Department+of+Chemical
-
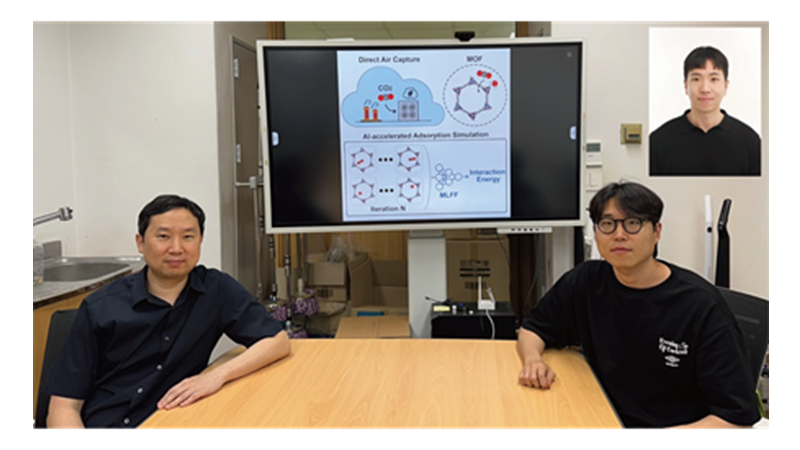 KAIST Develops AI to Easily Find Promising Materials That Capture Only CO₂
< Photo 1. (From left) Professor Jihan Kim, Ph.D. candidate Yunsung Lim and Dr. Hyunsoo Park of the Department of Chemical and Biomolecular Engineering >
In order to help prevent the climate crisis, actively reducing already-emitted CO₂ is essential. Accordingly, direct air capture (DAC) — a technology that directly extracts only CO₂ from the air — is gaining attention. However, effectively capturing pure CO₂ is not easy due to water vapor (H₂O) present in the air. KAIST researchers have successfully used AI-driven machine learning techniques to identify the most promising CO₂-capturing materials among metal-organic frameworks (MOFs), a key class of materials studied for this technology.
KAIST (President Kwang Hyung Lee) announced on the 29th of June that a research team led by Professor Jihan Kim from the Department of Chemical and Biomolecular Engineering, in collaboration with a team at Imperial College London, has developed a machine-learning-based simulation method that can quickly and accurately screen MOFs best suited for atmospheric CO₂ capture.
< Figure 1. Concept diagram of Direct Air Capture (DAC) technology and carbon capture using Metal-Organic Frameworks (MOFs). MOFs are promising porous materials capable of capturing carbon dioxide from the atmosphere, drawing attention as a core material for DAC technology. >
To overcome the difficulty of discovering high-performance materials due to the complexity of structures and the limitations of predicting intermolecular interactions, the research team developed a machine learning force field (MLFF) capable of precisely predicting the interactions between CO₂, water (H₂O), and MOFs. This new method enables calculations of MOF adsorption properties with quantum-mechanics-level accuracy at vastly faster speeds than before.
Using this system, the team screened over 8,000 experimentally synthesized MOF structures, identifying more than 100 promising candidates for CO₂ capture. Notably, this included new candidates that had not been uncovered by traditional force-field-based simulations. The team also analyzed the relationships between MOF chemical structure and adsorption performance, proposing seven key chemical features that will help in designing new materials for DAC.
< Figure 2. Concept diagram of adsorption simulation using Machine Learning Force Field (MLFF). The developed MLFF is applicable to various MOF structures and allows for precise calculation of adsorption properties by predicting interaction energies during repetitive Widom insertion simulations. It is characterized by simultaneously achieving high accuracy and low computational cost compared to conventional classical force fields. >
This research is recognized as a significant advance in the DAC field, greatly enhancing materials design and simulation by precisely predicting MOF-CO₂ and MOF-H₂O interactions.
The results of this research, with Ph.D. candidate Yunsung Lim and Dr. Hyunsoo Park of KAIST as co-first authors, were published in the international academic journal Matter on June 12.
※Paper Title: Accelerating CO₂ direct air capture screening for metal–organic frameworks with a transferable machine learning force field
※DOI: 10.1016/j.matt.2025.102203
This research was supported by the Saudi Aramco-KAIST CO₂ Management Center and the Ministry of Science and ICT's Global C.L.E.A.N. Project.
2025.06.29 View 351
KAIST Develops AI to Easily Find Promising Materials That Capture Only CO₂
< Photo 1. (From left) Professor Jihan Kim, Ph.D. candidate Yunsung Lim and Dr. Hyunsoo Park of the Department of Chemical and Biomolecular Engineering >
In order to help prevent the climate crisis, actively reducing already-emitted CO₂ is essential. Accordingly, direct air capture (DAC) — a technology that directly extracts only CO₂ from the air — is gaining attention. However, effectively capturing pure CO₂ is not easy due to water vapor (H₂O) present in the air. KAIST researchers have successfully used AI-driven machine learning techniques to identify the most promising CO₂-capturing materials among metal-organic frameworks (MOFs), a key class of materials studied for this technology.
KAIST (President Kwang Hyung Lee) announced on the 29th of June that a research team led by Professor Jihan Kim from the Department of Chemical and Biomolecular Engineering, in collaboration with a team at Imperial College London, has developed a machine-learning-based simulation method that can quickly and accurately screen MOFs best suited for atmospheric CO₂ capture.
< Figure 1. Concept diagram of Direct Air Capture (DAC) technology and carbon capture using Metal-Organic Frameworks (MOFs). MOFs are promising porous materials capable of capturing carbon dioxide from the atmosphere, drawing attention as a core material for DAC technology. >
To overcome the difficulty of discovering high-performance materials due to the complexity of structures and the limitations of predicting intermolecular interactions, the research team developed a machine learning force field (MLFF) capable of precisely predicting the interactions between CO₂, water (H₂O), and MOFs. This new method enables calculations of MOF adsorption properties with quantum-mechanics-level accuracy at vastly faster speeds than before.
Using this system, the team screened over 8,000 experimentally synthesized MOF structures, identifying more than 100 promising candidates for CO₂ capture. Notably, this included new candidates that had not been uncovered by traditional force-field-based simulations. The team also analyzed the relationships between MOF chemical structure and adsorption performance, proposing seven key chemical features that will help in designing new materials for DAC.
< Figure 2. Concept diagram of adsorption simulation using Machine Learning Force Field (MLFF). The developed MLFF is applicable to various MOF structures and allows for precise calculation of adsorption properties by predicting interaction energies during repetitive Widom insertion simulations. It is characterized by simultaneously achieving high accuracy and low computational cost compared to conventional classical force fields. >
This research is recognized as a significant advance in the DAC field, greatly enhancing materials design and simulation by precisely predicting MOF-CO₂ and MOF-H₂O interactions.
The results of this research, with Ph.D. candidate Yunsung Lim and Dr. Hyunsoo Park of KAIST as co-first authors, were published in the international academic journal Matter on June 12.
※Paper Title: Accelerating CO₂ direct air capture screening for metal–organic frameworks with a transferable machine learning force field
※DOI: 10.1016/j.matt.2025.102203
This research was supported by the Saudi Aramco-KAIST CO₂ Management Center and the Ministry of Science and ICT's Global C.L.E.A.N. Project.
2025.06.29 View 351 -
 Distinguished Professor Sang Yup Lee Wins 2025 Global Metabolic Engineering Award
< Distinguished Professor Sang Yup Lee (Senior Vice President for Research) from the Department of Chemical & Biomolecular Engineering >
KAIST announced on the 20th that Professor Sang Yup Lee, who serves as the Vice President for Research and a Distinguished Professor at our university, has been awarded the '2025 Gregory N. Stephanopoulos Award for Metabolic Engineering' by the International Metabolic Engineering Society (IMES). Professor Lee delivered his award lecture at the 16th Metabolic Engineering Conference (ME16), held in Copenhagen, Denmark, from June 15th to 19th.
This award was established through contributions from the American Institute of Chemical Engineers (AIChE) Foundation, as well as fellow colleagues and acquaintances, to honor the achievements of Dr. Gregory Stephanopoulos, widely recognized as one of the pioneers of metabolic engineering. Presented biennially, the award recognizes scientists who have successfully commercialized fundamental research in metabolic engineering or have made outstanding contributions to the quantitative analysis, design, and modeling of metabolic pathways.
Professor Sang Yup Lee boasts an impressive record of over 770 journal papers and more than 860 patents. His groundbreaking research in metabolic engineering and biochemical engineering is highly acclaimed globally.
Throughout his 31 years as a professor at KAIST, Professor Lee has developed various metabolic engineering-based technologies and strategies. These advancements have been transferred to industries, facilitating the production of bulk chemicals, polymers, natural products, pharmaceuticals, and health functional foods. He has also founded companies and actively engages in advisory roles with various enterprises.
The International Metabolic Engineering Society (IMES) defines metabolic engineering as the manipulation of metabolic pathways in microorganisms or cells to produce useful substances (such as pharmaceuticals, biofuels, and chemical products). It utilizes tools like systems biology, synthetic biology, and computational modeling with the aim of enhancing the economic viability and sustainability of bio-based processes.
Furthermore, Professor Lee previously received the Merck Metabolic Engineering Award, a prominent international award in the field, in 2008. In 2018, he was honored with the Eni Award, often referred to as the Nobel Prize in energy, presented by the President of Italy.
Professor Sang Yup Lee remarked, "Metabolic engineering is a discipline that leads the current and future of biotechnology. It is a tremendous honor to receive this meaningful award at a time when the transition to a bio-based economy is accelerating. Together with my students and fellow researchers, we have generated numerous patents and transferred technologies to industry, and also established startups in the fields of biofuels, wound healing, and cosmetics. I will continue to pursue research that encompasses both fundamental research and technological commercialization."
The 'International Metabolic Engineering Society (IMES)' is a specialized society under the American Institute of Chemical Engineers. Its mission is to enable the production of various bio-based products, including pharmaceuticals, food additives, chemicals, and fuels, through metabolic engineering. The society hosts the Metabolic Engineering Conference biennially, offering researchers opportunities for knowledge exchange and collaboration.
