Medicine
-
 KAIST Succeeds in the Real-time Observation of Organoids using Holotomography
Organoids, which are 3D miniature organs that mimic the structure and function of human organs, play an essential role in disease research and drug development. A Korean research team has overcome the limitations of existing imaging technologies, succeeding in the real-time, high-resolution observation of living organoids.
KAIST (represented by President Kwang Hyung Lee) announced on the 14th of October that Professor YongKeun Park’s research team from the Department of Physics, in collaboration with the Genome Editing Research Center (Director Bon-Kyoung Koo) of the Institute for Basic Science (IBS President Do-Young Noh) and Tomocube Inc., has developed an imaging technology using holotomography to observe live, small intestinal organoids in real time at a high resolution.
Existing imaging techniques have struggled to observe living organoids in high resolution over extended periods and often required additional treatments like fluorescent staining.
< Figure 1. Overview of the low-coherence HT workflow. Using holotomography, 3D morphological restoration and quantitative analysis of organoids can be performed. In order to improve the limited field of view, which is a limitation of the microscope, our research team utilized a large-area field of view combination algorithm and made a 3D restoration by acquiring multi-focus holographic images for 3D measurements. After that, the organoids were compartmentalized to divide the parts necessary for analysis and quantitatively evaluated the protein concentration measurable from the refractive index and the survival rate of the organoids. >
The research team introduced holotomography technology to address these issues, which provides high-resolution images without the need for fluorescent staining and allows for the long-term observation of dynamic changes in real time without causing cell damage.
The team validated this technology using small intestinal organoids from experimental mice and were able to observe various cell structures inside the organoids in detail. They also captured dynamic changes such as growth processes, cell division, and cell death in real time using holotomography.
Additionally, the technology allowed for the precise analysis of the organoids' responses to drug treatments, verifying the survival of the cells.
The researchers believe that this breakthrough will open new horizons in organoid research, enabling the greater utilization of organoids in drug development, personalized medicine, and regenerative medicine.
Future research is expected to more accurately replicate the in vivo environment of organoids, contributing significantly to a more detailed understanding of various life phenomena at the cellular level through more precise 3D imaging.
< Figure 2. Real-time organoid morphology analysis. Using holotomography, it is possible to observe the lumen and villus development process of intestinal organoids in real time, which was difficult to observe with a conventional microscope. In addition, various information about intestinal organoids can be obtained by quantifying the size and protein amount of intestinal organoids through image analysis. >
Dr. Mahn Jae Lee, a graduate of KAIST's Graduate School of Medical Science and Engineering, currently at Chungnam National University Hospital and the first author of the paper, commented, "This research represents a new imaging technology that surpasses previous limitations and is expected to make a major contribution to disease modeling, personalized treatments, and drug development research using organoids."
The research results were published online in the international journal Experimental & Molecular Medicine on October 1, 2024, and the technology has been recognized for its applicability in various fields of life sciences. (Paper title: “Long-term three-dimensional high-resolution imaging of live unlabeled small intestinal organoids via low-coherence holotomography”)
This research was supported by the National Research Foundation of Korea, KAIST Institutes, and the Institute for Basic Science.
2024.10.14 View 3494
KAIST Succeeds in the Real-time Observation of Organoids using Holotomography
Organoids, which are 3D miniature organs that mimic the structure and function of human organs, play an essential role in disease research and drug development. A Korean research team has overcome the limitations of existing imaging technologies, succeeding in the real-time, high-resolution observation of living organoids.
KAIST (represented by President Kwang Hyung Lee) announced on the 14th of October that Professor YongKeun Park’s research team from the Department of Physics, in collaboration with the Genome Editing Research Center (Director Bon-Kyoung Koo) of the Institute for Basic Science (IBS President Do-Young Noh) and Tomocube Inc., has developed an imaging technology using holotomography to observe live, small intestinal organoids in real time at a high resolution.
Existing imaging techniques have struggled to observe living organoids in high resolution over extended periods and often required additional treatments like fluorescent staining.
< Figure 1. Overview of the low-coherence HT workflow. Using holotomography, 3D morphological restoration and quantitative analysis of organoids can be performed. In order to improve the limited field of view, which is a limitation of the microscope, our research team utilized a large-area field of view combination algorithm and made a 3D restoration by acquiring multi-focus holographic images for 3D measurements. After that, the organoids were compartmentalized to divide the parts necessary for analysis and quantitatively evaluated the protein concentration measurable from the refractive index and the survival rate of the organoids. >
The research team introduced holotomography technology to address these issues, which provides high-resolution images without the need for fluorescent staining and allows for the long-term observation of dynamic changes in real time without causing cell damage.
The team validated this technology using small intestinal organoids from experimental mice and were able to observe various cell structures inside the organoids in detail. They also captured dynamic changes such as growth processes, cell division, and cell death in real time using holotomography.
Additionally, the technology allowed for the precise analysis of the organoids' responses to drug treatments, verifying the survival of the cells.
The researchers believe that this breakthrough will open new horizons in organoid research, enabling the greater utilization of organoids in drug development, personalized medicine, and regenerative medicine.
Future research is expected to more accurately replicate the in vivo environment of organoids, contributing significantly to a more detailed understanding of various life phenomena at the cellular level through more precise 3D imaging.
< Figure 2. Real-time organoid morphology analysis. Using holotomography, it is possible to observe the lumen and villus development process of intestinal organoids in real time, which was difficult to observe with a conventional microscope. In addition, various information about intestinal organoids can be obtained by quantifying the size and protein amount of intestinal organoids through image analysis. >
Dr. Mahn Jae Lee, a graduate of KAIST's Graduate School of Medical Science and Engineering, currently at Chungnam National University Hospital and the first author of the paper, commented, "This research represents a new imaging technology that surpasses previous limitations and is expected to make a major contribution to disease modeling, personalized treatments, and drug development research using organoids."
The research results were published online in the international journal Experimental & Molecular Medicine on October 1, 2024, and the technology has been recognized for its applicability in various fields of life sciences. (Paper title: “Long-term three-dimensional high-resolution imaging of live unlabeled small intestinal organoids via low-coherence holotomography”)
This research was supported by the National Research Foundation of Korea, KAIST Institutes, and the Institute for Basic Science.
