Immunotherapy
-
 KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences>
A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for developing innovative therapeutic technologies.
On the 22nd of January, KAIST (represented by President Kwang Hyung Lee) announced that the joint research team led by Professors Seyun Kim, Gwangrog Lee, and Won-Ki Cho from the Department of Biological Sciences had uncovered essential mechanisms controlling gene expression in animal cells.
Inositol phosphate metabolites produced by inositol metabolism enzymes serve as vital secondary messengers in eukaryotic cell signaling systems and are broadly implicated in cancer, obesity, diabetes, and neurological disorders.
The research team demonstrated that the inositol polyphosphate multikinase (IPMK) enzyme, a key player in the inositol metabolism system, acts as a critical transcriptional activator within the core gene expression networks of animal cells. Notably, although IPMK was previously reported to play an important role in the transcription process governed by serum response factor (SRF), a representative transcription factor in animal cells, the precise mechanism of its action was unclear.
SRF is a transcription factor directly controlling the expression of at least 200–300 genes, regulating cell growth, proliferation, apoptosis, and motility, and is indispensable for organ development, such as in the heart.
The team discovered that IPMK binds directly to SRF, altering the three-dimensional structure of the SRF protein. This interaction facilitates the transcriptional activity of various genes through the SRF activated by IPMK, demonstrating that IPMK acts as a critical regulatory switch to enhance SRF's protein activity.
< Figure 1. The serum response factor (SRF) protein, a key transcription factor in animal cells, directly binds to inositol polyphosphate multikinase (IPMK) enzyme and undergoes structural change to acquire DNA binding ability, and precisely regulates growth and differentiation of animal cells through transcriptional activation. >
The team further verified that disruptions in the direct interaction between IPMK and SRF lead to the reduced functionality and activity of SRF, causing severe impairments in gene expression.
By highlighting the significance of the intrinsically disordered region (IDR) in SRF, the researchers underscored the biological importance of intrinsically disordered proteins (IDPs). Unlike most proteins that adopt distinct structures through folding, IDPs, including those with IDRs, do not exhibit specific structures but play crucial biological roles, attracting significant attention in the scientific community.
Professor Seyun Kim commented, "This study provides a vital mechanism proving that IPMK, a key enzyme in the inositol metabolism system, is a major transcriptional activator in the core gene expression network of animal cells. By understanding fundamental processes such as cancer development and metastasis, tissue differentiation from stem cells, and neural activation through SRF, we hope this discovery will lead to the broad application of innovative therapeutic technologies."
The findings were published on January 7th in the international journal Nucleic Acids Research (IF=16.7, top 1.8% in Biochemistry and Molecular Biology), under the title “Single-molecule analysis reveals that IPMK enhances the DNA-binding activity of the transcription factor SRF" (DOI: 10.1093/nar/gkae1281).
This research was supported by the National Research Foundation of Korea's Mid-career Research Program, Leading Research Center Program, and Global Research Laboratory Program, as well as by the Suh Kyungbae Science Foundation and the Samsung Future Technology Development Program.
2025.01.24 View 6357
KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences>
A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for developing innovative therapeutic technologies.
On the 22nd of January, KAIST (represented by President Kwang Hyung Lee) announced that the joint research team led by Professors Seyun Kim, Gwangrog Lee, and Won-Ki Cho from the Department of Biological Sciences had uncovered essential mechanisms controlling gene expression in animal cells.
Inositol phosphate metabolites produced by inositol metabolism enzymes serve as vital secondary messengers in eukaryotic cell signaling systems and are broadly implicated in cancer, obesity, diabetes, and neurological disorders.
The research team demonstrated that the inositol polyphosphate multikinase (IPMK) enzyme, a key player in the inositol metabolism system, acts as a critical transcriptional activator within the core gene expression networks of animal cells. Notably, although IPMK was previously reported to play an important role in the transcription process governed by serum response factor (SRF), a representative transcription factor in animal cells, the precise mechanism of its action was unclear.
SRF is a transcription factor directly controlling the expression of at least 200–300 genes, regulating cell growth, proliferation, apoptosis, and motility, and is indispensable for organ development, such as in the heart.