2025.06.20 View 858
Distinguished Professor Sang Yup Lee Wins 2025 Global Metabolic Engineering Award
< Distinguished Professor Sang Yup Lee (Senior Vice President for Research) from the Department of Chemical & Biomolecular Engineering >
KAIST announced on the 20th that Professor Sang Yup Lee, who serves as the Vice President for Research and a Distinguished Professor at our university, has been awarded the '2025 Gregory N. Stephanopoulos Award for Metabolic Engineering' by the International Metabolic Engineering Society (IMES). Professor Lee delivered his award lecture at the 16th Metabolic Engineering Conference (ME16), held in Copenhagen, Denmark, from June 15th to 19th.
This award was established through contributions from the American Institute of Chemical Engineers (AIChE) Foundation, as well as fellow colleagues and acquaintances, to honor the achievements of Dr. Gregory Stephanopoulos, widely recognized as one of the pioneers of metabolic engineering. Presented biennially, the award recognizes scientists who have successfully commercialized fundamental research in metabolic engineering or have made outstanding contributions to the quantitative analysis, design, and modeling of metabolic pathways.
Professor Sang Yup Lee boasts an impressive record of over 770 journal papers and more than 860 patents. His groundbreaking research in metabolic engineering and biochemical engineering is highly acclaimed globally.
Throughout his 31 years as a professor at KAIST, Professor Lee has developed various metabolic engineering-based technologies and strategies. These advancements have been transferred to industries, facilitating the production of bulk chemicals, polymers, natural products, pharmaceuticals, and health functional foods. He has also founded companies and actively engages in advisory roles with various enterprises.
The International Metabolic Engineering Society (IMES) defines metabolic engineering as the manipulation of metabolic pathways in microorganisms or cells to produce useful substances (such as pharmaceuticals, biofuels, and chemical products). It utilizes tools like systems biology, synthetic biology, and computational modeling with the aim of enhancing the economic viability and sustainability of bio-based processes.
Furthermore, Professor Lee previously received the Merck Metabolic Engineering Award, a prominent international award in the field, in 2008. In 2018, he was honored with the Eni Award, often referred to as the Nobel Prize in energy, presented by the President of Italy.
Professor Sang Yup Lee remarked, "Metabolic engineering is a discipline that leads the current and future of biotechnology. It is a tremendous honor to receive this meaningful award at a time when the transition to a bio-based economy is accelerating. Together with my students and fellow researchers, we have generated numerous patents and transferred technologies to industry, and also established startups in the fields of biofuels, wound healing, and cosmetics. I will continue to pursue research that encompasses both fundamental research and technological commercialization."
The 'International Metabolic Engineering Society (IMES)' is a specialized society under the American Institute of Chemical Engineers. Its mission is to enable the production of various bio-based products, including pharmaceuticals, food additives, chemicals, and fuels, through metabolic engineering. The society hosts the Metabolic Engineering Conference biennially, offering researchers opportunities for knowledge exchange and collaboration.
2025.06.20 View 858 -
 KAIST Develops Glare-Free, Heat-Blocking 'Smart Window'... Applicable to Buildings and Vehicles
• Professor Hong Chul Moon of the Department of Chemical and Biomolecular Engineering develops RECM, a next-generation smart window technology, expecting cooling energy savings and effective indoor thermal management.
• When using the developed RECM, a significantly superior temperature reduction effect is observed compared to conventional windows.
• With a 'pedestrian-friendly smart window' design that eliminates glare by suppressing external reflections, it is expected to be adapted in architectural structures, transportation, and more.
< (From left) First author Hoy Jung Jo, Professor Hong Chul Moon >
In the building sector, which accounts for approximately 40% of global energy consumption, heat ingress through windows has been identified as a primary cause of wasted heating and cooling energy. Our research team has successfully developed a 'pedestrian-friendly smart window' technology capable of not only reducing heating and cooling energy in urban buildings but also resolving the persistent issue of 'light pollution' in urban living.
On the 17th of June, Professor Hong Chul Moon's research team at KAIST's Department of Chemical and Biomolecular Engineering announced the development of a 'smart window technology' that allows users to control the light and heat entering through windows according to their intent, and effectively neutralize glare from external sources.
Recently, 'active smart window' technology, which enables free adjustment of light and heat based on user operation, has garnered significant attention. Unlike conventional windows that passively react to changes in temperature or light, this is a next-generation window system that can be controlled in real-time via electrical signals.
The next-generation smart window technology developed by the research team, RECM (Reversible Electrodeposition and Electrochromic Mirror), is a smart window system based on a single-structured *electrochromic device that can actively control the transmittance of visible light and near-infrared (heat).
*Electrochromic device: A device whose optical properties change in response to an electrical signal.
In particular, by effectively suppressing the glare phenomenon caused by external reflected light—a problem previously identified in traditional metal *deposition smart windows—through the combined application of electrochromic materials, a 'pedestrian-friendly smart window' suitable for building facades has been realized.
*Deposition: A process involving the electrochemical reaction to coat metal ions, such as Ag+, onto an electrode surface in solid form.
The RECM system developed in this study operates in three modes depending on voltage control.
Mode I (Transparent Mode) is advantageous for allowing sunlight to enter the indoor space during winter, as it transmits both light and heat like ordinary glass.
In Mode II (Colored Mode), *Prussian Blue (PB) and **DHV+• chemical species are formed through a redox (oxidation-reduction) reaction, causing the window to turn a deep blue color. In this state, light is absorbed, and only a portion of the heat is transmitted, allowing for privacy while enabling appropriate indoor temperature control.
*Prussian Blue: An electrochromic material that transitions between colorless and blue upon electrical stimulation.
**DHV+•: A radical state colored molecule generated upon electrical stimulation.
Mode III (Colored and Deposition Mode) involves the reduction and deposition of silver (Ag+) ions on the electrode surface, reflecting both light and heat. Concurrently, the colored material absorbs the reflected light, effectively blocking glare for external pedestrians.
The research team validated the practical indoor temperature reduction effect of the RECM technology through experiments utilizing a miniature model house. When a conventional glass window was installed, the indoor temperature rose to 58.7°C within 45 minutes. Conversely, when RECM was operated in Mode III, the temperature reached 31.5°C, demonstrating a temperature reduction effect of approximately 27.2°C.
Furthermore, since each state transition is achievable solely by electrical signals, it is regarded as an active smart technology capable of instantaneous response according to season, time, and intended use.
< Figure 1. Operation mechanism of the RECM smart window. The RECM system can switch among three states—transparent, colored, and colored & deposition—via electrical stimulation. At -1.6 V, DHV•+ and Prussian Blue (PB) are formed, blocking visible light to provide privacy protection and heat blocking. At -2.0 V, silver (Ag) is deposited on the electrode surface, reflecting light and heat, while DHV•+ and Prussian Blue absorb reflected light, effectively suppressing external glare. Through this mechanism, it functions as an active smart window that simultaneously controls light, heat, and glare. >
Professor Hong Chul Moon of KAIST, the corresponding author of this study, stated, "This research goes beyond existing smart window technologies limited to visible light control, presenting a truly smart window platform that comprehensively considers not only active indoor thermal control but also the visual safety of pedestrians." He added, "Various applications are anticipated, from urban buildings to vehicles and trains."
< Figure 2. Analysis of glare suppression effect of conventional reflective smart windows and RECM. This figure presents the results comparing the glare phenomenon occurring during silver (Ag) deposition between conventional reflective smart windows and RECM Mode III. Conventional reflective devices resulted in strong reflected light on the desk surface due to their high reflectivity. In contrast, RECM Mode III, where the colored material absorbed reflected light, showed a 33% reduction in reflected light intensity, and no reflected light was observed from outside. This highlights the RECM system's distinctiveness and practicality as a 'pedestrian-friendly smart window' optimized for dense urban environments, extending beyond just heat blocking. >
The findings of this research were published on June 13, 2025, in Volume 10, Issue 6 of 'ACS Energy Letters'. The listed authors for this publication are Hoy Jung Jo, Yeon Jae Jang, Hyeon-Don Kim, Kwang-Seop Kim, and Hong Chul Moon.
※ Paper Title: Glare-Free, Energy-Efficient Smart Windows: A Pedestrian-Friendly System with Dynamically Tunable Light and Heat Regulation
※ DOI: 10.1021/acsenergylett.5c00637
< Figure 3. Temperature reduction performance verification in a miniature model house. The actual heat blocking effect was evaluated by applying RECM devices to a model building. Under identical conditions, the indoor temperature with ordinary glass rose to 58.7°C, whereas with RECM in Mode III, it reached 31.5°C, demonstrating a maximum temperature reduction effect of 27.2°C. The indoor temperature difference was also visually confirmed through thermal images, which proves the potential for indoor temperature control in urban buildings. >
This research was supported by the Nano & Material Technology Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT and the internal research program of the Korea Institute of Machinery and Materials.
2025.06.20 View 2322
KAIST Develops Glare-Free, Heat-Blocking 'Smart Window'... Applicable to Buildings and Vehicles
• Professor Hong Chul Moon of the Department of Chemical and Biomolecular Engineering develops RECM, a next-generation smart window technology, expecting cooling energy savings and effective indoor thermal management.
• When using the developed RECM, a significantly superior temperature reduction effect is observed compared to conventional windows.
• With a 'pedestrian-friendly smart window' design that eliminates glare by suppressing external reflections, it is expected to be adapted in architectural structures, transportation, and more.
< (From left) First author Hoy Jung Jo, Professor Hong Chul Moon >
In the building sector, which accounts for approximately 40% of global energy consumption, heat ingress through windows has been identified as a primary cause of wasted heating and cooling energy. Our research team has successfully developed a 'pedestrian-friendly smart window' technology capable of not only reducing heating and cooling energy in urban buildings but also resolving the persistent issue of 'light pollution' in urban living.
On the 17th of June, Professor Hong Chul Moon's research team at KAIST's Department of Chemical and Biomolecular Engineering announced the development of a 'smart window technology' that allows users to control the light and heat entering through windows according to their intent, and effectively neutralize glare from external sources.
Recently, 'active smart window' technology, which enables free adjustment of light and heat based on user operation, has garnered significant attention. Unlike conventional windows that passively react to changes in temperature or light, this is a next-generation window system that can be controlled in real-time via electrical signals.
The next-generation smart window technology developed by the research team, RECM (Reversible Electrodeposition and Electrochromic Mirror), is a smart window system based on a single-structured *electrochromic device that can actively control the transmittance of visible light and near-infrared (heat).
*Electrochromic device: A device whose optical properties change in response to an electrical signal.