2024.10.14 View 3494 -
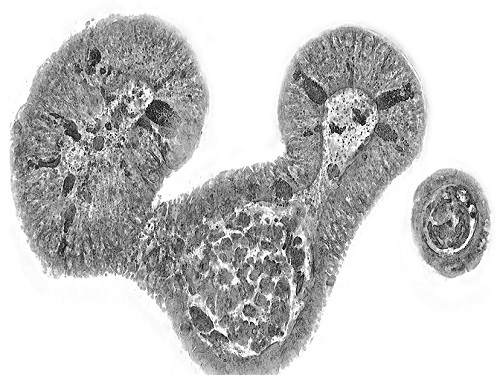 KAIST presents strategies for Holotomography in advanced bio research
Measuring and analyzing three-dimensional (3D) images of live cells and tissues is considered crucial in advanced fields of biology and medicine. Organoids, which are 3D structures that mimic organs, are particular examples that significantly benefits 3D live imaging. Organoids provide effective alternatives to animal testing in the drug development processes, and can rapidly determine personalized medicine. On the other hand, active researches are ongoing to utilize organoids for organ replacement.
< Figure 1. Schematic illustration of holotomography compared to X-ray CT. Similar to CT, they share the commonality of measuring the optical properties of an unlabeled specimen in three dimensions. Instead of X-rays, holotomography irradiates light in the visible range, and provides refractive index measurements of transparent specimens rather than absorptivity. While CT obtains three-dimensional information only through mechanical rotation of the irradiating light, holotomography can replace this by applying wavefront control technology in the visible range. >
Organelle-level observation of 3D biological specimens such as organoids and stem cell colonies without staining or preprocessing holds significant implications for both innovating basic research and bioindustrial applications related to regenerative medicine and bioindustrial applications.
Holotomography (HT) is a 3D optical microscopy that implements 3D reconstruction analogous to that of X-ray computed tomography (CT). Although HT and CT share a similar theoretical background, HT facilitates high-resolution examination inside cells and tissues, instead of the human body. HT obtains 3D images of cells and tissues at the organelle level without chemical or genetic labeling, thus overcomes various challenges of existing methods in bio research and industry. Its potential is highlighted in research fields where sample physiology must not be disrupted, such as regenerative medicine, personalized medicine, and infertility treatment.
< Figure 2. Label-free 3D imaging of diverse live cells. Time-lapse image of Hep3B cells illustrating subcellular morphology changes upon H2O2 treatment, followed by cellular recovery after returning to the regular cell culture medium. >
This paper introduces the advantages and broad applicability of HT to biomedical researchers, while presenting an overview of principles and future technical challenges to optical researchers. It showcases various cases of applying HT in studies such as 3D biology, regenerative medicine, and cancer research, as well as suggesting future optical development. Also, it categorizes HT based on the light source, to describe the principles, limitations, and improvements of each category in detail. Particularly, the paper addresses strategies for deepening cell and organoid studies by introducing artificial intelligence (AI) to HT.
Due to its potential to drive advanced bioindustry, HT is attracting interest and investment from universities and corporates worldwide. The KAIST research team has been leading this international field by developing core technologies and carrying out key application researches throughout the last decade.
< Figure 3. Various types of cells and organelles that make up the imaging barrier of a living intestinal organoid can be observed using holotomography. >
This paper, co-authored by Dr. Geon Kim from KAIST Research Center for Natural Sciences, Professor Ki-Jun Yoon's team from the Department of Biological Sciences, Director Bon-Kyoung Koo's team from the Institute for Basic Science (IBS) Center for Genome Engineering, and Dr. Seongsoo Lee's team from the Korea Basic Science Institute (KBSI), was published in 'Nature Reviews Methods Primers' on the 25th of July. This research was supported by the Leader Grant and Basic Science Research Program of the National Research Foundation, the Hologram Core Technology Development Grant of the Ministry of Science and ICT, the Nano and Material Technology Development Project, and the Health and Medical R&D Project of the Ministry of Health and Welfare.
2024.07.30 View 4169
KAIST presents strategies for Holotomography in advanced bio research
Measuring and analyzing three-dimensional (3D) images of live cells and tissues is considered crucial in advanced fields of biology and medicine. Organoids, which are 3D structures that mimic organs, are particular examples that significantly benefits 3D live imaging. Organoids provide effective alternatives to animal testing in the drug development processes, and can rapidly determine personalized medicine. On the other hand, active researches are ongoing to utilize organoids for organ replacement.
< Figure 1. Schematic illustration of holotomography compared to X-ray CT. Similar to CT, they share the commonality of measuring the optical properties of an unlabeled specimen in three dimensions. Instead of X-rays, holotomography irradiates light in the visible range, and provides refractive index measurements of transparent specimens rather than absorptivity. While CT obtains three-dimensional information only through mechanical rotation of the irradiating light, holotomography can replace this by applying wavefront control technology in the visible range. >
Organelle-level observation of 3D biological specimens such as organoids and stem cell colonies without staining or preprocessing holds significant implications for both innovating basic research and bioindustrial applications related to regenerative medicine and bioindustrial applications.
Holotomography (HT) is a 3D optical microscopy that implements 3D reconstruction analogous to that of X-ray computed tomography (CT). Although HT and CT share a similar theoretical background, HT facilitates high-resolution examination inside cells and tissues, instead of the human body. HT obtains 3D images of cells and tissues at the organelle level without chemical or genetic labeling, thus overcomes various challenges of existing methods in bio research and industry. Its potential is highlighted in research fields where sample physiology must not be disrupted, such as regenerative medicine, personalized medicine, and infertility treatment.
< Figure 2. Label-free 3D imaging of diverse live cells. Time-lapse image of Hep3B cells illustrating subcellular morphology changes upon H2O2 treatment, followed by cellular recovery after returning to the regular cell culture medium. >
This paper introduces the advantages and broad applicability of HT to biomedical researchers, while presenting an overview of principles and future technical challenges to optical researchers. It showcases various cases of applying HT in studies such as 3D biology, regenerative medicine, and cancer research, as well as suggesting future optical development. Also, it categorizes HT based on the light source, to describe the principles, limitations, and improvements of each category in detail. Particularly, the paper addresses strategies for deepening cell and organoid studies by introducing artificial intelligence (AI) to HT.
Due to its potential to drive advanced bioindustry, HT is attracting interest and investment from universities and corporates worldwide. The KAIST research team has been leading this international field by developing core technologies and carrying out key application researches throughout the last decade.