The team discovered that IPMK binds directly to SRF, altering the three-dimensional structure of the SRF protein. This interaction facilitates the transcriptional activity of various genes through the SRF activated by IPMK, demonstrating that IPMK acts as a critical regulatory switch to enhance SRF's protein activity.
< Figure 1. The serum response factor (SRF) protein, a key transcription factor in animal cells, directly binds to inositol polyphosphate multikinase (IPMK) enzyme and undergoes structural change to acquire DNA binding ability, and precisely regulates growth and differentiation of animal cells through transcriptional activation. >
The team further verified that disruptions in the direct interaction between IPMK and SRF lead to the reduced functionality and activity of SRF, causing severe impairments in gene expression.
By highlighting the significance of the intrinsically disordered region (IDR) in SRF, the researchers underscored the biological importance of intrinsically disordered proteins (IDPs). Unlike most proteins that adopt distinct structures through folding, IDPs, including those with IDRs, do not exhibit specific structures but play crucial biological roles, attracting significant attention in the scientific community.
Professor Seyun Kim commented, "This study provides a vital mechanism proving that IPMK, a key enzyme in the inositol metabolism system, is a major transcriptional activator in the core gene expression network of animal cells. By understanding fundamental processes such as cancer development and metastasis, tissue differentiation from stem cells, and neural activation through SRF, we hope this discovery will lead to the broad application of innovative therapeutic technologies."
The findings were published on January 7th in the international journal Nucleic Acids Research (IF=16.7, top 1.8% in Biochemistry and Molecular Biology), under the title “Single-molecule analysis reveals that IPMK enhances the DNA-binding activity of the transcription factor SRF" (DOI: 10.1093/nar/gkae1281).
This research was supported by the National Research Foundation of Korea's Mid-career Research Program, Leading Research Center Program, and Global Research Laboratory Program, as well as by the Suh Kyungbae Science Foundation and the Samsung Future Technology Development Program.
2025.01.24 View 6357 -
 KAIST Unveils New Possibilities for Treating Intractable Brain Tumors
< Photo 1. (From left) Professor Heung Kyu Lee, KAIST Department of Biological Sciences, and Dr. Keun Bon Ku >
Immunotherapy, which enhances the immune system's T cell response to eliminate cancer cells, has emerged as a key approach in cancer treatment. However, in the case of glioblastoma, an aggressive and treatment-resistant brain tumor, numerous clinical trials have failed to confirm their efficacy. Korean researchers have recently analyzed the mechanisms that cause T cell exhaustion, which is characterized by a loss of function or a weakened response following prolonged exposure to antigens in such intractable cancers, identifying key control factors in T cell activation and clarifying the mechanisms that enhance therapeutic effectiveness.
KAIST (represented by President Kwang Hyung Lee) announced on the 6th of November that Professor Heung Kyu Lee’s team from the Department of Biological Sciences, in collaboration with the Korea Research Institute of Chemical Technology (represented by President Young Kuk Lee), has confirmed improved survival rates in a glioblastoma mouse model. By removing the inhibitory Fc gamma receptor (FcγRIIB), the research team was able to restore the responsiveness of cytotoxic T cells to immune checkpoint inhibitors, leading to enhanced anticancer activity.
The research team examined the effect of FcγRIIB, an inhibitory receptor recently found in cytotoxic T cells, on tumor-infiltrating T cells and the therapeutic effectiveness of the anti-PD-1 immune checkpoint inhibitor.
< Figure 1. Study results on improved survival rate due to increased antitumor activity of anti-PD-1 treatment in inhibitory Fc gamma receptor(Fcgr2b) ablation mice with murine glioblastoma. >
Their findings showed that deleting FcγRIIB induced the increase of tumor antigen-specific memory T cells, which helps to suppress exhaustion, enhances stem-like qualities, and reactivates T cell-mediated antitumor immunity, particularly in response to anti-PD-1 treatment. Furthermore, FcγRIIB deletion led to an increase in antigen-specific memory T cells that maintained continuous infiltration into the tumor tissue.