In particular, by effectively suppressing the glare phenomenon caused by external reflected light—a problem previously identified in traditional metal *deposition smart windows—through the combined application of electrochromic materials, a 'pedestrian-friendly smart window' suitable for building facades has been realized.
*Deposition: A process involving the electrochemical reaction to coat metal ions, such as Ag+, onto an electrode surface in solid form.
The RECM system developed in this study operates in three modes depending on voltage control.
Mode I (Transparent Mode) is advantageous for allowing sunlight to enter the indoor space during winter, as it transmits both light and heat like ordinary glass.
In Mode II (Colored Mode), *Prussian Blue (PB) and **DHV+• chemical species are formed through a redox (oxidation-reduction) reaction, causing the window to turn a deep blue color. In this state, light is absorbed, and only a portion of the heat is transmitted, allowing for privacy while enabling appropriate indoor temperature control.
*Prussian Blue: An electrochromic material that transitions between colorless and blue upon electrical stimulation.
**DHV+•: A radical state colored molecule generated upon electrical stimulation.
Mode III (Colored and Deposition Mode) involves the reduction and deposition of silver (Ag+) ions on the electrode surface, reflecting both light and heat. Concurrently, the colored material absorbs the reflected light, effectively blocking glare for external pedestrians.
The research team validated the practical indoor temperature reduction effect of the RECM technology through experiments utilizing a miniature model house. When a conventional glass window was installed, the indoor temperature rose to 58.7°C within 45 minutes. Conversely, when RECM was operated in Mode III, the temperature reached 31.5°C, demonstrating a temperature reduction effect of approximately 27.2°C.
Furthermore, since each state transition is achievable solely by electrical signals, it is regarded as an active smart technology capable of instantaneous response according to season, time, and intended use.
< Figure 1. Operation mechanism of the RECM smart window. The RECM system can switch among three states—transparent, colored, and colored & deposition—via electrical stimulation. At -1.6 V, DHV•+ and Prussian Blue (PB) are formed, blocking visible light to provide privacy protection and heat blocking. At -2.0 V, silver (Ag) is deposited on the electrode surface, reflecting light and heat, while DHV•+ and Prussian Blue absorb reflected light, effectively suppressing external glare. Through this mechanism, it functions as an active smart window that simultaneously controls light, heat, and glare. >
Professor Hong Chul Moon of KAIST, the corresponding author of this study, stated, "This research goes beyond existing smart window technologies limited to visible light control, presenting a truly smart window platform that comprehensively considers not only active indoor thermal control but also the visual safety of pedestrians." He added, "Various applications are anticipated, from urban buildings to vehicles and trains."
< Figure 2. Analysis of glare suppression effect of conventional reflective smart windows and RECM. This figure presents the results comparing the glare phenomenon occurring during silver (Ag) deposition between conventional reflective smart windows and RECM Mode III. Conventional reflective devices resulted in strong reflected light on the desk surface due to their high reflectivity. In contrast, RECM Mode III, where the colored material absorbed reflected light, showed a 33% reduction in reflected light intensity, and no reflected light was observed from outside. This highlights the RECM system's distinctiveness and practicality as a 'pedestrian-friendly smart window' optimized for dense urban environments, extending beyond just heat blocking. >
The findings of this research were published on June 13, 2025, in Volume 10, Issue 6 of 'ACS Energy Letters'. The listed authors for this publication are Hoy Jung Jo, Yeon Jae Jang, Hyeon-Don Kim, Kwang-Seop Kim, and Hong Chul Moon.
※ Paper Title: Glare-Free, Energy-Efficient Smart Windows: A Pedestrian-Friendly System with Dynamically Tunable Light and Heat Regulation
※ DOI: 10.1021/acsenergylett.5c00637
< Figure 3. Temperature reduction performance verification in a miniature model house. The actual heat blocking effect was evaluated by applying RECM devices to a model building. Under identical conditions, the indoor temperature with ordinary glass rose to 58.7°C, whereas with RECM in Mode III, it reached 31.5°C, demonstrating a maximum temperature reduction effect of 27.2°C. The indoor temperature difference was also visually confirmed through thermal images, which proves the potential for indoor temperature control in urban buildings. >
This research was supported by the Nano & Material Technology Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT and the internal research program of the Korea Institute of Machinery and Materials.
2025.06.20 View 2322 -
 KAIST Successfully Develops High-Performance Water Electrolysis Without Platinum, Bringing Hydrogen Economy Closer
< Photo 1. (Front row, from left) Jeesoo Park (Ph.D. Candidate), Professor Hee-Tak Kim (Back row, from left) Kyunghwa Seok (Ph.D. Candidate), Dr. Gisu Doo, Euntaek Oh (Ph.D. Candidate) >
Hydrogen is gaining attention as a clean energy source that emits no carbon. Among various methods, water electrolysis, which splits water into hydrogen and oxygen using electricity, is recognized as an eco-friendly hydrogen production method. Specifically, proton exchange membrane water electrolysis (PEMWE) is considered a next-generation hydrogen production technology due to its ability to produce high-purity hydrogen at high pressure. However, existing PEMWE technology has faced limitations in commercialization due to its heavy reliance on expensive precious metal catalysts and coating materials. Korean researchers have now proposed a new solution to address these technical and economic bottlenecks.
KAIST (President Kwang Hyung Lee) announced on June 11th that a research team led by Professor Hee-Tak Kim of the Department of Chemical and Biomolecular Engineering, in a joint study with Dr. Gisu Doo of the Korea Institute of Energy Research (KIER, President Chang-keun Lee), has developed a next-generation water electrolysis technology that achieves high performance without the need for expensive platinum (Pt) coating.
The research team focused on the primary reason why 'iridium oxide (IrOx),' a highly active catalyst for water electrolysis electrodes, fails to perform optimally. They found that this is due to inefficient electron transfer and, for the first time in the world, demonstrated that performance can be maximized simply by controlling the catalyst particle size.
In this study, it was revealed that the reason iridium oxide catalysts do not exhibit excellent performance without platinum coating is due to 'electron transport resistance' that occurs at the interface between the catalyst, the ion conductor (hereinafter referred to as ionomer), and the Ti (titanium) substrate—core components inherently used together in water electrolysis electrodes.
Specifically, they identified that the 'pinch-off' phenomenon, where the electron pathway is blocked between the catalyst, ionomer, and titanium substrate, is the critical cause of reduced conductivity. The ionomer has properties close to an electron insulator, thereby hindering electron flow when it surrounds catalyst particles. Furthermore, when the ionomer comes into contact with the titanium substrate, an electron barrier forms on the surface oxide layer of the titanium substrate, significantly increasing resistance.
< Figure 1. Infographic related to electron transport resistance at the catalyst layer/diffusion layer interface >
To address this, the research team fabricated and compared catalysts of various particle sizes. Through single-cell evaluation and multiphysics simulations, they demonstrated, for the first time globally, that when iridium oxide catalyst particles with a size of 20 nanometers (nm) or larger are used, the ionomer mixed region decreases, ensuring an electron pathway and restoring conductivity.
Moreover, they successfully optimized the interfacial structure through precise design, simultaneously ensuring both reactivity and electron transport. This achievement demonstrated that the previously unavoidable trade-off between catalyst activity and conductivity can be overcome through meticulous interfacial design.
This breakthrough is expected to be a significant milestone not only for the development of high-performance catalyst materials but also for the future commercialization of proton exchange membrane water electrolysis systems that can achieve high efficiency while drastically reducing the amount of precious metals used.
Professor Hee-Tak Kim stated, "This research presents a new interface design strategy that can resolve the interfacial conductivity problem, which was a bottleneck in high-performance water electrolysis technology." He added, "By securing high performance even without expensive materials like platinum, it will be a stepping stone closer to realizing a hydrogen economy."
This research, with Jeesoo Park, a Ph.D. student from the Department of Chemical and Biomolecular Engineering at KAIST, as the first author, was published on June 7th in 'Energy & Environmental Science' (IF: 32.4, 2025), a leading international journal in the energy and environmental fields, and was recognized for its innovativeness and impact. (Paper title: On the interface electron transport problem of highly active IrOx catalysts, DOI: 10.1039/D4EE05816J).
This research was supported by the New and Renewable Energy Core Technology Development Project of the Ministry of Trade, Industry and Energy.
2025.06.11 View 1818
KAIST Successfully Develops High-Performance Water Electrolysis Without Platinum, Bringing Hydrogen Economy Closer
< Photo 1. (Front row, from left) Jeesoo Park (Ph.D. Candidate), Professor Hee-Tak Kim (Back row, from left) Kyunghwa Seok (Ph.D. Candidate), Dr. Gisu Doo, Euntaek Oh (Ph.D. Candidate) >
Hydrogen is gaining attention as a clean energy source that emits no carbon. Among various methods, water electrolysis, which splits water into hydrogen and oxygen using electricity, is recognized as an eco-friendly hydrogen production method. Specifically, proton exchange membrane water electrolysis (PEMWE) is considered a next-generation hydrogen production technology due to its ability to produce high-purity hydrogen at high pressure. However, existing PEMWE technology has faced limitations in commercialization due to its heavy reliance on expensive precious metal catalysts and coating materials. Korean researchers have now proposed a new solution to address these technical and economic bottlenecks.
KAIST (President Kwang Hyung Lee) announced on June 11th that a research team led by Professor Hee-Tak Kim of the Department of Chemical and Biomolecular Engineering, in a joint study with Dr. Gisu Doo of the Korea Institute of Energy Research (KIER, President Chang-keun Lee), has developed a next-generation water electrolysis technology that achieves high performance without the need for expensive platinum (Pt) coating.
The research team focused on the primary reason why 'iridium oxide (IrOx),' a highly active catalyst for water electrolysis electrodes, fails to perform optimally. They found that this is due to inefficient electron transfer and, for the first time in the world, demonstrated that performance can be maximized simply by controlling the catalyst particle size.
In this study, it was revealed that the reason iridium oxide catalysts do not exhibit excellent performance without platinum coating is due to 'electron transport resistance' that occurs at the interface between the catalyst, the ion conductor (hereinafter referred to as ionomer), and the Ti (titanium) substrate—core components inherently used together in water electrolysis electrodes.