< Figure 3. Various types of cells and organelles that make up the imaging barrier of a living intestinal organoid can be observed using holotomography. >
This paper, co-authored by Dr. Geon Kim from KAIST Research Center for Natural Sciences, Professor Ki-Jun Yoon's team from the Department of Biological Sciences, Director Bon-Kyoung Koo's team from the Institute for Basic Science (IBS) Center for Genome Engineering, and Dr. Seongsoo Lee's team from the Korea Basic Science Institute (KBSI), was published in 'Nature Reviews Methods Primers' on the 25th of July. This research was supported by the Leader Grant and Basic Science Research Program of the National Research Foundation, the Hologram Core Technology Development Grant of the Ministry of Science and ICT, the Nano and Material Technology Development Project, and the Health and Medical R&D Project of the Ministry of Health and Welfare.
2024.07.30 View 4169 -
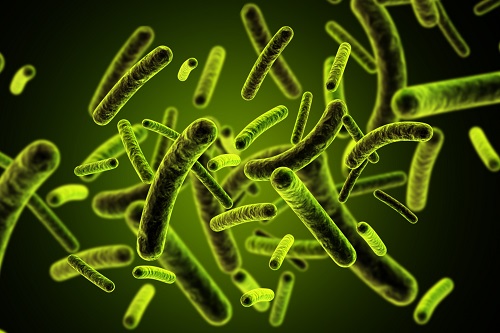 3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 13433
3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 13433 -
 Life After COVID-19: Big Questions on Medical and Bio-Engineering
KAIST GSI forum explores big questions in the medical and bio-engineering revolution caused by the COVID-19 in fight against infectious diseases and life quality
On September 9, the Global Strategy Institute at KAIST will delve into innovative future strategies for the medical and bio-engineering sectors that have been disrupted by COVID-19. The forum will live stream via YouTube, KTV, and Naver TV from 9:00 am Korean time.
The online forum features a speaker lineup of world-renowned scholars who will discuss an array of bio-engineering technologies that will improve our quality of life and even extend our life span. This is the GSI’s third online forum since the first one in April that covered the socio-economic implications of the global pandemic and the second one in June focusing on the education sector.
In hosting the third round of the GSI Forum series, KAIST President Sung-Chul Shin stressed the power of science and technology saying, “In this world full of uncertainties, one thing for sure is that only the advancement of science and technology will deliver us from this crisis.” Korean Prime Minister Sye-Kyun Chung will also deliver a speech explaining the government’s response to COVID-19 and vaccine development strategies.
The President of the National Academy of Medicine in the US will share ideal policies to back up the bio-engineering and medical sectors and Futurist Thomas Frey from the Davinci Institute will present his distinct perspectives on our future lives after COVID-19. His thought-provoking insights on advancements in the bioengineering sector will examine whether humanity can put an end to infectious diseases and find new ways to lengthen our lives.
Two distinguished professors in the field of genetic engineering technology will share their latest breakthroughs. Professor George McDonald Church from Harvard Medical School who developed genome sequencing will deliver a keynote speech on how the advancement of gene editing and genome technology will overcome diseases and contribute to extending human life spans.
Professor Kwang-Soo Kim, a KAIST alumnus from Harvard Medical School who recently reported new discoveries for Parkinson’s disease treatment by reprogramming a patient’s own skin cells to replace cells in the brain, will introduce the latest clinical cell treatment technologies based on personalized therapeutics.
Senior Vice President and Chief Product Officer of Illumina Susan Tousi, a leading genome sequencing solution provider, will describe genome analysis technology and explore the potential for disease prevention.
KAIST medical scientist Jeong Ho Lee, who was the first to identify the causes of intractable epilepsies and has identified the genes responsible for several developmental brain disorders. Professor Jin-Hyung Lee from Stanford University and Dr. David B. Resnik from the National Institute of Environmental Health Science will also join the speaker lineup to discuss genetics-based personalized solutions to extend human life spans.
The forum will also invite about 50 young scientists and medical researchers from around the world to participate in an online panel session. They will engage in a Q&A session and a discussion with the speakers.
(END)
2020.09.04 View 10728
Life After COVID-19: Big Questions on Medical and Bio-Engineering
KAIST GSI forum explores big questions in the medical and bio-engineering revolution caused by the COVID-19 in fight against infectious diseases and life quality
On September 9, the Global Strategy Institute at KAIST will delve into innovative future strategies for the medical and bio-engineering sectors that have been disrupted by COVID-19. The forum will live stream via YouTube, KTV, and Naver TV from 9:00 am Korean time.
The online forum features a speaker lineup of world-renowned scholars who will discuss an array of bio-engineering technologies that will improve our quality of life and even extend our life span. This is the GSI’s third online forum since the first one in April that covered the socio-economic implications of the global pandemic and the second one in June focusing on the education sector.
In hosting the third round of the GSI Forum series, KAIST President Sung-Chul Shin stressed the power of science and technology saying, “In this world full of uncertainties, one thing for sure is that only the advancement of science and technology will deliver us from this crisis.” Korean Prime Minister Sye-Kyun Chung will also deliver a speech explaining the government’s response to COVID-19 and vaccine development strategies.
The President of the National Academy of Medicine in the US will share ideal policies to back up the bio-engineering and medical sectors and Futurist Thomas Frey from the Davinci Institute will present his distinct perspectives on our future lives after COVID-19. His thought-provoking insights on advancements in the bioengineering sector will examine whether humanity can put an end to infectious diseases and find new ways to lengthen our lives.
Two distinguished professors in the field of genetic engineering technology will share their latest breakthroughs. Professor George McDonald Church from Harvard Medical School who developed genome sequencing will deliver a keynote speech on how the advancement of gene editing and genome technology will overcome diseases and contribute to extending human life spans.
Professor Kwang-Soo Kim, a KAIST alumnus from Harvard Medical School who recently reported new discoveries for Parkinson’s disease treatment by reprogramming a patient’s own skin cells to replace cells in the brain, will introduce the latest clinical cell treatment technologies based on personalized therapeutics.
Senior Vice President and Chief Product Officer of Illumina Susan Tousi, a leading genome sequencing solution provider, will describe genome analysis technology and explore the potential for disease prevention.