This study presents a new therapeutic target for tumors unresponsive to immune checkpoint inhibitors and demonstrates that combining FcγRIIB inhibition with anti-PD-1 treatment can produce synergistic effects, potentially improving therapeutic outcomes for tumors like glioblastoma, which typically show resistance to anti-PD-1 therapy.
< Figure 2. Overview of the study on the enhanced response to anti-PD-1 therapy for glioblastoma brain tumors upon deletion of the inhibitory Fc gamma receptor (FcγRIIB) in tumor microenvironment. When the inhibitory Fc gamma receptor (FcγRIIB) of cytotoxic T cells is deleted, an increase in tumor-specific memory T cells (Ttsms) was observed. In addition, this T cell subset is identified as originating from the tumor-draining lymph nodes(TdLNs) and leads to persistent infiltration into the tumor tissue. Anti-PD-1 therapy leads to an increased anti-tumor immune response via Ttsms, which is confirmed by increased tumor cell toxicity and increased cell division and decreased cell de-migration indices. Ultimately, the increased cytotoxic T cell immune response leads to an increase in the survival rate of glioblastoma. >
Professor Heung Kyu Lee explained, "This study offers a way to overcome clinical failures in treating brain tumors with immune checkpoint therapy and opens possibilities for broader applications to other intractable cancers. It also highlights the potential of utilizing cytotoxic T cells for tumor cell therapy."
The study, led by Dr. Keun Bon Ku of KAIST (currently a senior researcher at the Korea Research Institute of Chemical Technology's Center for Infectious Disease Diagnosis and Prevention), along with Chae Won Kim, Yumin Kim, Byeong Hoon Kang, Jeongwoo La, In Kang, Won Hyung Park, Stephen Ahn, and Sung Ki Lee, was published online on October 26 in the Journal for ImmunoTherapy of Cancer, an international journal in tumor immunology and therapy from the Society for Immunotherapy of Cancer. (Paper title: “Inhibitory Fcγ receptor deletion enhances CD8 T cell stemness increasing anti-PD-1 therapy responsiveness against glioblastoma,” http://dx.doi.org/10.1136/jitc-2024-009449).
This research received support from the National Research Foundation of Korea, the Bio & Medical Technology Development Program, and the Samsung Science & Technology Foundation.
2024.11.15 View 2793
KAIST Unveils New Possibilities for Treating Intractable Brain Tumors
< Photo 1. (From left) Professor Heung Kyu Lee, KAIST Department of Biological Sciences, and Dr. Keun Bon Ku >
Immunotherapy, which enhances the immune system's T cell response to eliminate cancer cells, has emerged as a key approach in cancer treatment. However, in the case of glioblastoma, an aggressive and treatment-resistant brain tumor, numerous clinical trials have failed to confirm their efficacy. Korean researchers have recently analyzed the mechanisms that cause T cell exhaustion, which is characterized by a loss of function or a weakened response following prolonged exposure to antigens in such intractable cancers, identifying key control factors in T cell activation and clarifying the mechanisms that enhance therapeutic effectiveness.
KAIST (represented by President Kwang Hyung Lee) announced on the 6th of November that Professor Heung Kyu Lee’s team from the Department of Biological Sciences, in collaboration with the Korea Research Institute of Chemical Technology (represented by President Young Kuk Lee), has confirmed improved survival rates in a glioblastoma mouse model. By removing the inhibitory Fc gamma receptor (FcγRIIB), the research team was able to restore the responsiveness of cytotoxic T cells to immune checkpoint inhibitors, leading to enhanced anticancer activity.
The research team examined the effect of FcγRIIB, an inhibitory receptor recently found in cytotoxic T cells, on tumor-infiltrating T cells and the therapeutic effectiveness of the anti-PD-1 immune checkpoint inhibitor.
