic algorithms often make mistakes,” explained Professor YongKeun Park, “particularly with respect to adjoining segmentation, which of course is exactly what is occurring here in the immune response we’re most interested in.”
So, the researchers applied a deep learning framework to the HT segmentation problem. Deep learning is a type of machine learning in which artificial neural networks based on the human brain recognize patterns in a way that is similar to how humans do this. Regular machine learning requires data as an input that has already been labelled. The AI “learns” by understanding the labeled data and then recognizes the concept that has been labelled when it is fed novel data. For example, AI trained on a thousand images of cats labelled “cat” should be able to recognize a cat the next time it encounters an image with a cat in it. Deep learning involves multiple layers of artificial neural networks attacking much larger, but unlabeled datasets, in which the AI develops its own ‘labels’ for concepts it encounters.
In essence, the deep learning framework that KAIST researchers developed, called DeepIS, came up with its own concepts by which it distinguishes the different parts of the IS junction process. To validate this method, the research team applied it to the dynamics of a particular IS junction formed between chimeric antigen receptor (CAR) T-cells and target cancer cells. They then compared the results to what they would normally have done: the laborious process of performing the segmentation manually. They found not only that DeepIS was able to define areas within the IS with high accuracy, but that the technique was even able to capture information about the total distribution of proteins within the IS that may not have been easily measured using conventional techniques.
“In addition to allowing us to avoid the drudgery of manual segmentation and the problems of photo-bleaching and photo-toxicity, we found that the AI actually did a better job,” Professor Park added.
The next step will be to combine the technique with methods of measuring how much physical force is applied by different parts of the IS junction, such as holographic optical tweezers or traction force microscopy.
-Profile
Professor YongKeun Park
Department of Physics
Biomedical Optics Laboratory
http://bmol.kaist.ac.kr
KAIST
2021.02.24 View 12667 -
 DeepTFactor Predicts Transcription Factors
A deep learning-based tool predicts transcription factors using protein sequences as inputs
A joint research team from KAIST and UCSD has developed a deep neural network named DeepTFactor that predicts transcription factors from protein sequences. DeepTFactor will serve as a useful tool for understanding the regulatory systems of organisms, accelerating the use of deep learning for solving biological problems.
A transcription factor is a protein that specifically binds to DNA sequences to control the transcription initiation. Analyzing transcriptional regulation enables the understanding of how organisms control gene expression in response to genetic or environmental changes. In this regard, finding the transcription factor of an organism is the first step in the analysis of the transcriptional regulatory system of an organism.
Previously, transcription factors have been predicted by analyzing sequence homology with already characterized transcription factors or by data-driven approaches such as machine learning. Conventional machine learning models require a rigorous feature selection process that relies on domain expertise such as calculating the physicochemical properties of molecules or analyzing the homology of biological sequences. Meanwhile, deep learning can inherently learn latent features for the specific task.
A joint research team comprised of Ph.D. candidate Gi Bae Kim and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Ye Gao and Professor Bernhard O. Palsson of the Department of Biochemical Engineering at UCSD reported a deep learning-based tool for the prediction of transcription factors. Their research paper “DeepTFactor: A deep learning-based tool for the prediction of transcription factors” was published online in PNAS.
Their article reports the development of DeepTFactor, a deep learning-based tool that predicts whether a given protein sequence is a transcription factor using three parallel convolutional neural networks. The joint research team predicted 332 transcription factors of Escherichia coli K-12 MG1655 using DeepTFactor and the performance of DeepTFactor by experimentally confirming the genome-wide binding sites of three predicted transcription factors (YqhC, YiaU, and YahB).
The joint research team further used a saliency method to understand the reasoning process of DeepTFactor. The researchers confirmed that even though information on the DNA binding domains of the transcription factor was not explicitly given the training process, DeepTFactor implicitly learned and used them for prediction. Unlike previous transcription factor prediction tools that were developed only for protein sequences of specific organisms, DeepTFactor is expected to be used in the analysis of the transcription systems of all organisms at a high level of performance.