Specifically, they identified that the 'pinch-off' phenomenon, where the electron pathway is blocked between the catalyst, ionomer, and titanium substrate, is the critical cause of reduced conductivity. The ionomer has properties close to an electron insulator, thereby hindering electron flow when it surrounds catalyst particles. Furthermore, when the ionomer comes into contact with the titanium substrate, an electron barrier forms on the surface oxide layer of the titanium substrate, significantly increasing resistance.
< Figure 1. Infographic related to electron transport resistance at the catalyst layer/diffusion layer interface >
To address this, the research team fabricated and compared catalysts of various particle sizes. Through single-cell evaluation and multiphysics simulations, they demonstrated, for the first time globally, that when iridium oxide catalyst particles with a size of 20 nanometers (nm) or larger are used, the ionomer mixed region decreases, ensuring an electron pathway and restoring conductivity.
Moreover, they successfully optimized the interfacial structure through precise design, simultaneously ensuring both reactivity and electron transport. This achievement demonstrated that the previously unavoidable trade-off between catalyst activity and conductivity can be overcome through meticulous interfacial design.
This breakthrough is expected to be a significant milestone not only for the development of high-performance catalyst materials but also for the future commercialization of proton exchange membrane water electrolysis systems that can achieve high efficiency while drastically reducing the amount of precious metals used.
Professor Hee-Tak Kim stated, "This research presents a new interface design strategy that can resolve the interfacial conductivity problem, which was a bottleneck in high-performance water electrolysis technology." He added, "By securing high performance even without expensive materials like platinum, it will be a stepping stone closer to realizing a hydrogen economy."
This research, with Jeesoo Park, a Ph.D. student from the Department of Chemical and Biomolecular Engineering at KAIST, as the first author, was published on June 7th in 'Energy & Environmental Science' (IF: 32.4, 2025), a leading international journal in the energy and environmental fields, and was recognized for its innovativeness and impact. (Paper title: On the interface electron transport problem of highly active IrOx catalysts, DOI: 10.1039/D4EE05816J).
This research was supported by the New and Renewable Energy Core Technology Development Project of the Ministry of Trade, Industry and Energy.
2025.06.11 View 1818 -
 KAIST Discovers Protein Switch that Turns Anti-Viral Immune Response On and Off
Even after the COVID-19 pandemic, various new infectious diseases continue to emerge, posing ongoing viral threats that demand robust and sustained immune defenses. However, excessive immune reactions can also harm body tissues, causing significant health issues. KAIST and an international research team have discovered a critical protein that acts as a 'switch' regulating immune responses to viruses. This breakthrough is expected to lay the groundwork for future infectious disease responses and autoimmune disease treatment strategies.
KAIST (President Kwang-Hyung Lee) announced on May 14 that a joint research team led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering at KAIST and Professor Seunghee Cha from University of Florida has discovered the mechanism by which double-stranded RNA derived from mitochondria amplifies immune responses. They identified the protein SLIRP as an 'immune switch' that regulates this process, playing a crucial role in both viral infections and autoimmune diseases.
< (From left) Master's candidate Yewon Yang, Professor Yoosik Kim and Ph.D. candidate Doyeong Ku of the Department of Chemical and Biomolecular Engineering >
Autoimmune diseases arise when the immune system fails to differentiate between external pathogens and the body's own molecules, leading to self-directed attacks. Despite extensive research, the precise causes of excessive inflammatory conditions like Sjögren’s syndrome and systemic lupus erythematosus remain unclear, and effective treatments are still limited.
To uncover the molecular mechanisms driving immune hyperactivation and to identify potential regulatory factors, the research team led by Professor Yoosik Kim focused on mitochondrial double-stranded RNA (mt-dsRNA), a genetic immunogenic material produced within cellular organelles. Since mt-dsRNA structurally resembles viral RNA, it can mistakenly trigger immune responses even in the absence of an actual viral infection.
The team discovered that SLIRP, a key regulator of mt-dsRNA, amplifies immune responses by stabilizing the RNA. They confirmed that SLIRP expression increases in experimental models simulating the tissues of autoimmune disease patients and viral infections. Conversely, suppressing SLIRP significantly reduced the immune response, underscoring its role as a critical factor in immune amplification.
This study also demonstrated the dual function of SLIRP in different contexts. In cells infected with human beta coronavirus OC43 and encephalomyocarditis virus (EMCV), SLIRP suppression led to reduced antiviral responses and increased viral replication. Meanwhile, in the blood and salivary gland cells of Sjögren’s syndrome patients, where both SLIRP and mt-dsRNA levels were elevated, suppressing SLIRP alleviated the abnormal immune response.
These findings highlight SLIRP as a key molecular switch that regulates immune responses in both infections and autoimmune diseases.
< Figure 1. Schematic diagram of antiviral signal amplification by SLIRP: SLIRP-based mt-dsRNA induction, cytoplasmic accumulation, and strong interferon response induction by positive feedback of immune response activation. Confirmation of the immune regulatory function of SLIRP in defense against autoimmune diseases Sjögren's syndrome, coronavirus, and encephalomyocarditis virus infection. >
Professor Yoosik Kim remarked, "Through this study, we have identified SLIRP as a crucial protein that drives immune amplification via mt-dsRNAs. Given its dual role in autoimmune diseases and viral infections, SLIRP presents a promising target for immune regulation therapies across various inflammatory disease contexts."
The study, with Ph.D. student Do-Young Ku (first author) and M.S. student Ye-Won Yang (second author) from the Department of Chemical and Biomolecular Engineering at KAIST as primary contributors, was published online in the journal Cell Reports on April 19, 2025.
※ Paper title: SLIRP amplifies antiviral signaling via positive feedback regulation and contributes to autoimmune diseases※ Main authors: Do-Young Ku (KAIST, first author), Ye-Won Yang (KAIST, second author), Seunghee Cha (University of Florida, corresponding author), Yoosik Kim (KAIST, corresponding author)
This study was supported by the Ministry of Health and Welfare's Public Health Technology Research Program and the National Institutes of Health (NIH) through Research Project (R01) funding.
2025.05.14 View 2638
KAIST Discovers Protein Switch that Turns Anti-Viral Immune Response On and Off
Even after the COVID-19 pandemic, various new infectious diseases continue to emerge, posing ongoing viral threats that demand robust and sustained immune defenses. However, excessive immune reactions can also harm body tissues, causing significant health issues. KAIST and an international research team have discovered a critical protein that acts as a 'switch' regulating immune responses to viruses. This breakthrough is expected to lay the groundwork for future infectious disease responses and autoimmune disease treatment strategies.
KAIST (President Kwang-Hyung Lee) announced on May 14 that a joint research team led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering at KAIST and Professor Seunghee Cha from University of Florida has discovered the mechanism by which double-stranded RNA derived from mitochondria amplifies immune responses. They identified the protein SLIRP as an 'immune switch' that regulates this process, playing a crucial role in both viral infections and autoimmune diseases.
< (From left) Master's candidate Yewon Yang, Professor Yoosik Kim and Ph.D. candidate Doyeong Ku of the Department of Chemical and Biomolecular Engineering >
Autoimmune diseases arise when the immune system fails to differentiate between external pathogens and the body's own molecules, leading to self-directed attacks. Despite extensive research, the precise causes of excessive inflammatory conditions like Sjögren’s syndrome and systemic lupus erythematosus remain unclear, and effective treatments are still limited.
To uncover the molecular mechanisms driving immune hyperactivation and to identify potential regulatory factors, the research team led by Professor Yoosik Kim focused on mitochondrial double-stranded RNA (mt-dsRNA), a genetic immunogenic material produced within cellular organelles. Since mt-dsRNA structurally resembles viral RNA, it can mistakenly trigger immune responses even in the absence of an actual viral infection.
The team discovered that SLIRP, a key regulator of mt-dsRNA, amplifies immune responses by stabilizing the RNA. They confirmed that SLIRP expression increases in experimental models simulating the tissues of autoimmune disease patients and viral infections. Conversely, suppressing SLIRP significantly reduced the immune response, underscoring its role as a critical factor in immune amplification.
This study also demonstrated the dual function of SLIRP in different contexts. In cells infected with human beta coronavirus OC43 and encephalomyocarditis virus (EMCV), SLIRP suppression led to reduced antiviral responses and increased viral replication. Meanwhile, in the blood and salivary gland cells of Sjögren’s syndrome patients, where both SLIRP and mt-dsRNA levels were elevated, suppressing SLIRP alleviated the abnormal immune response.
These findings highlight SLIRP as a key molecular switch that regulates immune responses in both infections and autoimmune diseases.
< Figure 1. Schematic diagram of antiviral signal amplification by SLIRP: SLIRP-based mt-dsRNA induction, cytoplasmic accumulation, and strong interferon response induction by positive feedback of immune response activation. Confirmation of the immune regulatory function of SLIRP in defense against autoimmune diseases Sjögren's syndrome, coronavirus, and encephalomyocarditis virus infection. >
Professor Yoosik Kim remarked, "Through this study, we have identified SLIRP as a crucial protein that drives immune amplification via mt-dsRNAs. Given its dual role in autoimmune diseases and viral infections, SLIRP presents a promising target for immune regulation therapies across various inflammatory disease contexts."
The study, with Ph.D. student Do-Young Ku (first author) and M.S. student Ye-Won Yang (second author) from the Department of Chemical and Biomolecular Engineering at KAIST as primary contributors, was published online in the journal Cell Reports on April 19, 2025.
※ Paper title: SLIRP amplifies antiviral signaling via positive feedback regulation and contributes to autoimmune diseases※ Main authors: Do-Young Ku (KAIST, first author), Ye-Won Yang (KAIST, second author), Seunghee Cha (University of Florida, corresponding author), Yoosik Kim (KAIST, corresponding author)
This study was supported by the Ministry of Health and Welfare's Public Health Technology Research Program and the National Institutes of Health (NIH) through Research Project (R01) funding.
2025.05.14 View 2638 -
 KAIST Develops Novel Catalyst With 100-Fold Platinum Efficiency
Propylene, a key building block used in producing plastics, textiles, automotive components, and electronics, is essential to the petrochemical industry. A KAIST research team has developed a novel catalyst that dramatically enhances the efficiency of propylene production while significantly reducing costs.