< Figure 1. Study results on improved survival rate due to increased antitumor activity of anti-PD-1 treatment in inhibitory Fc gamma receptor(Fcgr2b) ablation mice with murine glioblastoma. >
Their findings showed that deleting FcγRIIB induced the increase of tumor antigen-specific memory T cells, which helps to suppress exhaustion, enhances stem-like qualities, and reactivates T cell-mediated antitumor immunity, particularly in response to anti-PD-1 treatment. Furthermore, FcγRIIB deletion led to an increase in antigen-specific memory T cells that maintained continuous infiltration into the tumor tissue.
This study presents a new therapeutic target for tumors unresponsive to immune checkpoint inhibitors and demonstrates that combining FcγRIIB inhibition with anti-PD-1 treatment can produce synergistic effects, potentially improving therapeutic outcomes for tumors like glioblastoma, which typically show resistance to anti-PD-1 therapy.
< Figure 2. Overview of the study on the enhanced response to anti-PD-1 therapy for glioblastoma brain tumors upon deletion of the inhibitory Fc gamma receptor (FcγRIIB) in tumor microenvironment. When the inhibitory Fc gamma receptor (FcγRIIB) of cytotoxic T cells is deleted, an increase in tumor-specific memory T cells (Ttsms) was observed. In addition, this T cell subset is identified as originating from the tumor-draining lymph nodes(TdLNs) and leads to persistent infiltration into the tumor tissue. Anti-PD-1 therapy leads to an increased anti-tumor immune response via Ttsms, which is confirmed by increased tumor cell toxicity and increased cell division and decreased cell de-migration indices. Ultimately, the increased cytotoxic T cell immune response leads to an increase in the survival rate of glioblastoma. >
Professor Heung Kyu Lee explained, "This study offers a way to overcome clinical failures in treating brain tumors with immune checkpoint therapy and opens possibilities for broader applications to other intractable cancers. It also highlights the potential of utilizing cytotoxic T cells for tumor cell therapy."
The study, led by Dr. Keun Bon Ku of KAIST (currently a senior researcher at the Korea Research Institute of Chemical Technology's Center for Infectious Disease Diagnosis and Prevention), along with Chae Won Kim, Yumin Kim, Byeong Hoon Kang, Jeongwoo La, In Kang, Won Hyung Park, Stephen Ahn, and Sung Ki Lee, was published online on October 26 in the Journal for ImmunoTherapy of Cancer, an international journal in tumor immunology and therapy from the Society for Immunotherapy of Cancer. (Paper title: “Inhibitory Fcγ receptor deletion enhances CD8 T cell stemness increasing anti-PD-1 therapy responsiveness against glioblastoma,” http://dx.doi.org/10.1136/jitc-2024-009449).
This research received support from the National Research Foundation of Korea, the Bio & Medical Technology Development Program, and the Samsung Science & Technology Foundation.
2024.11.15 View 2793 -
 KAIST team develops smart immune system that can pin down on malignant tumors
A joint research team led by Professor Jung Kyoon Choi of the KAIST Department of Bio and Brain Engineering and Professor Jong-Eun Park of the KAIST Graduate School of Medical Science and Engineering (GSMSE) announced the development of the key technologies to treat cancers using smart immune cells designed based on AI and big data analysis. This technology is expected to be a next-generation immunotherapy that allows precision targeting of tumor cells by having the chimeric antigen receptors (CARs) operate through a logical circuit. Professor Hee Jung An of CHA Bundang Medical Center and Professor Hae-Ock Lee of the Catholic University of Korea also participated in this research to contribute joint effort.
Professor Jung Kyoon Choi’s team built a gene expression database from millions of cells, and used this to successfully develop and verify a deep-learning algorithm that could detect the differences in gene expression patterns between tumor cells and normal cells through a logical circuit. CAR immune cells that were fitted with the logic circuits discovered through this methodology could distinguish between tumorous and normal cells as a computer would, and therefore showed potentials to strike only on tumor cells accurately without causing unwanted side effects.
This research, conducted by co-first authors Dr. Joonha Kwon of the KAIST Department of Bio and Brain Engineering and Ph.D. candidate Junho Kang of KAIST GSMSE, was published by Nature Biotechnology on February 16, under the title Single-cell mapping of combinatorial target antigens for CAR switches using logic gates.