KAIST medical scientist Jeong Ho Lee, who was the first to identify the causes of intractable epilepsies and has identified the genes responsible for several developmental brain disorders. Professor Jin-Hyung Lee from Stanford University and Dr. David B. Resnik from the National Institute of Environmental Health Science will also join the speaker lineup to discuss genetics-based personalized solutions to extend human life spans.
The forum will also invite about 50 young scientists and medical researchers from around the world to participate in an online panel session. They will engage in a Q&A session and a discussion with the speakers.
(END)
2020.09.04 View 10728 -
 Breastfeeding Helps Prevent Mothers from Developing Diabetes after Childbirth
A team of South Korean researchers found that lactation can lower the incidence and reduce the risk of maternal postpartum diabetes. The researchers identified that lactation increases the mass and function of pancreatic beta cells through serotonin production. The team suggested that sustained improvements in pancreatic beta cells, which can last for years even after the cessation of lactation, improve mothers’ metabolic health in addition to providing health benefits for infants.
Pregnancy imposes a substantial metabolic burden on women through weight gain and increased insulin resistance. Various other factors, including a history of gestational diabetes, maternal age, and obesity, further affect women’s risk of progressing to diabetes after delivery, and the risk of postpartum diabetes increases more in women who have had gestational diabetes and/or repeated deliveries.
Diabetes-related complications include damage to blood vessels, which can lead to cardiovascular and cerebrovascular diseases such as heart attack and stroke, and problems with the nerves, eyes, kidneys, and many more. Since diabetes can pose a serious threat to mothers’ metabolic health, the management of maternal metabolic risk factors is important, especially in the peripartum period. Previous epidemiological studies have reported that lactation reduces the risk of postpartum diabetes, but the mechanisms underlying this benefit have remained elusive.
The study, published in Science Translational Medicine on April 29, explains the biology underpinning this observation on the beneficial effects of lactation. Professor Hail Kim from the Graduate School of Medical Science and Engineering at KAIST led and jointly conducted the study in conjunction with researchers from the Seoul National University Bundang Hospital (SNUBH) and Chungnam National University (CNU) in Korea, and the University of California, San Francisco (UCSF) in the US.
In their study, the team observed that the milk-secreting hormone ‘prolactin’ in lactating mothers not only promotes milk production, but also plays a major role in stimulating insulin-secreting pancreatic beta cells that regulate blood glucose in the body.
The researchers also found that ‘serotonin’, known as a chemical that contributes to wellbeing and happiness, is produced in pancreatic beta cells during lactation. Serotonin in pancreatic beta cells act as an antioxidant and reduce oxidative stress, making mothers’ beta cells healthier. Serotonin also induces the proliferation of beta cells, thereby increasing the beta cell mass and helping maintain proper glucose levels.
The research team conducted follow-up examinations on a total of 174 postpartum women, 85 lactated and 99 non-lactated, at two months postpartum and annually thereafter for at least three years. The results demonstrated that mothers who had undergone lactation improved pancreatic beta cell mass and function, and showed improved glucose homeostasis with approximately 20mg/dL lower glucose levels, thereby reducing the risk of postpartum diabetes in women. Surprisingly, this beneficial effect was maintained after the cessation of lactation, for more than three years after delivery.
Professor Kim said, “We are happy to prove that lactation benefits female metabolic health by improving beta cell mass and function as well as glycemic control.”
“Our future studies on the modulation of the molecular serotonergic pathway in accordance with the management of maternal metabolic risk factors may lead to new therapeutics to help prevent mothers from developing metabolic disorders,” he added.
This work was supported by grants from the National Research Foundation (NRF) and the National Research Council of Science and Technology (NST) of Korea, the National Institutes of Health (NIH), the Larry L. Hillblom Foundation, and the Health Fellowship Foundation.
Image credit: Professor Hail Kim, KAIST
Image usage restrictions: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Publication:
Moon, J. H et al. (2020) ‘Lactation improves pancreatic β cell mass and function through serotonin production.’ Science Translational Medicine, 12, eaay0455. Available online at https://doi.org/10.1126/scitranslmed.aay0455
Profile: Hail Kim, MD, PhD
hailkim@kaist.edu
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Profile: Hak Chul Jang, MD, PhD
janghak@snu.ac.kr
Professor
Division of Endocrinology and Metabolism
Seoul National University Bundang Hospital (SNUBH)
President
Korean Diabetes Association
Profile: Joon Ho Moon, MD, PhD
moonjoonho@gmail.com
Clinical Fellow
Division of Endocrinology and Metabolism
SNUBH
Profile: Hyeongseok Kim, MD, PhD
hskim85kor@gmail.com
Assistant Professor
Chungnam National University (CNU)
Profile: Professor Michael S. German, MD
Michael.German@ucsf.edu
Professor
Diabetes Center
University of California, San Francisco (UCSF)
(END)
2020.04.29 View 19667
Breastfeeding Helps Prevent Mothers from Developing Diabetes after Childbirth
A team of South Korean researchers found that lactation can lower the incidence and reduce the risk of maternal postpartum diabetes. The researchers identified that lactation increases the mass and function of pancreatic beta cells through serotonin production. The team suggested that sustained improvements in pancreatic beta cells, which can last for years even after the cessation of lactation, improve mothers’ metabolic health in addition to providing health benefits for infants.
Pregnancy imposes a substantial metabolic burden on women through weight gain and increased insulin resistance. Various other factors, including a history of gestational diabetes, maternal age, and obesity, further affect women’s risk of progressing to diabetes after delivery, and the risk of postpartum diabetes increases more in women who have had gestational diabetes and/or repeated deliveries.
Diabetes-related complications include damage to blood vessels, which can lead to cardiovascular and cerebrovascular diseases such as heart attack and stroke, and problems with the nerves, eyes, kidneys, and many more. Since diabetes can pose a serious threat to mothers’ metabolic health, the management of maternal metabolic risk factors is important, especially in the peripartum period. Previous epidemiological studies have reported that lactation reduces the risk of postpartum diabetes, but the mechanisms underlying this benefit have remained elusive.
The study, published in Science Translational Medicine on April 29, explains the biology underpinning this observation on the beneficial effects of lactation. Professor Hail Kim from the Graduate School of Medical Science and Engineering at KAIST led and jointly conducted the study in conjunction with researchers from the Seoul National University Bundang Hospital (SNUBH) and Chungnam National University (CNU) in Korea, and the University of California, San Francisco (UCSF) in the US.