Distinguished Professor Sang Yup Lee said, “DeepTFactor can be used to discover unknown transcription factors from numerous protein sequences that have not yet been characterized. It is expected that DeepTFactor will serve as an important tool for analyzing the regulatory systems of organisms of interest.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea.
-Publication
Gi Bae Kim, Ye Gao, Bernhard O. Palsson, and Sang Yup Lee. DeepTFactor: A deep learning-based tool for the prediction of transcription factors. (https://doi.org/10.1073/pnas202117118)
-Profile
Distinguished Professor Sang Yup Lee
leesy@kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
http://mbel.kaist.ac.kr
Department of Chemical and Biomolecular Engineering
KAIST
2021.01.05 View 9865
DeepTFactor Predicts Transcription Factors
A deep learning-based tool predicts transcription factors using protein sequences as inputs
A joint research team from KAIST and UCSD has developed a deep neural network named DeepTFactor that predicts transcription factors from protein sequences. DeepTFactor will serve as a useful tool for understanding the regulatory systems of organisms, accelerating the use of deep learning for solving biological problems.
A transcription factor is a protein that specifically binds to DNA sequences to control the transcription initiation. Analyzing transcriptional regulation enables the understanding of how organisms control gene expression in response to genetic or environmental changes. In this regard, finding the transcription factor of an organism is the first step in the analysis of the transcriptional regulatory system of an organism.
Previously, transcription factors have been predicted by analyzing sequence homology with already characterized transcription factors or by data-driven approaches such as machine learning. Conventional machine learning models require a rigorous feature selection process that relies on domain expertise such as calculating the physicochemical properties of molecules or analyzing the homology of biological sequences. Meanwhile, deep learning can inherently learn latent features for the specific task.
A joint research team comprised of Ph.D. candidate Gi Bae Kim and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Ye Gao and Professor Bernhard O. Palsson of the Department of Biochemical Engineering at UCSD reported a deep learning-based tool for the prediction of transcription factors. Their research paper “DeepTFactor: A deep learning-based tool for the prediction of transcription factors” was published online in PNAS.
Their article reports the development of DeepTFactor, a deep learning-based tool that predicts whether a given protein sequence is a transcription factor using three parallel convolutional neural networks. The joint research team predicted 332 transcription factors of Escherichia coli K-12 MG1655 using DeepTFactor and the performance of DeepTFactor by experimentally confirming the genome-wide binding sites of three predicted transcription factors (YqhC, YiaU, and YahB).
The joint research team further used a saliency method to understand the reasoning process of DeepTFactor. The researchers confirmed that even though information on the DNA binding domains of the transcription factor was not explicitly given the training process, DeepTFactor implicitly learned and used them for prediction. Unlike previous transcription factor prediction tools that were developed only for protein sequences of specific organisms, DeepTFactor is expected to be used in the analysis of the transcription systems of all organisms at a high level of performance.
Distinguished Professor Sang Yup Lee said, “DeepTFactor can be used to discover unknown transcription factors from numerous protein sequences that have not yet been characterized. It is expected that DeepTFactor will serve as an important tool for analyzing the regulatory systems of organisms of interest.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea.
-Publication
Gi Bae Kim, Ye Gao, Bernhard O. Palsson, and Sang Yup Lee. DeepTFactor: A deep learning-based tool for the prediction of transcription factors. (https://doi.org/10.1073/pnas202117118)
-Profile
Distinguished Professor Sang Yup Lee
leesy@kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
http://mbel.kaist.ac.kr
Department of Chemical and Biomolecular Engineering
KAIST
2021.01.05 View 9865 -
 Deep Learning Helps Explore the Structural and Strategic Bases of Autism
Psychiatrists typically diagnose autism spectrum disorders (ASD) by observing a person’s behavior and by leaning on the Diagnostic and Statistical Manual of Mental Disorders (DSM-5), widely considered the “bible” of mental health diagnosis.
However, there are substantial differences amongst individuals on the spectrum and a great deal remains unknown by science about the causes of autism, or even what autism is. As a result, an accurate diagnosis of ASD and a prognosis prediction for patients can be extremely difficult.