An area in cancer research where the most attempts and advances have been made in recent years is immunotherapy. This field of treatment, which utilizes the patient’s own immune system in order to overcome cancer, has several methods including immune checkpoint inhibitors, cancer vaccines and cellular treatments. Immune cells like CAR-T or CAR-NK equipped with chimera antigen receptors, in particular, can recognize cancer antigens and directly destroy cancer cells.
Starting with its success in blood cancer treatment, scientists have been trying to expand the application of CAR cell therapy to treat solid cancer. But there have been difficulties to develop CAR cells with effective killing abilities against solid cancer cells with minimized side effects. Accordingly, in recent years, the development of smarter CAR engineering technologies, i.e., computational logic gates such as AND, OR, and NOT, to effectively target cancer cells has been underway.
At this point in time, the research team built a large-scale database for cancer and normal cells to discover the exact genes that are expressed only from cancer cells at a single-cell level. The team followed this up by developing an AI algorithm that could search for a combination of genes that best distinguishes cancer cells from normal cells. This algorithm, in particular, has been used to find a logic circuit that can specifically target cancer cells through cell-level simulations of all gene combinations. CAR-T cells equipped with logic circuits discovered through this methodology are expected to distinguish cancerous cells from normal cells like computers, thereby minimizing side effects and maximizing the effects of chemotherapy.
Dr. Joonha Kwon, who is the first author of this paper, said, “this research suggests a new method that hasn’t been tried before. What’s particularly noteworthy is the process in which we found the optimal CAR cell circuit through simulations of millions of individual tumors and normal cells.” He added, “This is an innovative technology that can apply AI and computer logic circuits to immune cell engineering. It would contribute greatly to expanding CAR therapy, which is being successfully used for blood cancer, to solid cancers as well.”
This research was funded by the Original Technology Development Project and Research Program for Next Generation Applied Omic of the Korea Research Foundation.
Figure 1. A schematic diagram of manufacturing and administration process of CAR therapy and of cancer cell-specific dual targeting using CAR.
Figure 2. Deep learning (convolutional neural networks, CNNs) algorithm for selection of dual targets based on gene combination (left) and algorithm for calculating expressing cell fractions by gene combination according to logical circuit (right).
2023.03.09 View 8394
KAIST team develops smart immune system that can pin down on malignant tumors
A joint research team led by Professor Jung Kyoon Choi of the KAIST Department of Bio and Brain Engineering and Professor Jong-Eun Park of the KAIST Graduate School of Medical Science and Engineering (GSMSE) announced the development of the key technologies to treat cancers using smart immune cells designed based on AI and big data analysis. This technology is expected to be a next-generation immunotherapy that allows precision targeting of tumor cells by having the chimeric antigen receptors (CARs) operate through a logical circuit. Professor Hee Jung An of CHA Bundang Medical Center and Professor Hae-Ock Lee of the Catholic University of Korea also participated in this research to contribute joint effort.
Professor Jung Kyoon Choi’s team built a gene expression database from millions of cells, and used this to successfully develop and verify a deep-learning algorithm that could detect the differences in gene expression patterns between tumor cells and normal cells through a logical circuit. CAR immune cells that were fitted with the logic circuits discovered through this methodology could distinguish between tumorous and normal cells as a computer would, and therefore showed potentials to strike only on tumor cells accurately without causing unwanted side effects.
This research, conducted by co-first authors Dr. Joonha Kwon of the KAIST Department of Bio and Brain Engineering and Ph.D. candidate Junho Kang of KAIST GSMSE, was published by Nature Biotechnology on February 16, under the title Single-cell mapping of combinatorial target antigens for CAR switches using logic gates.
An area in cancer research where the most attempts and advances have been made in recent years is immunotherapy. This field of treatment, which utilizes the patient’s own immune system in order to overcome cancer, has several methods including immune checkpoint inhibitors, cancer vaccines and cellular treatments. Immune cells like CAR-T or CAR-NK equipped with chimera antigen receptors, in particular, can recognize cancer antigens and directly destroy cancer cells.