In their study, the team observed that the milk-secreting hormone ‘prolactin’ in lactating mothers not only promotes milk production, but also plays a major role in stimulating insulin-secreting pancreatic beta cells that regulate blood glucose in the body.
The researchers also found that ‘serotonin’, known as a chemical that contributes to wellbeing and happiness, is produced in pancreatic beta cells during lactation. Serotonin in pancreatic beta cells act as an antioxidant and reduce oxidative stress, making mothers’ beta cells healthier. Serotonin also induces the proliferation of beta cells, thereby increasing the beta cell mass and helping maintain proper glucose levels.
The research team conducted follow-up examinations on a total of 174 postpartum women, 85 lactated and 99 non-lactated, at two months postpartum and annually thereafter for at least three years. The results demonstrated that mothers who had undergone lactation improved pancreatic beta cell mass and function, and showed improved glucose homeostasis with approximately 20mg/dL lower glucose levels, thereby reducing the risk of postpartum diabetes in women. Surprisingly, this beneficial effect was maintained after the cessation of lactation, for more than three years after delivery.
Professor Kim said, “We are happy to prove that lactation benefits female metabolic health by improving beta cell mass and function as well as glycemic control.”
“Our future studies on the modulation of the molecular serotonergic pathway in accordance with the management of maternal metabolic risk factors may lead to new therapeutics to help prevent mothers from developing metabolic disorders,” he added.
This work was supported by grants from the National Research Foundation (NRF) and the National Research Council of Science and Technology (NST) of Korea, the National Institutes of Health (NIH), the Larry L. Hillblom Foundation, and the Health Fellowship Foundation.
Image credit: Professor Hail Kim, KAIST
Image usage restrictions: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Publication:
Moon, J. H et al. (2020) ‘Lactation improves pancreatic β cell mass and function through serotonin production.’ Science Translational Medicine, 12, eaay0455. Available online at https://doi.org/10.1126/scitranslmed.aay0455
Profile: Hail Kim, MD, PhD
hailkim@kaist.edu
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Profile: Hak Chul Jang, MD, PhD
janghak@snu.ac.kr
Professor
Division of Endocrinology and Metabolism
Seoul National University Bundang Hospital (SNUBH)
President
Korean Diabetes Association
Profile: Joon Ho Moon, MD, PhD
moonjoonho@gmail.com
Clinical Fellow
Division of Endocrinology and Metabolism
SNUBH
Profile: Hyeongseok Kim, MD, PhD
hskim85kor@gmail.com
Assistant Professor
Chungnam National University (CNU)
Profile: Professor Michael S. German, MD
Michael.German@ucsf.edu
Professor
Diabetes Center
University of California, San Francisco (UCSF)
(END)
2020.04.29 View 19667 -
 Professor Jong Chul Ye Appointed as Distinguished Lecturer of IEEE EMBS
Professor Jong Chul Ye from the Department of Bio and Brain Engineering was appointed as a distinguished lecturer by the International Association of Electrical and Electronic Engineers (IEEE) Engineering in Medicine and Biology Society (EMBS). Professor Ye was invited to deliver a lecture on his leading research on artificial intelligence (AI) technology in medical video restoration. He will serve a term of two years beginning in 2020.
IEEE EMBS's distinguished lecturer program is designed to educate researchers around the world on the latest trends and technology in biomedical engineering. Sponsored by IEEE, its members can attend lectures on the distinguished professor's research subject.
Professor Ye said, "We are at a time where the importance of AI in medical imaging is increasing.” He added, “I am proud to be appointed as a distinguished lecturer of the IEEE EMBS in recognition of my contributions to this field.”
(END)
2020.02.27 View 11031
Professor Jong Chul Ye Appointed as Distinguished Lecturer of IEEE EMBS
Professor Jong Chul Ye from the Department of Bio and Brain Engineering was appointed as a distinguished lecturer by the International Association of Electrical and Electronic Engineers (IEEE) Engineering in Medicine and Biology Society (EMBS). Professor Ye was invited to deliver a lecture on his leading research on artificial intelligence (AI) technology in medical video restoration. He will serve a term of two years beginning in 2020.
IEEE EMBS's distinguished lecturer program is designed to educate researchers around the world on the latest trends and technology in biomedical engineering. Sponsored by IEEE, its members can attend lectures on the distinguished professor's research subject.
Professor Ye said, "We are at a time where the importance of AI in medical imaging is increasing.” He added, “I am proud to be appointed as a distinguished lecturer of the IEEE EMBS in recognition of my contributions to this field.”
(END)
2020.02.27 View 11031 -
 Professor Youngseok Ju Awarded the 13th ASAN Award for Young Medical Scientists
Professor Youngseok Ju from the Graduate School of Medical Science and Engineering was selected for the 13th ASAN Award for Young Medical Scientists under the age of 40. Professor Ju will receive 50 million won in prize money.
The ASAN Foundation established this Award in 2007 to encourage young medical scientists who accomplished outstanding achievements in basic and clinical medicine.
The winners are chosen based on a comprehensive assessment of consistency and originality, domestic and international impact, and contributions to medical development and fostering future generations.
Professor Ju is known for having identified the generation principle of cancer genome mutations. In particular, he is recognized for his contributions to the development of cancer prevention, diagnosis, and treatment, by having proven that some cases of lung cancer can occur from destructive changes in chromosomes in lung cells regardless of smoking.
The award ceremony will be held on March 19 in Seoul. The other award will be given to Professor Yong-Ho Lee from the Yonsei University College of Medicine.
2020.01.31 View 4574
Professor Youngseok Ju Awarded the 13th ASAN Award for Young Medical Scientists
Professor Youngseok Ju from the Graduate School of Medical Science and Engineering was selected for the 13th ASAN Award for Young Medical Scientists under the age of 40. Professor Ju will receive 50 million won in prize money.
The ASAN Foundation established this Award in 2007 to encourage young medical scientists who accomplished outstanding achievements in basic and clinical medicine.
The winners are chosen based on a comprehensive assessment of consistency and originality, domestic and international impact, and contributions to medical development and fostering future generations.
Professor Ju is known for having identified the generation principle of cancer genome mutations. In particular, he is recognized for his contributions to the development of cancer prevention, diagnosis, and treatment, by having proven that some cases of lung cancer can occur from destructive changes in chromosomes in lung cells regardless of smoking.