But what if artificial intelligence (AI) could help? Deep learning, a type of AI, deploys artificial neural networks based on the human brain to recognize patterns in a way that is akin to, and in some cases can surpass, human ability. The technique, or rather suite of techniques, has enjoyed remarkable success in recent years in fields as diverse as voice recognition, translation, autonomous vehicles, and drug discovery.
A group of researchers from KAIST in collaboration with the Yonsei University College of Medicine has applied these deep learning techniques to autism diagnosis. Their findings were published on August 14 in the journal IEEE Access.
Magnetic resonance imaging (MRI) scans of brains of people known to have autism have been used by researchers and clinicians to try to identify structures of the brain they believed were associated with ASD. These researchers have achieved considerable success in identifying abnormal grey and white matter volume and irregularities in cerebral cortex activation and connections as being associated with the condition.
These findings have subsequently been deployed in studies attempting more consistent diagnoses of patients than has been achieved via psychiatrist observations during counseling sessions. While such studies have reported high levels of diagnostic accuracy, the number of participants in these studies has been small, often under 50, and diagnostic performance drops markedly when applied to large sample sizes or on datasets that include people from a wide variety of populations and locations.
“There was something as to what defines autism that human researchers and clinicians must have been overlooking,” said Keun-Ah Cheon, one of the two corresponding authors and a professor in Department of Child and Adolescent Psychiatry at Severance Hospital of the Yonsei University College of Medicine.
“And humans poring over thousands of MRI scans won’t be able to pick up on what we’ve been missing,” she continued. “But we thought AI might be able to.”
So the team applied five different categories of deep learning models to an open-source dataset of more than 1,000 MRI scans from the Autism Brain Imaging Data Exchange (ABIDE) initiative, which has collected brain imaging data from laboratories around the world, and to a smaller, but higher-resolution MRI image dataset (84 images) taken from the Child Psychiatric Clinic at Severance Hospital, Yonsei University College of Medicine. In both cases, the researchers used both structural MRIs (examining the anatomy of the brain) and functional MRIs (examining brain activity in different regions).
The models allowed the team to explore the structural bases of ASD brain region by brain region, focusing in particular on many structures below the cerebral cortex, including the basal ganglia, which are involved in motor function (movement) as well as learning and memory.
Crucially, these specific types of deep learning models also offered up possible explanations of how the AI had come up with its rationale for these findings.
“Understanding the way that the AI has classified these brain structures and dynamics is extremely important,” said Sang Wan Lee, the other corresponding author and an associate professor at KAIST. “It’s no good if a doctor can tell a patient that the computer says they have autism, but not be able to say why the computer knows that.”
The deep learning models were also able to describe how much a particular aspect contributed to ASD, an analysis tool that can assist psychiatric physicians during the diagnosis process to identify the severity of the autism.
“Doctors should be able to use this to offer a personalized diagnosis for patients, including a prognosis of how the condition could develop,” Lee said.
“Artificial intelligence is not going to put psychiatrists out of a job,” he explained. “But using AI as a tool should enable doctors to better understand and diagnose complex disorders than they could do on their own.”
-ProfileProfessor Sang Wan LeeDepartment of Bio and Brain EngineeringLaboratory for Brain and Machine Intelligence https://aibrain.kaist.ac.kr/
KAIST
2020.09.23 View 11561
Deep Learning Helps Explore the Structural and Strategic Bases of Autism
Psychiatrists typically diagnose autism spectrum disorders (ASD) by observing a person’s behavior and by leaning on the Diagnostic and Statistical Manual of Mental Disorders (DSM-5), widely considered the “bible” of mental health diagnosis.
However, there are substantial differences amongst individuals on the spectrum and a great deal remains unknown by science about the causes of autism, or even what autism is. As a result, an accurate diagnosis of ASD and a prognosis prediction for patients can be extremely difficult.
But what if artificial intelligence (AI) could help? Deep learning, a type of AI, deploys artificial neural networks based on the human brain to recognize patterns in a way that is akin to, and in some cases can surpass, human ability. The technique, or rather suite of techniques, has enjoyed remarkable success in recent years in fields as diverse as voice recognition, translation, autonomous vehicles, and drug discovery.