< Photo. Professor Minkee Choi (left), and Ph.D. Candidate Susung Lee (right) of the Department of Chemical and Biomolecular Engineering >
KAIST (represented by President Kwang-Hyung Lee) announced on the 12th of May that a research group led by Professor Minkee Choi from the Department of Chemical and Biomolecular Engineering has successfully developed a new catalyst using inexpensive metals—gallium (Ga) and alumina (Al₂O₃)—with only a trace amount of platinum (100 ppm, or 0.01%). Remarkably, this new catalyst outperforms conventional industrial catalysts that use high concentrations of platinum (10,000 ppm).
Propylene is commonly produced through the propane dehydrogenation (PDH) process, which removes hydrogen from propane. Platinum has long been used as a catalyst in PDH due to its high efficiency in breaking carbon-hydrogen bonds and facilitating hydrogen removal. However, platinum is costly and suffers from performance degradation over repeated use.
To address this, the KAIST team engineered a catalyst that incorporates only a minimal amount of platinum, relying on gallium and alumina as the primary components.
< Figure 1. Schematic diagram showing the catalytic cooperation between gallium (Ga) and platinum (Pt) >
The core mechanism of the catalyst involves a cooperative function between the metals: gallium activates the C–H bond in propane to produce propylene, while platinum bonds the residual hydrogen atoms on the surface to form hydrogen gas (H₂), which is then released. This division of roles allows for high catalytic efficiency despite the drastic reduction in platinum content.
The researchers identified an optimal platinum-to-gallium ratio that delivered peak performance and provided a scientific rationale and quantitative metrics to predict this ideal composition.
Additionally, the team addressed a major limitation of traditional platinum catalysts: sintering—the agglomeration of platinum particles during repeated use, which causes performance loss. By adding a small amount of cerium (Ce), the researchers successfully suppressed this aggregation. As a result, the new catalyst maintained stable performance even after more than 20 reaction-regeneration cycles.
< Figure 2. Performance comparison of KAIST's newly developed catalyst (100 ppm platinum) and existing commercial platinum catalyst (10,000 ppm platinum) >
Professor Choi stated, “This research demonstrates the possibility of reducing platinum usage to 1/100th of current levels without compromising, and even enhancing, performance. It presents significant economic and environmental advantages, including lower catalyst costs, extended replacement intervals, and reduced catalyst waste.”
He added, “We are planning to evaluate this technology for large-scale process demonstration and commercialization. If adopted in industry, it could greatly improve the economic viability and efficiency of propylene production.”
The study was led by Professor Minkee Choi as corresponding author, with Ph.D. candidate Susung Lee as the first author. The findings were published in the Journal of the American Chemical Society (JACS), a leading journal in chemistry and chemical engineering, on February 13.※ Paper title: Ideal Bifunctional Catalysis for Propane Dehydrogenation over Pt-Promoted Gallia-Alumina and Minimized Use of Precious Elements※ DOI: https://pubs.acs.org/doi/10.1021/jacs.4c13787
The research was supported by the National Research Foundation of Korea and Hanwha Solutions Corporation.
2025.05.12 View 1669
KAIST Develops Novel Catalyst With 100-Fold Platinum Efficiency
Propylene, a key building block used in producing plastics, textiles, automotive components, and electronics, is essential to the petrochemical industry. A KAIST research team has developed a novel catalyst that dramatically enhances the efficiency of propylene production while significantly reducing costs.
< Photo. Professor Minkee Choi (left), and Ph.D. Candidate Susung Lee (right) of the Department of Chemical and Biomolecular Engineering >
KAIST (represented by President Kwang-Hyung Lee) announced on the 12th of May that a research group led by Professor Minkee Choi from the Department of Chemical and Biomolecular Engineering has successfully developed a new catalyst using inexpensive metals—gallium (Ga) and alumina (Al₂O₃)—with only a trace amount of platinum (100 ppm, or 0.01%). Remarkably, this new catalyst outperforms conventional industrial catalysts that use high concentrations of platinum (10,000 ppm).
Propylene is commonly produced through the propane dehydrogenation (PDH) process, which removes hydrogen from propane. Platinum has long been used as a catalyst in PDH due to its high efficiency in breaking carbon-hydrogen bonds and facilitating hydrogen removal. However, platinum is costly and suffers from performance degradation over repeated use.
To address this, the KAIST team engineered a catalyst that incorporates only a minimal amount of platinum, relying on gallium and alumina as the primary components.
< Figure 1. Schematic diagram showing the catalytic cooperation between gallium (Ga) and platinum (Pt) >
The core mechanism of the catalyst involves a cooperative function between the metals: gallium activates the C–H bond in propane to produce propylene, while platinum bonds the residual hydrogen atoms on the surface to form hydrogen gas (H₂), which is then released. This division of roles allows for high catalytic efficiency despite the drastic reduction in platinum content.
The researchers identified an optimal platinum-to-gallium ratio that delivered peak performance and provided a scientific rationale and quantitative metrics to predict this ideal composition.
Additionally, the team addressed a major limitation of traditional platinum catalysts: sintering—the agglomeration of platinum particles during repeated use, which causes performance loss. By adding a small amount of cerium (Ce), the researchers successfully suppressed this aggregation. As a result, the new catalyst maintained stable performance even after more than 20 reaction-regeneration cycles.
< Figure 2. Performance comparison of KAIST's newly developed catalyst (100 ppm platinum) and existing commercial platinum catalyst (10,000 ppm platinum) >
Professor Choi stated, “This research demonstrates the possibility of reducing platinum usage to 1/100th of current levels without compromising, and even enhancing, performance. It presents significant economic and environmental advantages, including lower catalyst costs, extended replacement intervals, and reduced catalyst waste.”
He added, “We are planning to evaluate this technology for large-scale process demonstration and commercialization. If adopted in industry, it could greatly improve the economic viability and efficiency of propylene production.”
The study was led by Professor Minkee Choi as corresponding author, with Ph.D. candidate Susung Lee as the first author. The findings were published in the Journal of the American Chemical Society (JACS), a leading journal in chemistry and chemical engineering, on February 13.※ Paper title: Ideal Bifunctional Catalysis for Propane Dehydrogenation over Pt-Promoted Gallia-Alumina and Minimized Use of Precious Elements※ DOI: https://pubs.acs.org/doi/10.1021/jacs.4c13787
The research was supported by the National Research Foundation of Korea and Hanwha Solutions Corporation.
2025.05.12 View 1669 -
 KAIST Accelerates Synthetic Microbe Design by Discovering Novel Enzymes Using AI
< (From left) Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering (top), Hongkeun Ji, PhD candidate of the Department of Chemical and Biomolecular Engineering (top), Ha Rim Kim, PhD candidate of the Department of Chemical and Biomolecular Engineering, and Dr. Gi Bae Kim of the BioProcess Engineering Research Center >
Enzymes are proteins that catalyze biochemical reactions within cells and play a pivotal role in metabolic processes. Accordingly, identifying the functions of novel enzymes is a critical task in the construction of microbial cell factories.
A KAIST research team has leveraged artificial intelligence (AI) to design novel enzymes that do not exist in nature, significantly accelerating microbial cell factory development and boosting the potential for next-generation biotechnological applications such as drug development and biofuel production.
KAIST (represented by President Kwang-Hyung Lee) announced on the 21st of April that Distinguished Professor Sang Yup Lee and his team from the Department of Chemical and Biomolecular Engineering have published a review titled “Enzyme Functional Classification Using Artificial Intelligence,” which outlines the advancement of AI-based enzyme function prediction technologies and analyzes how AI has contributed to the discovery and design of new enzymes.
Professor Lee’s team systematically reviewed the development of enzyme function prediction technologies utilizing machine learning and deep learning, offering a comprehensive analysis.
From sequence similarity-based prediction methods to the integration of convolutional neural networks (CNNs), recurrent neural networks (RNNs), graph neural networks (GNNs), and transformer-based large language models, the paper covers a broad range of AI applications. It analyzes how these technologies extract meaningful information from protein sequences and enhance prediction accuracy.
In particular, enzyme function prediction using deep learning goes beyond simple sequence similarity analysis. By automatically extracting structural and evolutionary features embedded in amino acid sequences, deep learning enables more precise predictions of catalytic functions.
This highlights the unique advantages of AI models compared to traditional bioinformatics approaches.
Moreover, the review suggests that the advancement of generative AI will move future research beyond predicting existing functions to generating entirely new enzymes with functions not found in nature. This shift is expected to profoundly impact the trajectory of biotechnology and synthetic biology.
< Figure 1. Extraction of enzyme characteristics and function prediction using various deep learning structures >
Ha Rim Kim, a Ph.D. candidate and co-first author from the Department of Chemical and Biomolecular Engineering, stated, “AI-based enzyme function prediction and enzyme design are highly important across various fields including metabolic engineering, synthetic biology, and healthcare.”
Distinguished Professor Sang Yup Lee added, “AI-powered enzyme function prediction shows the potential to solve diverse biological problems and will significantly contribute to accelerating research across the entire field.”
The review was published on March 28 in Trends in Biotechnology, a leading biotechnology journal issued by Cell Press.
※ Title: Enzyme Functional Classification Using Artificial Intelligence
※DOI: https://doi.org/10.1016/j.tibtech.2025.03.003
※ Author Information: Ha Rim Kim (KAIST, Co-first author), Hongkeun Ji (KAIST, Co-first author), Gi Bae Kim (KAIST, Third author), Sang Yup Lee (KAIST, Corresponding author)
This research was supported by the Ministry of Science and ICT under the project Development of Core Technologies for Advanced Synthetic Biology to Lead the Bio-Manufacturing Industry (aimed at replacing petroleum-based chemicals), and also by joint support from the Ministry of Science and ICT and the Ministry of Health and Welfare for the project Development of Novel Antibiotic Structures Using Deep Learning-Based Synthetic Biology.
2025.04.07 View 3395
KAIST Accelerates Synthetic Microbe Design by Discovering Novel Enzymes Using AI
< (From left) Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering (top), Hongkeun Ji, PhD candidate of the Department of Chemical and Biomolecular Engineering (top), Ha Rim Kim, PhD candidate of the Department of Chemical and Biomolecular Engineering, and Dr. Gi Bae Kim of the BioProcess Engineering Research Center >
Enzymes are proteins that catalyze biochemical reactions within cells and play a pivotal role in metabolic processes. Accordingly, identifying the functions of novel enzymes is a critical task in the construction of microbial cell factories.
A KAIST research team has leveraged artificial intelligence (AI) to design novel enzymes that do not exist in nature, significantly accelerating microbial cell factory development and boosting the potential for next-generation biotechnological applications such as drug development and biofuel production.