The award ceremony will be held on March 19 in Seoul. The other award will be given to Professor Yong-Ho Lee from the Yonsei University College of Medicine.
2020.01.31 View 4574 -
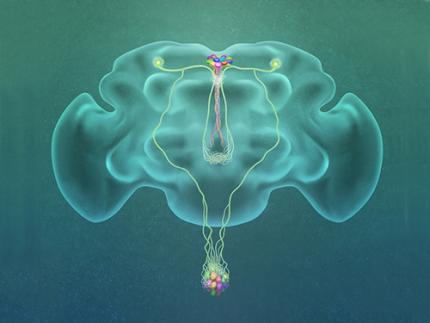 A Single, Master Switch for Sugar Levels?
When a fly eats sugar, a single brain cell sends simultaneous messages to stimulate one hormone and inhibit another to control glucose levels in the body. Further research into this control system with remarkable precision could shed light on the neural mechanisms of diabetes and obesity in humans .
A single neuron appears to monitor and control sugar levels in the fly body, according to research published this week in Nature. This new insight into the mechanisms in the fly brain that maintain a balance of two key hormones controlling glucose levels, insulin and glucagon, can provide a framework for understanding diabetes and obesity in humans.
Neurons that sense and respond to glucose were identified more than 50 years ago, but what they do in our body has remained unclear. Researchers at the Korea Advanced Institute of Science and Technology (KAIST) and New York University School of Medicine have now found a single “glucose-sensing neuron” that appears to be the master controller in Drosophila, the vinegar fly, for maintaining an ideal glucose balance, called homeostasis.
Professor Greg Seong-Bae Suh, Dr. Yangkyun Oh and colleagues identified a key neuron that is excited by glucose, which they called CN neuron. This CN neuron has a unique shape – it has an axon (which is used to transmit information to downstream cells) that is bifurcated. One branch projects to insulin-producing cells, and sends a signal triggering the secretion of the insulin equivalent in flies. The other branch projects to glucagon-producing cells and sends a signal inhibiting the secretion of the glucagon equivalent.
When flies consume food, the levels of glucose in their body increase; this excites the CN neuron, which fires the simultaneous signals to stimulate insulin and inhibit glucagon secretion, thereby maintaining the appropriate balance between the hormones and sugar in the blood. The researchers were able to see this happening in the brain in real time by using a combination of cutting-edge fluorescent calcium imaging technology, as well as measuring hormone and sugar levels and applying highly sophisticated molecular genetic techniques.
When flies were not fed, however, the researchers observed a reduction in the activity of CN neuron, a reduction in insulin secretion and an increase in glucagon secretion. These findings indicate that these key hormones are under the direct control of the glucose-sensing neuron. Furthermore, when they silenced the CN neuron rendering dysfunctional CN neuron in flies, these animals experienced an imbalance, resulting in hyperglycemia – high levels of sugars in the blood, similar to what is observed in diabetes in humans. This further suggests that the CN neuron is critical to maintaining glucose homeostasis in animals.
While further research is required to investigate this process in humans, Suh notes this is a significant step forward in the fields of both neurobiology and endocrinology.
“This work lays the foundation for translational research to better understand how this delicate regulatory process is affected by diabetes, obesity, excessive nutrition and diets high in sugar,” Suh said.
Profile: Greg Seong-Bae Suh
seongbaesuh@kaist.ac.kr
Professor Department of Biological Sciences
KAIST
(Figure: A single glucose-excited CN neuron extends bifurcated axonal branches,
one of which innervates insulin producing cells and stimulates their activity an the other axonal branch projects to glucagon producing cells and inhibits their activity.)
2019.10.24 View 17847
A Single, Master Switch for Sugar Levels?
When a fly eats sugar, a single brain cell sends simultaneous messages to stimulate one hormone and inhibit another to control glucose levels in the body. Further research into this control system with remarkable precision could shed light on the neural mechanisms of diabetes and obesity in humans .
A single neuron appears to monitor and control sugar levels in the fly body, according to research published this week in Nature. This new insight into the mechanisms in the fly brain that maintain a balance of two key hormones controlling glucose levels, insulin and glucagon, can provide a framework for understanding diabetes and obesity in humans.
Neurons that sense and respond to glucose were identified more than 50 years ago, but what they do in our body has remained unclear. Researchers at the Korea Advanced Institute of Science and Technology (KAIST) and New York University School of Medicine have now found a single “glucose-sensing neuron” that appears to be the master controller in Drosophila, the vinegar fly, for maintaining an ideal glucose balance, called homeostasis.
Professor Greg Seong-Bae Suh, Dr. Yangkyun Oh and colleagues identified a key neuron that is excited by glucose, which they called CN neuron. This CN neuron has a unique shape – it has an axon (which is used to transmit information to downstream cells) that is bifurcated. One branch projects to insulin-producing cells, and sends a signal triggering the secretion of the insulin equivalent in flies. The other branch projects to glucagon-producing cells and sends a signal inhibiting the secretion of the glucagon equivalent.
When flies consume food, the levels of glucose in their body increase; this excites the CN neuron, which fires the simultaneous signals to stimulate insulin and inhibit glucagon secretion, thereby maintaining the appropriate balance between the hormones and sugar in the blood. The researchers were able to see this happening in the brain in real time by using a combination of cutting-edge fluorescent calcium imaging technology, as well as measuring hormone and sugar levels and applying highly sophisticated molecular genetic techniques.
When flies were not fed, however, the researchers observed a reduction in the activity of CN neuron, a reduction in insulin secretion and an increase in glucagon secretion. These findings indicate that these key hormones are under the direct control of the glucose-sensing neuron. Furthermore, when they silenced the CN neuron rendering dysfunctional CN neuron in flies, these animals experienced an imbalance, resulting in hyperglycemia – high levels of sugars in the blood, similar to what is observed in diabetes in humans. This further suggests that the CN neuron is critical to maintaining glucose homeostasis in animals.
While further research is required to investigate this process in humans, Suh notes this is a significant step forward in the fields of both neurobiology and endocrinology.
“This work lays the foundation for translational research to better understand how this delicate regulatory process is affected by diabetes, obesity, excessive nutrition and diets high in sugar,” Suh said.