Starting with its success in blood cancer treatment, scientists have been trying to expand the application of CAR cell therapy to treat solid cancer. But there have been difficulties to develop CAR cells with effective killing abilities against solid cancer cells with minimized side effects. Accordingly, in recent years, the development of smarter CAR engineering technologies, i.e., computational logic gates such as AND, OR, and NOT, to effectively target cancer cells has been underway.
At this point in time, the research team built a large-scale database for cancer and normal cells to discover the exact genes that are expressed only from cancer cells at a single-cell level. The team followed this up by developing an AI algorithm that could search for a combination of genes that best distinguishes cancer cells from normal cells. This algorithm, in particular, has been used to find a logic circuit that can specifically target cancer cells through cell-level simulations of all gene combinations. CAR-T cells equipped with logic circuits discovered through this methodology are expected to distinguish cancerous cells from normal cells like computers, thereby minimizing side effects and maximizing the effects of chemotherapy.
Dr. Joonha Kwon, who is the first author of this paper, said, “this research suggests a new method that hasn’t been tried before. What’s particularly noteworthy is the process in which we found the optimal CAR cell circuit through simulations of millions of individual tumors and normal cells.” He added, “This is an innovative technology that can apply AI and computer logic circuits to immune cell engineering. It would contribute greatly to expanding CAR therapy, which is being successfully used for blood cancer, to solid cancers as well.”
This research was funded by the Original Technology Development Project and Research Program for Next Generation Applied Omic of the Korea Research Foundation.
Figure 1. A schematic diagram of manufacturing and administration process of CAR therapy and of cancer cell-specific dual targeting using CAR.
Figure 2. Deep learning (convolutional neural networks, CNNs) algorithm for selection of dual targets based on gene combination (left) and algorithm for calculating expressing cell fractions by gene combination according to logical circuit (right).
2023.03.09 View 8394 -
 Deep-Learning and 3D Holographic Microscopy Beats Scientists at Analyzing Cancer Immunotherapy
Live tracking and analyzing of the dynamics of chimeric antigen receptor (CAR) T-cells targeting cancer cells can open new avenues for the development of cancer immunotherapy. However, imaging via conventional microscopy approaches can result in cellular damage, and assessments of cell-to-cell interactions are extremely difficult and labor-intensive. When researchers applied deep learning and 3D holographic microscopy to the task, however, they not only avoided these difficultues but found that AI was better at it than humans were.
Artificial intelligence (AI) is helping researchers decipher images from a new holographic microscopy technique needed to investigate a key process in cancer immunotherapy “live” as it takes place. The AI transformed work that, if performed manually by scientists, would otherwise be incredibly labor-intensive and time-consuming into one that is not only effortless but done better than they could have done it themselves. The research, conducted by the team of Professor YongKeun Park from the Department of Physics, appeared in the journal eLife last December.
A critical stage in the development of the human immune system’s ability to respond not just generally to any invader (such as pathogens or cancer cells) but specifically to that particular type of invader and remember it should it attempt to invade again is the formation of a junction between an immune cell called a T-cell and a cell that presents the antigen, or part of the invader that is causing the problem, to it. This process is like when a picture of a suspect is sent to a police car so that the officers can recognize the criminal they are trying to track down. The junction between the two cells, called the immunological synapse, or IS, is the key process in teaching the immune system how to recognize a specific type of invader.
Since the formation of the IS junction is such a critical step for the initiation of an antigen-specific immune response, various techniques allowing researchers to observe the process as it happens have been used to study its dynamics. Most of these live imaging techniques rely on fluorescence microscopy, where genetic tweaking causes part of a protein from a cell to fluoresce, in turn allowing the subject to be tracked via fluorescence rather than via the reflected light used in many conventional microscopy techniques.
However, fluorescence-based imaging can suffer from effects such as photo-bleaching and photo-toxicity, preventing the assessment of dynamic changes in the IS junction process over the long term. Fluorescence-based imaging still involves illumination, whereupon the fluorophores (chemical compounds that cause the fluorescence) emit light of a different color. Photo-bleaching or photo-toxicity occur when the subject is exposed to too much illumination, resulting in chemical alteration or cellular damage.