KAIST (represented by President Kwang-Hyung Lee) announced on the 21st of April that Distinguished Professor Sang Yup Lee and his team from the Department of Chemical and Biomolecular Engineering have published a review titled “Enzyme Functional Classification Using Artificial Intelligence,” which outlines the advancement of AI-based enzyme function prediction technologies and analyzes how AI has contributed to the discovery and design of new enzymes.
Professor Lee’s team systematically reviewed the development of enzyme function prediction technologies utilizing machine learning and deep learning, offering a comprehensive analysis.
From sequence similarity-based prediction methods to the integration of convolutional neural networks (CNNs), recurrent neural networks (RNNs), graph neural networks (GNNs), and transformer-based large language models, the paper covers a broad range of AI applications. It analyzes how these technologies extract meaningful information from protein sequences and enhance prediction accuracy.
In particular, enzyme function prediction using deep learning goes beyond simple sequence similarity analysis. By automatically extracting structural and evolutionary features embedded in amino acid sequences, deep learning enables more precise predictions of catalytic functions.
This highlights the unique advantages of AI models compared to traditional bioinformatics approaches.
Moreover, the review suggests that the advancement of generative AI will move future research beyond predicting existing functions to generating entirely new enzymes with functions not found in nature. This shift is expected to profoundly impact the trajectory of biotechnology and synthetic biology.
< Figure 1. Extraction of enzyme characteristics and function prediction using various deep learning structures >
Ha Rim Kim, a Ph.D. candidate and co-first author from the Department of Chemical and Biomolecular Engineering, stated, “AI-based enzyme function prediction and enzyme design are highly important across various fields including metabolic engineering, synthetic biology, and healthcare.”
Distinguished Professor Sang Yup Lee added, “AI-powered enzyme function prediction shows the potential to solve diverse biological problems and will significantly contribute to accelerating research across the entire field.”
The review was published on March 28 in Trends in Biotechnology, a leading biotechnology journal issued by Cell Press.
※ Title: Enzyme Functional Classification Using Artificial Intelligence
※DOI: https://doi.org/10.1016/j.tibtech.2025.03.003
※ Author Information: Ha Rim Kim (KAIST, Co-first author), Hongkeun Ji (KAIST, Co-first author), Gi Bae Kim (KAIST, Third author), Sang Yup Lee (KAIST, Corresponding author)
This research was supported by the Ministry of Science and ICT under the project Development of Core Technologies for Advanced Synthetic Biology to Lead the Bio-Manufacturing Industry (aimed at replacing petroleum-based chemicals), and also by joint support from the Ministry of Science and ICT and the Ministry of Health and Welfare for the project Development of Novel Antibiotic Structures Using Deep Learning-Based Synthetic Biology.
2025.04.07 View 3395 -
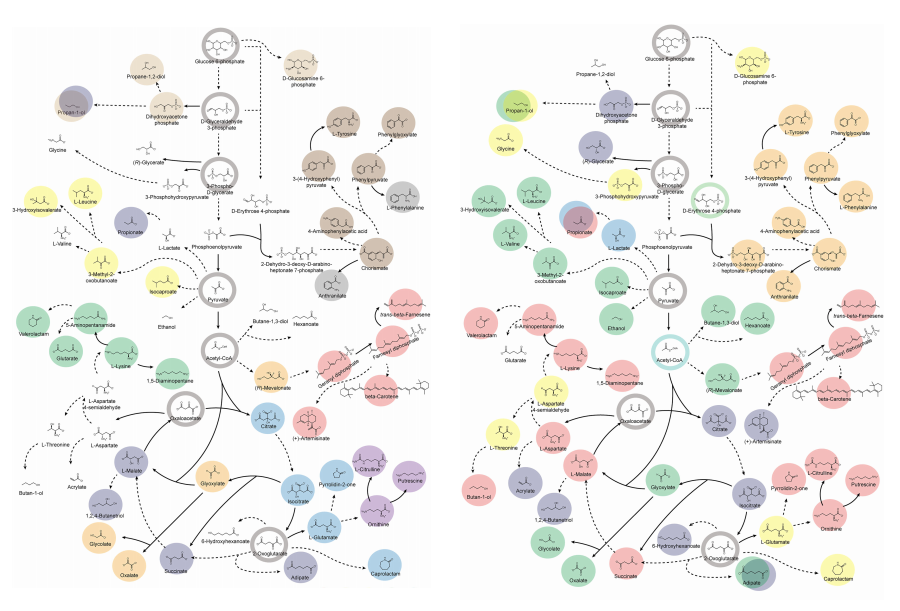 KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3854
KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3854 -
 KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 5069
KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 5069 -
 KAIST Develops World-Leading Ammonia Catalyst for Hydrogen Economy
Hydrogen production using renewable energy is a key technology for eco-friendly energy and chemical production. However, storing and transporting hydrogen remains a challenge. To address this, researchers worldwide are investigating methods to store hydrogen in the form of ammonia (NH₃), which is carbon-free and easier to liquify. A research team at KAIST has successfully developed a high-performance catalyst that enables ammonia synthesis at very low temperatures and pressures without energy loss.
KAIST (represented by President Kwang Hyung Lee) announced on the 11th of March that a research team led by Professor Minkee Choi from the Department of Chemical and Biomolecular Engineering has developed an innovative catalytic system that significantly enhances ammonia production while drastically reducing energy consumption and CO₂ emissions.
< (From left) Baek Ye-jun, Ph.D. candidate in the Department of Biochemical Engineering, Professor Choi Min-ki >
Currently, ammonia is produced using the Haber-Bosch process, a technology over a century old that relies on iron (Fe)-based catalysts. This method requires extreme conditions—temperatures above 500°C and pressures exceeding 100 atmospheres—resulting in enormous energy consumption and contributing significantly to global CO₂ emissions. Additionally, ammonia is primarily produced in large-scale industrial plants, leading to high distribution costs.
As an alternative, there is growing interest in an eco-friendly process that synthesizes ammonia using green hydrogen—produced via water electrolysis—under mild conditions (300°C, 10 atmospheres). However, developing catalysts that can achieve high ammonia productivity at such low temperatures and pressures is essential, as current technologies struggle to maintain efficiency under these conditions.
The research team developed a novel catalyst by incorporating ruthenium (Ru) nanoparticles and highly basic barium oxide (BaO) particles onto a conductive carbon surface, allowing it to function like a chemical capacitor*.
*Capacitor: A device that stores electrical energy by separating positive and negative charges.
During ammonia synthesis, hydrogen molecules (H₂) first dissociate into hydrogen atoms (H) on the ruthenium catalyst. These hydrogen atoms are further split into protons (H⁺) and electrons (e⁻). The study revealed that the acidic protons are stored in the strongly basic BaO, while the remaining electrons are separated and stored in ruthenium and carbon.
This unique chemical capacitor effect significantly enhances the ruthenium catalyst's electron density, accelerating nitrogen (N₂) dissociation—the rate-limiting step of ammonia synthesis—thereby dramatically increasing catalytic activity.
Furthermore, the team discovered that optimizing the nanostructure of the carbon material further boosts the electron density of ruthenium, maximizing catalytic performance. As a result, the new catalyst demonstrated over seven times higher ammonia synthesis performance compared to state-of-the-art catalysts under mild conditions (300°C, 10 atm).
< Schematic diagram showing the mechanism of ruthenium catalyst activity enhancement by barium oxide cocatalyst >
Professor Minkee Choi stated, “This research has garnered significant attention for demonstrating that catalytic activity can be greatly enhanced by controlling electron transfer within a thermal catalytic reaction system, not just in electrochemical processes.”
He further explained, “Our findings confirm that high-performance catalysts can enable efficient ammonia synthesis under low-temperature and low-pressure conditions. This could shift ammonia production from centralized, large-scale industrial plants to decentralized, small-scale production, making the hydrogen economy more sustainable and flexible.”
The study was led by Professor Minkee Choi as corresponding author and Yaejun Baik, a Ph.D. candidate, as first author. The research findings were published in Nature Catalysis on February 24.
(Paper title: “Electron and proton storage on separate Ru and BaO domains mediated by conductive low-work-function carbon to accelerate ammonia synthesis,” https://doi.org/10.1038/s41929-025-01302-z)
This research was supported by the Korea Institute of Energy Research and the National Research Foundation of Korea.
2025.03.11 View 3154
KAIST Develops World-Leading Ammonia Catalyst for Hydrogen Economy
Hydrogen production using renewable energy is a key technology for eco-friendly energy and chemical production. However, storing and transporting hydrogen remains a challenge. To address this, researchers worldwide are investigating methods to store hydrogen in the form of ammonia (NH₃), which is carbon-free and easier to liquify. A research team at KAIST has successfully developed a high-performance catalyst that enables ammonia synthesis at very low temperatures and pressures without energy loss.
KAIST (represented by President Kwang Hyung Lee) announced on the 11th of March that a research team led by Professor Minkee Choi from the Department of Chemical and Biomolecular Engineering has developed an innovative catalytic system that significantly enhances ammonia production while drastically reducing energy consumption and CO₂ emissions.
< (From left) Baek Ye-jun, Ph.D. candidate in the Department of Biochemical Engineering, Professor Choi Min-ki >
Currently, ammonia is produced using the Haber-Bosch process, a technology over a century old that relies on iron (Fe)-based catalysts. This method requires extreme conditions—temperatures above 500°C and pressures exceeding 100 atmospheres—resulting in enormous energy consumption and contributing significantly to global CO₂ emissions. Additionally, ammonia is primarily produced in large-scale industrial plants, leading to high distribution costs.
As an alternative, there is growing interest in an eco-friendly process that synthesizes ammonia using green hydrogen—produced via water electrolysis—under mild conditions (300°C, 10 atmospheres). However, developing catalysts that can achieve high ammonia productivity at such low temperatures and pressures is essential, as current technologies struggle to maintain efficiency under these conditions.
The research team developed a novel catalyst by incorporating ruthenium (Ru) nanoparticles and highly basic barium oxide (BaO) particles onto a conductive carbon surface, allowing it to function like a chemical capacitor*.
*Capacitor: A device that stores electrical energy by separating positive and negative charges.