Profile: Greg Seong-Bae Suh
seongbaesuh@kaist.ac.kr
Professor Department of Biological Sciences
KAIST
(Figure: A single glucose-excited CN neuron extends bifurcated axonal branches,
one of which innervates insulin producing cells and stimulates their activity an the other axonal branch projects to glucagon producing cells and inhibits their activity.)
2019.10.24 View 17847 -
 Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 37733
Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 37733 -
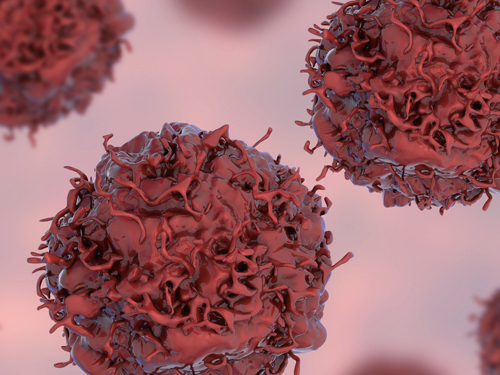 Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke
(Professor Young Seok Ju at the Graduate School of Medical Science and Engineering)
Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancers develop.
Researchers at KAIST, Seoul National University and their collaborators confirmed that gene fusions in non-smokers mostly occur early on, sometimes as early as childhood or adolescence, and on average about three decades before cancer is diagnosed. The study showed that these mutant lung cells, harboring oncogenic seeds, remain dormant for several decades until a number of further mutations accumulate sufficiently for progression into cancer. This is the first study to reveal the landscape of genome structural variations in lung adenocarcinoma.
Lung cancer is the leading cause of cancer-related deaths worldwide, and lung adenocarcinoma is its most common type. Most lung adenocarcinomas are associated with chronic smoking, but about a fourth develop in non-smokers. Precisely what happens in non-smokers for this cancer to develop is not clearly understood.
Researchers analyzed the genomes of 138 lung adenocarcinoma patients, including smokers and non-smokers, with whole-genome sequencing technologies. They explored DNA damage that induced neoplastic transformation.
Lung adenocarcinomas that originated from chronic smoking, referred to as signature 4-high (S4-high) cancers in the study, showed several distinguishing features compared to smoking-unrelated cancers (S4-low).
People in the S4-high group were largely older, men and had more frequent mutations in a cancer-related gene called KRAS. Cancer genomes in the S4-high group were hypermutated with simple mutational classes, such as the substitution, insertion, or deletion of a single base, the building block of DNA.
But the story was very different in the S4-low group. Generally, mutational profiles in this group were much more silent than the S4-high group. However, all cancer-related gene fusions, which are abnormally activated from the merging of two originally separate genes, were exclusively observed in the S4-low group.
The patterns of genomic structural changes underlying gene fusions suggest that about three in four cases of gene fusions emerged from a single cellular crisis causing massive genomic fragmentation and subsequent imprecise repair in normal lung epithelium.
Most strikingly, these major genomic rearrangements, which led to the development of lung adenocarcinoma, are very likely to be acquired decades before cancer diagnosis. The researchers used genomic archaeology techniques to trace the timing of when the catastrophes took place.
Researchers started this study seven years ago when they discovered the expression of the KIF5B-RET gene fusion in lung adenocarcinoma for the first time. Professor Young-Seok Ju, co-lead author from the Graduate School of Medical Science and Engineering at KAIST says, “It is remarkable that oncogenesis can begin by a massive shattering of chromosomes early in life. Our study immediately raises a new question: What induces the mutational catastrophe in our normal lung epithelium.”
Professor Young Tae Kim, co-lead author from Seoul National University says, “We hope this work will help us get one step closer to precision medicine for lung cancer patients.”
The research team plans to further focus on the molecular mechanisms that stimulate complex rearrangements in the body, through screening the genomic structures of fusion genes in other cancer types.
This study was supported by the National Research Foundation of Korea (NRF), Korea Health Industry Development Institute (KHIDI), Suh Kyungbae Foundation, the College of Medicine Research Foundations at Seoul National University and others.
Figure.
(Smoking-unrelated oncogenesis of lung cancers by gene fusions)
Publication.
Jake June-Koo Lee, Seongyeol Park et al., Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma
Cell 177, June 13 2019, online publication ahead of print at May 30, 2019
https://doi.org/10.1016/j.cell.2019.05.013
Profile: Prof Young Seok Ju, MD, PhD
ysju@kaist.ac.kr
http://julab.kaist.ac.kr
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon 34141, Korea
Profile: Prof Young Tae Kim, MD, PhD
ytkim@snu.ac.kr
Professor
Seoul National University Cancer Research Institute
Department of Thoracic and Cardiovascular Surgery
Seoul National University Hospital Seoul 03080, Korea
2019.05.31 View 57123
Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke
(Professor Young Seok Ju at the Graduate School of Medical Science and Engineering)
Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancers develop.
Researchers at KAIST, Seoul National University and their collaborators confirmed that gene fusions in non-smokers mostly occur early on, sometimes as early as childhood or adolescence, and on average about three decades before cancer is diagnosed. The study showed that these mutant lung cells, harboring oncogenic seeds, remain dormant for several decades until a number of further mutations accumulate sufficiently for progression into cancer. This is the first study to reveal the landscape of genome structural variations in lung adenocarcinoma.
Lung cancer is the leading cause of cancer-related deaths worldwide, and lung adenocarcinoma is its most common type. Most lung adenocarcinomas are associated with chronic smoking, but about a fourth develop in non-smokers. Precisely what happens in non-smokers for this cancer to develop is not clearly understood.
Researchers analyzed the genomes of 138 lung adenocarcinoma patients, including smokers and non-smokers, with whole-genome sequencing technologies. They explored DNA damage that induced neoplastic transformation.
Lung adenocarcinomas that originated from chronic smoking, referred to as signature 4-high (S4-high) cancers in the study, showed several distinguishing features compared to smoking-unrelated cancers (S4-low).
People in the S4-high group were largely older, men and had more frequent mutations in a cancer-related gene called KRAS. Cancer genomes in the S4-high group were hypermutated with simple mutational classes, such as the substitution, insertion, or deletion of a single base, the building block of DNA.
But the story was very different in the S4-low group. Generally, mutational profiles in this group were much more silent than the S4-high group. However, all cancer-related gene fusions, which are abnormally activated from the merging of two originally separate genes, were exclusively observed in the S4-low group.