One recent option that does away with fluorescent labelling and thereby avoids such problems is 3D holographic microscopy or holotomography (HT). In this technique, the refractive index (the way that light changes direction when encountering a substance with a different density—why a straw looks like it bends in a glass of water) is recorded in 3D as a hologram.
Until now, HT has been used to study single cells, but never cell-cell interactions involved in immune responses. One of the main reasons is the difficulty of “segmentation,” or distinguishing the different parts of a cell and thus distinguishing between the interacting cells; in other words, deciphering which part belongs to which cell.
Manual segmentation, or marking out the different parts manually, is one option, but it is difficult and time-consuming, especially in three dimensions. To overcome this problem, automatic segmentation has been developed in which simple computer algorithms perform the identification.
“But these basic algorithms often make mistakes,” explained Professor YongKeun Park, “particularly with respect to adjoining segmentation, which of course is exactly what is occurring here in the immune response we’re most interested in.”
So, the researchers applied a deep learning framework to the HT segmentation problem. Deep learning is a type of machine learning in which artificial neural networks based on the human brain recognize patterns in a way that is similar to how humans do this. Regular machine learning requires data as an input that has already been labelled. The AI “learns” by understanding the labeled data and then recognizes the concept that has been labelled when it is fed novel data. For example, AI trained on a thousand images of cats labelled “cat” should be able to recognize a cat the next time it encounters an image with a cat in it. Deep learning involves multiple layers of artificial neural networks attacking much larger, but unlabeled datasets, in which the AI develops its own ‘labels’ for concepts it encounters.
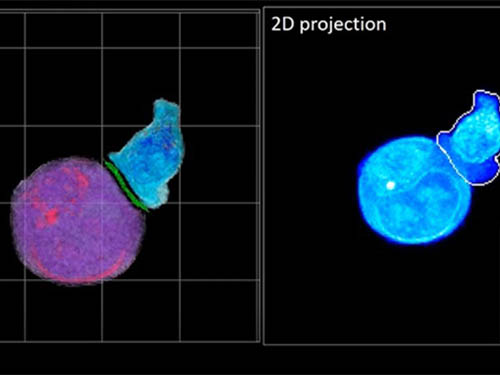
In essence, the deep learning framework that KAIST researchers developed, called DeepIS, came up with its own concepts by which it distinguishes the different parts of the IS junction process. To validate this method, the research team applied it to the dynamics of a particular IS junction formed between chimeric antigen receptor (CAR) T-cells and target cancer cells. They then compared the results to what they would normally have done: the laborious process of performing the segmentation manually. They found not only that DeepIS was able to define areas within the IS with high accuracy, but that the technique was even able to capture information about the total distribution of proteins within the IS that may not have been easily measured using conventional techniques.
“In addition to allowing us to avoid the drudgery of manual segmentation and the problems of photo-bleaching and photo-toxicity, we found that the AI actually did a better job,” Professor Park added.
The next step will be to combine the technique with methods of measuring how much physical force is applied by different parts of the IS junction, such as holographic optical tweezers or traction force microscopy.
-Profile
Professor YongKeun Park
Department of Physics
Biomedical Optics Laboratory
http://bmol.kaist.ac.kr
KAIST
2021.02.24 View 11951
Deep-Learning and 3D Holographic Microscopy Beats Scientists at Analyzing Cancer Immunotherapy
Live tracking and analyzing of the dynamics of chimeric antigen receptor (CAR) T-cells targeting cancer cells can open new avenues for the development of cancer immunotherapy. However, imaging via conventional microscopy approaches can result in cellular damage, and assessments of cell-to-cell interactions are extremely difficult and labor-intensive. When researchers applied deep learning and 3D holographic microscopy to the task, however, they not only avoided these difficultues but found that AI was better at it than humans were.
Artificial intelligence (AI) is helping researchers decipher images from a new holographic microscopy technique needed to investigate a key process in cancer immunotherapy “live” as it takes place. The AI transformed work that, if performed manually by scientists, would otherwise be incredibly labor-intensive and time-consuming into one that is not only effortless but done better than they could have done it themselves. The research, conducted by the team of Professor YongKeun Park from the Department of Physics, appeared in the journal eLife last December.