A group of researchers from KAIST in collaboration with the Yonsei University College of Medicine has applied these deep learning techniques to autism diagnosis. Their findings were published on August 14 in the journal IEEE Access.
Magnetic resonance imaging (MRI) scans of brains of people known to have autism have been used by researchers and clinicians to try to identify structures of the brain they believed were associated with ASD. These researchers have achieved considerable success in identifying abnormal grey and white matter volume and irregularities in cerebral cortex activation and connections as being associated with the condition.
These findings have subsequently been deployed in studies attempting more consistent diagnoses of patients than has been achieved via psychiatrist observations during counseling sessions. While such studies have reported high levels of diagnostic accuracy, the number of participants in these studies has been small, often under 50, and diagnostic performance drops markedly when applied to large sample sizes or on datasets that include people from a wide variety of populations and locations.
“There was something as to what defines autism that human researchers and clinicians must have been overlooking,” said Keun-Ah Cheon, one of the two corresponding authors and a professor in Department of Child and Adolescent Psychiatry at Severance Hospital of the Yonsei University College of Medicine.
“And humans poring over thousands of MRI scans won’t be able to pick up on what we’ve been missing,” she continued. “But we thought AI might be able to.”
So the team applied five different categories of deep learning models to an open-source dataset of more than 1,000 MRI scans from the Autism Brain Imaging Data Exchange (ABIDE) initiative, which has collected brain imaging data from laboratories around the world, and to a smaller, but higher-resolution MRI image dataset (84 images) taken from the Child Psychiatric Clinic at Severance Hospital, Yonsei University College of Medicine. In both cases, the researchers used both structural MRIs (examining the anatomy of the brain) and functional MRIs (examining brain activity in different regions).
The models allowed the team to explore the structural bases of ASD brain region by brain region, focusing in particular on many structures below the cerebral cortex, including the basal ganglia, which are involved in motor function (movement) as well as learning and memory.
Crucially, these specific types of deep learning models also offered up possible explanations of how the AI had come up with its rationale for these findings.
“Understanding the way that the AI has classified these brain structures and dynamics is extremely important,” said Sang Wan Lee, the other corresponding author and an associate professor at KAIST. “It’s no good if a doctor can tell a patient that the computer says they have autism, but not be able to say why the computer knows that.”
The deep learning models were also able to describe how much a particular aspect contributed to ASD, an analysis tool that can assist psychiatric physicians during the diagnosis process to identify the severity of the autism.
“Doctors should be able to use this to offer a personalized diagnosis for patients, including a prognosis of how the condition could develop,” Lee said.
“Artificial intelligence is not going to put psychiatrists out of a job,” he explained. “But using AI as a tool should enable doctors to better understand and diagnose complex disorders than they could do on their own.”
-ProfileProfessor Sang Wan LeeDepartment of Bio and Brain EngineeringLaboratory for Brain and Machine Intelligence https://aibrain.kaist.ac.kr/
KAIST
2020.09.23 View 11561 -
 Seong-Tae Kim Wins Robert-Wagner All-Conference Best Paper Award
(Ph.D. candidate Seong-Tae Kim)
Ph.D. candidate Seong-Tae Kim from the School of Electrical Engineering won the Robert Wagner All-Conference Best Student Paper Award during the 2018 International Society for Optics and Photonics (SPIE) Medical Imaging Conference, which was held in Houston last month.
Kim, supervised by Professor Yong Man Ro, received the award for his paper in the category of computer-aided diagnosis. His paper, titled “ICADx: Interpretable Computer-Aided Diagnosis of Breast Masses”, was selected as the best paper out of 900 submissions. The conference selects the best paper in nine different categories. His research provides new insights on diagnostic technology to detect breast cancer powered by deep learning.
2018.03.15 View 10972
Seong-Tae Kim Wins Robert-Wagner All-Conference Best Paper Award
(Ph.D. candidate Seong-Tae Kim)
Ph.D. candidate Seong-Tae Kim from the School of Electrical Engineering won the Robert Wagner All-Conference Best Student Paper Award during the 2018 International Society for Optics and Photonics (SPIE) Medical Imaging Conference, which was held in Houston last month.