The patterns of genomic structural changes underlying gene fusions suggest that about three in four cases of gene fusions emerged from a single cellular crisis causing massive genomic fragmentation and subsequent imprecise repair in normal lung epithelium.
Most strikingly, these major genomic rearrangements, which led to the development of lung adenocarcinoma, are very likely to be acquired decades before cancer diagnosis. The researchers used genomic archaeology techniques to trace the timing of when the catastrophes took place.
Researchers started this study seven years ago when they discovered the expression of the KIF5B-RET gene fusion in lung adenocarcinoma for the first time. Professor Young-Seok Ju, co-lead author from the Graduate School of Medical Science and Engineering at KAIST says, “It is remarkable that oncogenesis can begin by a massive shattering of chromosomes early in life. Our study immediately raises a new question: What induces the mutational catastrophe in our normal lung epithelium.”
Professor Young Tae Kim, co-lead author from Seoul National University says, “We hope this work will help us get one step closer to precision medicine for lung cancer patients.”
The research team plans to further focus on the molecular mechanisms that stimulate complex rearrangements in the body, through screening the genomic structures of fusion genes in other cancer types.
This study was supported by the National Research Foundation of Korea (NRF), Korea Health Industry Development Institute (KHIDI), Suh Kyungbae Foundation, the College of Medicine Research Foundations at Seoul National University and others.
Figure.
(Smoking-unrelated oncogenesis of lung cancers by gene fusions)
Publication.
Jake June-Koo Lee, Seongyeol Park et al., Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma
Cell 177, June 13 2019, online publication ahead of print at May 30, 2019
https://doi.org/10.1016/j.cell.2019.05.013
Profile: Prof Young Seok Ju, MD, PhD
ysju@kaist.ac.kr
http://julab.kaist.ac.kr
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon 34141, Korea
Profile: Prof Young Tae Kim, MD, PhD
ytkim@snu.ac.kr
Professor
Seoul National University Cancer Research Institute
Department of Thoracic and Cardiovascular Surgery
Seoul National University Hospital Seoul 03080, Korea
2019.05.31 View 57123 -
 Professor Gou Young Koh, 2018 Laureate of Ho-Am Prize
Distinguished Professor Gou Young Koh from the Graduate School of Medical Science and Engineering was appointed a 2018 laureate in medicine of the Ho-Am Prize by the Ho-Am Foundation. Professor Koh is a renowned expert in the field of tumor angiogenesis by exploring the hidden nature of capillary and lymphatic vessels in human organs.
He was recognized for demonstrating the effective reduction of tumor progression and metastasis via tumor vessel normalization. This counterintuitive study result is regarded as a stepping stone for a drug discovery to prevent microvascular diseases.
Besides Professor Koh, Professor Hee Oh from Yale University (Science), Professor Nam-Gyu Park from Sungkyunkwan University (Engineering), Opera Singer Kwangchul Youn (The Arts) and Sister Carla Kang (Community Service) received awards.
The Ho-Am Prize is presented to individuals who have contributed to academics, the arts, and social development, or furthered the welfare of humanity, and commemorates the noble spirit of public service espoused by the late Chairman Byung-chull Lee, who used the pen name Ho-Am.
It was established in 1990 by Kun-Hee Lee, the chairman of Samsung. Awards have been presented to 143 individuals worth a total of 24.4 billion KRW.
2018.04.11 View 8679
Professor Gou Young Koh, 2018 Laureate of Ho-Am Prize
Distinguished Professor Gou Young Koh from the Graduate School of Medical Science and Engineering was appointed a 2018 laureate in medicine of the Ho-Am Prize by the Ho-Am Foundation. Professor Koh is a renowned expert in the field of tumor angiogenesis by exploring the hidden nature of capillary and lymphatic vessels in human organs.
He was recognized for demonstrating the effective reduction of tumor progression and metastasis via tumor vessel normalization. This counterintuitive study result is regarded as a stepping stone for a drug discovery to prevent microvascular diseases.
Besides Professor Koh, Professor Hee Oh from Yale University (Science), Professor Nam-Gyu Park from Sungkyunkwan University (Engineering), Opera Singer Kwangchul Youn (The Arts) and Sister Carla Kang (Community Service) received awards.
The Ho-Am Prize is presented to individuals who have contributed to academics, the arts, and social development, or furthered the welfare of humanity, and commemorates the noble spirit of public service espoused by the late Chairman Byung-chull Lee, who used the pen name Ho-Am.
It was established in 1990 by Kun-Hee Lee, the chairman of Samsung. Awards have been presented to 143 individuals worth a total of 24.4 billion KRW.
2018.04.11 View 8679 -
 Two Professors Receive the Asan Medical Award
(Professor Ho Min Kim and Chair Profesor Eunjoon Kim (from far right)
Chair Professor Eunjoon Kim of the Department of Biological Sciences and Professor Ho Min Kim from the Graduate School of Medical Science & Engineering won the 11th Asan Medical Award in the areas of basic medicine and young medical scholar on March 21.
The Asan Medical Award has been recognizing the most distinguished scholars in the areas of basic and clinical medicines annually since 2007.
Chair Professor Kim won the 300 million KRW award in recognition of his research in the mechanism of synaptic brain dysfunction and its relation with neural diseases.
The young medical scholar’s award recognizes a promising scholar under the age of 40. Professor Kim won the award for identifying the key protein structure and molecular mechanism controlling immunocytes and neurons. He earned a 50 million KRW prize.
2018.03.26 View 9479
Two Professors Receive the Asan Medical Award
(Professor Ho Min Kim and Chair Profesor Eunjoon Kim (from far right)
Chair Professor Eunjoon Kim of the Department of Biological Sciences and Professor Ho Min Kim from the Graduate School of Medical Science & Engineering won the 11th Asan Medical Award in the areas of basic medicine and young medical scholar on March 21.
The Asan Medical Award has been recognizing the most distinguished scholars in the areas of basic and clinical medicines annually since 2007.
Chair Professor Kim won the 300 million KRW award in recognition of his research in the mechanism of synaptic brain dysfunction and its relation with neural diseases.
The young medical scholar’s award recognizes a promising scholar under the age of 40. Professor Kim won the award for identifying the key protein structure and molecular mechanism controlling immunocytes and neurons. He earned a 50 million KRW prize.
2018.03.26 View 9479