A critical stage in the development of the human immune system’s ability to respond not just generally to any invader (such as pathogens or cancer cells) but specifically to that particular type of invader and remember it should it attempt to invade again is the formation of a junction between an immune cell called a T-cell and a cell that presents the antigen, or part of the invader that is causing the problem, to it. This process is like when a picture of a suspect is sent to a police car so that the officers can recognize the criminal they are trying to track down. The junction between the two cells, called the immunological synapse, or IS, is the key process in teaching the immune system how to recognize a specific type of invader.
Since the formation of the IS junction is such a critical step for the initiation of an antigen-specific immune response, various techniques allowing researchers to observe the process as it happens have been used to study its dynamics. Most of these live imaging techniques rely on fluorescence microscopy, where genetic tweaking causes part of a protein from a cell to fluoresce, in turn allowing the subject to be tracked via fluorescence rather than via the reflected light used in many conventional microscopy techniques.
However, fluorescence-based imaging can suffer from effects such as photo-bleaching and photo-toxicity, preventing the assessment of dynamic changes in the IS junction process over the long term. Fluorescence-based imaging still involves illumination, whereupon the fluorophores (chemical compounds that cause the fluorescence) emit light of a different color. Photo-bleaching or photo-toxicity occur when the subject is exposed to too much illumination, resulting in chemical alteration or cellular damage.
One recent option that does away with fluorescent labelling and thereby avoids such problems is 3D holographic microscopy or holotomography (HT). In this technique, the refractive index (the way that light changes direction when encountering a substance with a different density—why a straw looks like it bends in a glass of water) is recorded in 3D as a hologram.
Until now, HT has been used to study single cells, but never cell-cell interactions involved in immune responses. One of the main reasons is the difficulty of “segmentation,” or distinguishing the different parts of a cell and thus distinguishing between the interacting cells; in other words, deciphering which part belongs to which cell.
Manual segmentation, or marking out the different parts manually, is one option, but it is difficult and time-consuming, especially in three dimensions. To overcome this problem, automatic segmentation has been developed in which simple computer algorithms perform the identification.
“But these basic algorithms often make mistakes,” explained Professor YongKeun Park, “particularly with respect to adjoining segmentation, which of course is exactly what is occurring here in the immune response we’re most interested in.”
So, the researchers applied a deep learning framework to the HT segmentation problem. Deep learning is a type of machine learning in which artificial neural networks based on the human brain recognize patterns in a way that is similar to how humans do this. Regular machine learning requires data as an input that has already been labelled. The AI “learns” by understanding the labeled data and then recognizes the concept that has been labelled when it is fed novel data. For example, AI trained on a thousand images of cats labelled “cat” should be able to recognize a cat the next time it encounters an image with a cat in it. Deep learning involves multiple layers of artificial neural networks attacking much larger, but unlabeled datasets, in which the AI develops its own ‘labels’ for concepts it encounters.
In essence, the deep learning framework that KAIST researchers developed, called DeepIS, came up with its own concepts by which it distinguishes the different parts of the IS junction process. To validate this method, the research team applied it to the dynamics of a particular IS junction formed between chimeric antigen receptor (CAR) T-cells and target cancer cells. They then compared the results to what they would normally have done: the laborious process of performing the segmentation manually. They found not only that DeepIS was able to define areas within the IS with high accuracy, but that the technique was even able to capture information about the total distribution of proteins within the IS that may not have been easily measured using conventional techniques.
“In addition to allowing us to avoid the drudgery of manual segmentation and the problems of photo-bleaching and photo-toxicity, we found that the AI actually did a better job,” Professor Park added.
The next step will be to combine the technique with methods of measuring how much physical force is applied by different parts of the IS junction, such as holographic optical tweezers or traction force microscopy.
-Profile
Professor YongKeun Park
Department of Physics
Biomedical Optics Laboratory
http://bmol.kaist.ac.kr
KAIST
2021.02.24 View 11951