Kim, supervised by Professor Yong Man Ro, received the award for his paper in the category of computer-aided diagnosis. His paper, titled “ICADx: Interpretable Computer-Aided Diagnosis of Breast Masses”, was selected as the best paper out of 900 submissions. The conference selects the best paper in nine different categories. His research provides new insights on diagnostic technology to detect breast cancer powered by deep learning.
2018.03.15 View 10972 -
 KAIST AI Academy for LG CNS Employees
The Department of Industrial & Systems Engineering (Graduate School of Knowledge Service Engineering) at KAIST has collaborated with LG CNS to start a full-fledged KAIST AI Academy course after the two-week pilot course for employees of LG CNS, a Korean company specializing in IT services.
Approximately 100 employees participated in the first KAIST AI Academy course held over two weeks from August 24 to September 1. LG CNS is planning to enroll a total of 500 employees in this course by the end of the year.
Artificial intelligence is widely recognized as essential technology in various industries. In that sense, the KAIST AI Academy course was established to reinforce both the AI technology and the business ability of the company. In addition, it aims at leading employees to develop new business using novel technologies. The main contents of this course are as follows: i) discussing AI technology development and its influence on industries; ii) understanding AI technologies and acquiring the major technologies applicable to business; and iii) introducing cases of AI applications and deep learning.
During the course, seven professors with expertise in AI deep learning from the Department of Industrial & Systems Engineering (Graduate School of Knowledge Service Engineering), including Jae-Gil Lee and Jinkyoo Park will be leading the class, including practical on-site educational programs.
Based on the accumulated business experience integrated with the latest AI technology, LG CNS has been making an effort to find new business opportunities to support companies that are hoping to make digital innovations.
The company aims to reinforce the AI capabilities of its employees and is planning to upgrade the course in a sustainable manner. It will also foster outside manpower by expanding the AI education to its clients who pursue manufacturing reinforcement and innovation in digital marketing.
Seong Wook Lee, the Director of the AI and Big Data Business Unit said, “As AI plays an important role in business services, LG CNS decided to open the KAIST AI Academy course to deliver better value to our clients by incorporating our AI-based business cases and KAIST’s up-to-date knowledge.”
2017.09.06 View 7714
KAIST AI Academy for LG CNS Employees
The Department of Industrial & Systems Engineering (Graduate School of Knowledge Service Engineering) at KAIST has collaborated with LG CNS to start a full-fledged KAIST AI Academy course after the two-week pilot course for employees of LG CNS, a Korean company specializing in IT services.
Approximately 100 employees participated in the first KAIST AI Academy course held over two weeks from August 24 to September 1. LG CNS is planning to enroll a total of 500 employees in this course by the end of the year.
Artificial intelligence is widely recognized as essential technology in various industries. In that sense, the KAIST AI Academy course was established to reinforce both the AI technology and the business ability of the company. In addition, it aims at leading employees to develop new business using novel technologies. The main contents of this course are as follows: i) discussing AI technology development and its influence on industries; ii) understanding AI technologies and acquiring the major technologies applicable to business; and iii) introducing cases of AI applications and deep learning.
During the course, seven professors with expertise in AI deep learning from the Department of Industrial & Systems Engineering (Graduate School of Knowledge Service Engineering), including Jae-Gil Lee and Jinkyoo Park will be leading the class, including practical on-site educational programs.
Based on the accumulated business experience integrated with the latest AI technology, LG CNS has been making an effort to find new business opportunities to support companies that are hoping to make digital innovations.
The company aims to reinforce the AI capabilities of its employees and is planning to upgrade the course in a sustainable manner. It will also foster outside manpower by expanding the AI education to its clients who pursue manufacturing reinforcement and innovation in digital marketing.
Seong Wook Lee, the Director of the AI and Big Data Business Unit said, “As AI plays an important role in business services, LG CNS decided to open the KAIST AI Academy course to deliver better value to our clients by incorporating our AI-based business cases and KAIST’s up-to-date knowledge.”
2017.09.06 View 7714