During ammonia synthesis, hydrogen molecules (H₂) first dissociate into hydrogen atoms (H) on the ruthenium catalyst. These hydrogen atoms are further split into protons (H⁺) and electrons (e⁻). The study revealed that the acidic protons are stored in the strongly basic BaO, while the remaining electrons are separated and stored in ruthenium and carbon.
This unique chemical capacitor effect significantly enhances the ruthenium catalyst's electron density, accelerating nitrogen (N₂) dissociation—the rate-limiting step of ammonia synthesis—thereby dramatically increasing catalytic activity.
Furthermore, the team discovered that optimizing the nanostructure of the carbon material further boosts the electron density of ruthenium, maximizing catalytic performance. As a result, the new catalyst demonstrated over seven times higher ammonia synthesis performance compared to state-of-the-art catalysts under mild conditions (300°C, 10 atm).
< Schematic diagram showing the mechanism of ruthenium catalyst activity enhancement by barium oxide cocatalyst >
Professor Minkee Choi stated, “This research has garnered significant attention for demonstrating that catalytic activity can be greatly enhanced by controlling electron transfer within a thermal catalytic reaction system, not just in electrochemical processes.”
He further explained, “Our findings confirm that high-performance catalysts can enable efficient ammonia synthesis under low-temperature and low-pressure conditions. This could shift ammonia production from centralized, large-scale industrial plants to decentralized, small-scale production, making the hydrogen economy more sustainable and flexible.”
The study was led by Professor Minkee Choi as corresponding author and Yaejun Baik, a Ph.D. candidate, as first author. The research findings were published in Nature Catalysis on February 24.
(Paper title: “Electron and proton storage on separate Ru and BaO domains mediated by conductive low-work-function carbon to accelerate ammonia synthesis,” https://doi.org/10.1038/s41929-025-01302-z)
This research was supported by the Korea Institute of Energy Research and the National Research Foundation of Korea.
2025.03.11 View 3154 -
 KAIST perfectly reproduces Joseon-era Irworobongdo without pigments
Typically, chemical pigments that absorb specific wavelengths of light within the visible spectrum are required to produce colors. However, KAIST researchers have successfully reproduced the Joseon-era Irworobongdo [일월오봉도] painting using ultra-precise color graphics without any chemical pigments, allowing for the permanent and eco-friendly preservation of color graphics without fading or discoloration.
< (From left) Chaerim Son, a graduate of the Department of Biochemical Engineering (lead author), Seong Kyeong Nam, a graduate of the PhD program, Jiwoo Lee, a PhD student, and Professor Shin-Hyun Kim >
KAIST (represented by President Kwang Hyung Lee) announced on the 26th of February that a research team led by Professor Shinhyun Kim from the Department of Biological and Chemical Engineering had developed a technology that enables high-resolution color graphics without using any chemical pigments by employing hemisphere-shaped microstructures.
Morpho butterflies that are brilliant blue in color or Panther chameleons that change skin color exhibit coloration without chemical pigments, as ordered nanostructures within a material reflect visible light through optical interference. Since structural colors arise from physical structures rather than chemical substances, a single material can produce a wide range of colors.
However, the artificial implementation of structural coloration is highly challenging due to the complexity of creating ordered nanostructures. Additionally, it is difficult to produce a variety of colors and to pattern them precisely into complex designs.
< Figure 1. Principle of structural color expression using micro-hemispheres (left) and method of forming micro-hemisphere patterns based on photolithography (right) >
Professor Kim’s team overcame these challenges by using smooth-surfaced hemispherical microstructures instead of ordered nanostructures, enabling the high-precision patterning of diverse structural colors.
When light enters the inverted hemispherical microstructures, the portion of light entering from the sides undergoes total internal reflection along the curved surface, creating retroreflection. When the hemisphere diameter is approximately 10 micrometers (about one-tenth the thickness of a human hair), light traveling along different reflection paths interferes within the visible spectrum, producing structural coloration.
< Figure 2. “Irworobongdo”, the Painting of the Sun, Moon, and the Five Peaks, reproduced in fingernail size without pigment using approximately 200,000 micro-hemispheres >
The structural color can be tuned by adjusting the size of the hemispheres. By arranging hemispheres of varying sizes, much like mixing paints on a palette, an infinite range of colors can be generated.
To precisely pattern microscale hemispheres of different sizes, the research team employed photolithography* using positive photoresists** commonly used in semiconductor processing. They first patterned photoresists into micropillar structures, then induced reflow*** by heating the material, forming hemispherical microstructures.
*Photolithography: A technique used in semiconductor fabrication to pattern microscale structures.
**Positive photoresist: A photosensitive polymer that dissolves more easily in a developer solution after exposure to ultraviolet light.
***Reflow: A process in which a polymer material softens and reshapes into a curved structure when heated.
This method enables the formation of hemisphere-shaped microstructures with the desired sizes and colors in a single-step fabrication process. It also allows for the reproduction of arbitrary color graphics using a single material without any pigments.
The ultra-precise color graphics created with this technique can exhibit color variations depending on the angle of incident light or the viewing perspective. The pattern appears colored from one direction while remaining transparent from the opposite side, exhibiting a Janus effect. These structural color graphics achieve resolution comparable to cutting-edge LED displays, allowing complex color images to be captured within a fingernail-sized area and projected onto large screens.
< Figure 3. “Irworobongdo” that displays different shades depending on the angle of light and viewing direction >
Professor Shinhyun Kim, who led the research, stated, “Our newly developed pigment-free color graphics technology can serve as an innovative method for artistic expression, merging art with advanced materials. Additionally, it holds broad application potential in optical devices and sensors, anti-counterfeiting materials, aesthetic photocard printing, and many other fields.”
This research, with KAIST researcher Chaerim Son as the first author, was published in the prestigious materials science journal Advanced Materials on February 5.
(Paper title: “Retroreflective Multichrome Microdome Arrays Created by Single-Step Reflow”, DOI: 10.1002/adma.202413143 )
< Figure 4. Famous paintings reproduced without pigment: “Impression, Sunrise” (left), “Girl with a Pearl Earring” (right) >
The study was supported by the National Research Foundation of Korea through the Pioneer Converging Technology R&D Program and the Mid-Career Researcher Program.
2025.02.26 View 4365
KAIST perfectly reproduces Joseon-era Irworobongdo without pigments
Typically, chemical pigments that absorb specific wavelengths of light within the visible spectrum are required to produce colors. However, KAIST researchers have successfully reproduced the Joseon-era Irworobongdo [일월오봉도] painting using ultra-precise color graphics without any chemical pigments, allowing for the permanent and eco-friendly preservation of color graphics without fading or discoloration.
< (From left) Chaerim Son, a graduate of the Department of Biochemical Engineering (lead author), Seong Kyeong Nam, a graduate of the PhD program, Jiwoo Lee, a PhD student, and Professor Shin-Hyun Kim >
KAIST (represented by President Kwang Hyung Lee) announced on the 26th of February that a research team led by Professor Shinhyun Kim from the Department of Biological and Chemical Engineering had developed a technology that enables high-resolution color graphics without using any chemical pigments by employing hemisphere-shaped microstructures.
Morpho butterflies that are brilliant blue in color or Panther chameleons that change skin color exhibit coloration without chemical pigments, as ordered nanostructures within a material reflect visible light through optical interference. Since structural colors arise from physical structures rather than chemical substances, a single material can produce a wide range of colors.
However, the artificial implementation of structural coloration is highly challenging due to the complexity of creating ordered nanostructures. Additionally, it is difficult to produce a variety of colors and to pattern them precisely into complex designs.
< Figure 1. Principle of structural color expression using micro-hemispheres (left) and method of forming micro-hemisphere patterns based on photolithography (right) >
Professor Kim’s team overcame these challenges by using smooth-surfaced hemispherical microstructures instead of ordered nanostructures, enabling the high-precision patterning of diverse structural colors.
When light enters the inverted hemispherical microstructures, the portion of light entering from the sides undergoes total internal reflection along the curved surface, creating retroreflection. When the hemisphere diameter is approximately 10 micrometers (about one-tenth the thickness of a human hair), light traveling along different reflection paths interferes within the visible spectrum, producing structural coloration.
< Figure 2. “Irworobongdo”, the Painting of the Sun, Moon, and the Five Peaks, reproduced in fingernail size without pigment using approximately 200,000 micro-hemispheres >
The structural color can be tuned by adjusting the size of the hemispheres. By arranging hemispheres of varying sizes, much like mixing paints on a palette, an infinite range of colors can be generated.
To precisely pattern microscale hemispheres of different sizes, the research team employed photolithography* using positive photoresists** commonly used in semiconductor processing. They first patterned photoresists into micropillar structures, then induced reflow*** by heating the material, forming hemispherical microstructures.
*Photolithography: A technique used in semiconductor fabrication to pattern microscale structures.
**Positive photoresist: A photosensitive polymer that dissolves more easily in a developer solution after exposure to ultraviolet light.
***Reflow: A process in which a polymer material softens and reshapes into a curved structure when heated.
This method enables the formation of hemisphere-shaped microstructures with the desired sizes and colors in a single-step fabrication process. It also allows for the reproduction of arbitrary color graphics using a single material without any pigments.
The ultra-precise color graphics created with this technique can exhibit color variations depending on the angle of incident light or the viewing perspective. The pattern appears colored from one direction while remaining transparent from the opposite side, exhibiting a Janus effect. These structural color graphics achieve resolution comparable to cutting-edge LED displays, allowing complex color images to be captured within a fingernail-sized area and projected onto large screens.
< Figure 3. “Irworobongdo” that displays different shades depending on the angle of light and viewing direction >
Professor Shinhyun Kim, who led the research, stated, “Our newly developed pigment-free color graphics technology can serve as an innovative method for artistic expression, merging art with advanced materials. Additionally, it holds broad application potential in optical devices and sensors, anti-counterfeiting materials, aesthetic photocard printing, and many other fields.”
This research, with KAIST researcher Chaerim Son as the first author, was published in the prestigious materials science journal Advanced Materials on February 5.
(Paper title: “Retroreflective Multichrome Microdome Arrays Created by Single-Step Reflow”, DOI: 10.1002/adma.202413143 )
< Figure 4. Famous paintings reproduced without pigment: “Impression, Sunrise” (left), “Girl with a Pearl Earring” (right) >
The study was supported by the National Research Foundation of Korea through the Pioneer Converging Technology R&D Program and the Mid-Career Researcher Program.
2025.02.26 View 4365 -
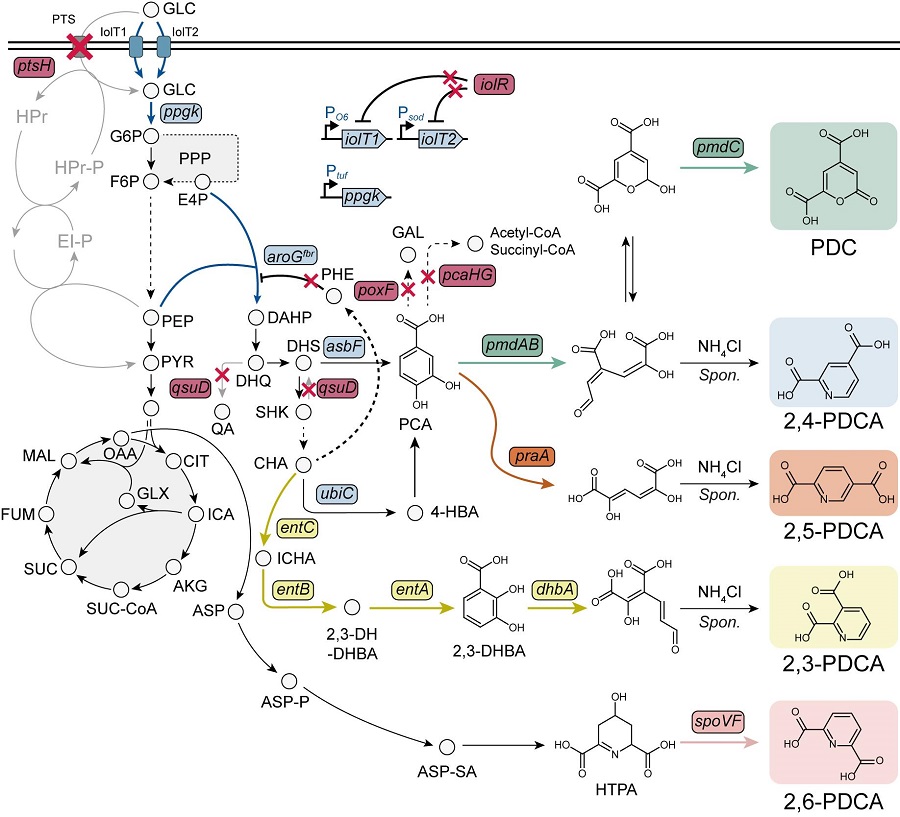 KAIST Researchers Suggest an Extraordinary Alternative to Petroleum-based PET - Bacteria!
< (From left) Dr. Cindy Pricilia, Ph.D. Candidate Cheon Woo Moon, Distinguished Professor Sang Yup Lee >
Currently, the world is suffering from environmental problems caused by plastic waste. The KAIST research team has succeeded in producing a microbial-based plastic that is biodegradable and can replace existing PET bottles, making it a hot topic.
Our university announced on the 7th of November that the research team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering has succeeded in developing a microbial strain that efficiently produces pseudoaromatic polyester monomer to replace polyethylene terephthalate (PET) using systems metabolic engineering.
Pseudoaromatic dicarboxylic acids have better physical properties and higher biodegradability than aromatic polyester (PET) when synthesized as polymers, and are attracting attention as an eco-friendly monomer* that can be synthesized into polymers. The production of pseudoaromatic dicarboxylic acids through chemical methods has the problems of low yield and selectivity, complex reaction conditions, and the generation of hazardous waste.
*Monomer: A material for making polymers, which is used to synthesize polymers by polymerizing monomers together
< Figure. Overview of pseudoaromatic dicarboxylic acid production using metabolically engineered C. glutamicum. >
To solve this problem, Professor Sang Yup Lee's research team used metabolic engineering to develop a microbial strain that efficiently produces five types of pseudoaromatic dicarboxylic acids, including 2-pyrone-4,6-dicarboxylic acid and four types of pyridine dicarboxylic acids (2,3-, 2,4-, 2,5-, 2,6-pyridine dicarboxylic acids), in Corynebacterium, a bacterium mainly used for amino acid production.
The research team used metabolic engineering techniques to build a platform microbial strain that enhances the metabolic flow of protocatechuic acid, which is used as a precursor for several pseudoaromatic dicarboxylic acids, and prevents the loss of precursors.
Based on this, the genetic manipulation target was discovered through transcriptome analysis, producing 76.17 g/L of 2-pyrone-4,6-dicarboxylic acid, and by newly discovering and constructing three types of pyridine dicarboxylic acid production metabolic pathways, successfully producing 2.79 g/L of 2,3-pyridine dicarboxylic acid, 0.49 g/L of 2,4-pyridine dicarboxylic acid, and 1.42 g/L of 2,5-pyridine dicarboxylic acid.
In addition, the research team confirmed the production of 15.01 g/L through the construction and reinforcement of the 2,6-pyridine dicarboxylic acid biosynthesis pathway, successfully producing a total of five similar aromatic dicarboxylic acids with high efficiency.
In conclusion, the team succeeded in producing 2,4-, 2,5-, and 2,6-pyridine dicarboxylic acids at the world's highest concentration. In particular, 2,4-, 2,5-pyridine dicarboxylic acid achieved production on the scale of g/L, which was previously produced in extremely small amounts (mg/L).
Based on this study, it is expected that it will be applied to various polyester production industrial processes, and it is also expected that it will be actively utilized in research on the production of similar aromatic polyesters.
Professor Sang Yup Lee, the corresponding author, said, “The significance lies in the fact that we have developed an eco-friendly technology that efficiently produces similar aromatic polyester monomers based on microorganisms,” and “This study will help the microorganism-based bio-monomer industry replace the petrochemical-based chemical industry in the future.”
The results of this study were published in the international academic journal, the Proceedings of the National Academy of Sciences of United States of America (PNAS) on October 30th.
※ Paper title: Metabolic engineering of Corynebacterium glutamicum for the production of pyrone and pyridine dicarboxylic acids
※ Author information: Jae Sung Cho (co-first author), Zi Wei Luo (co-first author), Cheon Woo Moon (co-first author), Cindy Prabowo (co-author), Sang Yup Lee (corresponding author) - a total of 5 people
This study was conducted with the support of the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project leader: Professor Sang Yup Lee) from the National Research Foundation supported by the Ministry of Science and Technology and ICT of Korea.
2024.11.08 View 9215
KAIST Researchers Suggest an Extraordinary Alternative to Petroleum-based PET - Bacteria!
< (From left) Dr. Cindy Pricilia, Ph.D. Candidate Cheon Woo Moon, Distinguished Professor Sang Yup Lee >
Currently, the world is suffering from environmental problems caused by plastic waste. The KAIST research team has succeeded in producing a microbial-based plastic that is biodegradable and can replace existing PET bottles, making it a hot topic.
Our university announced on the 7th of November that the research team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering has succeeded in developing a microbial strain that efficiently produces pseudoaromatic polyester monomer to replace polyethylene terephthalate (PET) using systems metabolic engineering.
Pseudoaromatic dicarboxylic acids have better physical properties and higher biodegradability than aromatic polyester (PET) when synthesized as polymers, and are attracting attention as an eco-friendly monomer* that can be synthesized into polymers. The production of pseudoaromatic dicarboxylic acids through chemical methods has the problems of low yield and selectivity, complex reaction conditions, and the generation of hazardous waste.
*Monomer: A material for making polymers, which is used to synthesize polymers by polymerizing monomers together
< Figure. Overview of pseudoaromatic dicarboxylic acid production using metabolically engineered C. glutamicum. >
To solve this problem, Professor Sang Yup Lee's research team used metabolic engineering to develop a microbial strain that efficiently produces five types of pseudoaromatic dicarboxylic acids, including 2-pyrone-4,6-dicarboxylic acid and four types of pyridine dicarboxylic acids (2,3-, 2,4-, 2,5-, 2,6-pyridine dicarboxylic acids), in Corynebacterium, a bacterium mainly used for amino acid production.
The research team used metabolic engineering techniques to build a platform microbial strain that enhances the metabolic flow of protocatechuic acid, which is used as a precursor for several pseudoaromatic dicarboxylic acids, and prevents the loss of precursors.
Based on this, the genetic manipulation target was discovered through transcriptome analysis, producing 76.17 g/L of 2-pyrone-4,6-dicarboxylic acid, and by newly discovering and constructing three types of pyridine dicarboxylic acid production metabolic pathways, successfully producing 2.79 g/L of 2,3-pyridine dicarboxylic acid, 0.49 g/L of 2,4-pyridine dicarboxylic acid, and 1.42 g/L of 2,5-pyridine dicarboxylic acid.
In addition, the research team confirmed the production of 15.01 g/L through the construction and reinforcement of the 2,6-pyridine dicarboxylic acid biosynthesis pathway, successfully producing a total of five similar aromatic dicarboxylic acids with high efficiency.
In conclusion, the team succeeded in producing 2,4-, 2,5-, and 2,6-pyridine dicarboxylic acids at the world's highest concentration. In particular, 2,4-, 2,5-pyridine dicarboxylic acid achieved production on the scale of g/L, which was previously produced in extremely small amounts (mg/L).
Based on this study, it is expected that it will be applied to various polyester production industrial processes, and it is also expected that it will be actively utilized in research on the production of similar aromatic polyesters.
Professor Sang Yup Lee, the corresponding author, said, “The significance lies in the fact that we have developed an eco-friendly technology that efficiently produces similar aromatic polyester monomers based on microorganisms,” and “This study will help the microorganism-based bio-monomer industry replace the petrochemical-based chemical industry in the future.”
The results of this study were published in the international academic journal, the Proceedings of the National Academy of Sciences of United States of America (PNAS) on October 30th.
※ Paper title: Metabolic engineering of Corynebacterium glutamicum for the production of pyrone and pyridine dicarboxylic acids
※ Author information: Jae Sung Cho (co-first author), Zi Wei Luo (co-first author), Cheon Woo Moon (co-first author), Cindy Prabowo (co-author), Sang Yup Lee (corresponding author) - a total of 5 people
This study was conducted with the support of the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project leader: Professor Sang Yup Lee) from the National Research Foundation supported by the Ministry of Science and Technology and ICT of Korea.
2024.11.08 View 9215