Bioengineering
-
 Formosa Group of Taiwan to Establish Bio R&D Center at KAIST Investing 12.5 M USD
KAIST (President Kwang-Hyung Lee) announced on February 17th that it signed an agreement for cooperation in the bio-medical field with Formosa Group, one of the three largest companies in Taiwan.
< Formosa Group Chairman Sandy Wang and KAIST President Kwang-Hyung Lee at the signing ceremony >
Formosa Group Executive Committee member and Chairman Sandy Wang, who leads the group's bio and eco-friendly energy sectors, decided to establish a bio-medical research center within KAIST and invest approximately KRW 18 billion or more over 5 years. In addition, to commercialize the research results, KAIST and Formosa Group will establish a joint venture in Korea with KAIST Holdings, a KAIST-funded company.
The cooperation between the two organizations began in early 2023 when KAIST signed a comprehensive exchange and cooperation agreement (MOU) with Ming Chi University of Science and Technology (明志科技大學), Chang Gung University (長庚大學), and Chang Gung Memorial Hospital (長庚記念醫院), which are established and supported by Formosa Group. Afterwards, Chairman Sandy Wang visited KAIST in May 2024 and signed a more specific business agreement (MOA).
KAIST Holdings is a holding company established by KAIST, a government-funded organization, to attract investment and conduct business, and will pursue the establishment of a joint venture with a 50:50 equity structure in cooperation with Formosa Group. KAIST Holdings will invest KAIST’s intellectual property rights, and Formosa Group will invest a corresponding amount of funds.
The KAIST-Formosa joint venture will provide research funds to the KAIST-Formosa Bio-Medical Research Center to be established in the future, secure the right to implement the intellectual property rights generated, and promote full-scale business.
The KAIST-Formosa Bio-Medical Research Center will establish a ‘brain organoid bank’ created by obtaining tissues from hundreds of patients with degenerative brain diseases, thereby securing high-dimensional data that will reveal the fundamental causes of aging and disease. It is expected that KAIST’s world-class artificial intelligence technology will analyze large-scale patient data to find the causes of aging and disease.
Through this business, it is expected that by 2030, five years from now, it will discover more than 10 types of intractable brain disease treatments and expand to more than 20 businesses, including human cell-centered diagnostics and preclinical businesses, and secure infrastructure and intellectual property rights that can create value worth approximately KRW 250 billion.
The Chang Gung Memorial Hospital in Taiwan has 10,000 beds and handles 35,000 patients per day, and systematically accumulates patient tissue and clinical data. Chang Gung Memorial Hospital will differentiate the tissues of patients with degenerative brain diseases and send them to the KAIST-Formosa Bio-Medical Research Center, which will then produce brain organoids to be used for disease research and new drug development. This will allow the world’s largest patient tissue data bank to be established.
Dean Daesoo Kim of the College of Life Science and Bioengineering at KAIST said, “This collaboration between KAIST and Formosa Group is a new research collaboration model that goes beyond joint research to establish a joint venture and global commercialization of developed technologies, and it is significant in that it can serve as an opportunity to promote biomedical research and development.”
With this agreement, KAIST, which has been promoting the KAIST Advanced Regenerative Medicine Engineering Center in Osong K-Bio Square, has secured a practical global partner.
< Representatives of the Formosa Group and KAIST >
KAIST’s Senior Vice President for Planning and Budget, Professor Kyung-Soo Kim emphasized, “KAIST has made great efforts to secure an edge in state-of-the-art biomedical fields such as stem cells and gene editing technology, by attracting the world’s best experts and discovering global cooperation partners, and these results can ultimately be linked to the Osong K-Bio Square project.”
SVP Kim then predicted, “In particular, the practical cooperation with Taiwan’s best Formosa Chang Gung Memorial Hospital, which has abundant clinical experience in stem cell treatment, will be an important axis of KAIST’s bio innovation strategy.”
Formosa Chairman Sandy Wang emphasized that this investment and cooperation is built on trust in KAIST’s R&D capabilities and the passion of its researchers. And added that through this, the Formosa Group will practice corporate social responsibility and take an important first step together with KAIST to protect the welfare and health of humanity. She also went on the say that she expects to see the cooperation expanded to various fields such as mobility and semiconductors based on the successes begotten from the cooperation in the bio field.
KAIST President Kwang-Hyung Lee said, “I evaluate this agreement as one of the most important events that will spearhead KAIST into overseas biotechnology stages,” and added, “I expect that this cooperation will be an opportunity for Taiwan and Korea, both of which have IT industry-centered structures, to create new growth engines in the bio industry.” Meanwhile, Formosa Group is a company founded by Chairman Sandy Wang’s father, Chairman Yung-Ching Wang. It is the world’s No. 1 plastic PVC producer and is leading core industries of the Taiwanese economy, including semiconductors, steel, heavy industry, bio, and batteries. Chairman Yung-Ching Wang was respected by the Taiwanese people for his exemplary return of wealth to society under the belief that the companies and assets he founded “belong to the people.”
2025.02.17 View 1874
Formosa Group of Taiwan to Establish Bio R&D Center at KAIST Investing 12.5 M USD
KAIST (President Kwang-Hyung Lee) announced on February 17th that it signed an agreement for cooperation in the bio-medical field with Formosa Group, one of the three largest companies in Taiwan.
< Formosa Group Chairman Sandy Wang and KAIST President Kwang-Hyung Lee at the signing ceremony >
Formosa Group Executive Committee member and Chairman Sandy Wang, who leads the group's bio and eco-friendly energy sectors, decided to establish a bio-medical research center within KAIST and invest approximately KRW 18 billion or more over 5 years. In addition, to commercialize the research results, KAIST and Formosa Group will establish a joint venture in Korea with KAIST Holdings, a KAIST-funded company.
The cooperation between the two organizations began in early 2023 when KAIST signed a comprehensive exchange and cooperation agreement (MOU) with Ming Chi University of Science and Technology (明志科技大學), Chang Gung University (長庚大學), and Chang Gung Memorial Hospital (長庚記念醫院), which are established and supported by Formosa Group. Afterwards, Chairman Sandy Wang visited KAIST in May 2024 and signed a more specific business agreement (MOA).
KAIST Holdings is a holding company established by KAIST, a government-funded organization, to attract investment and conduct business, and will pursue the establishment of a joint venture with a 50:50 equity structure in cooperation with Formosa Group. KAIST Holdings will invest KAIST’s intellectual property rights, and Formosa Group will invest a corresponding amount of funds.
The KAIST-Formosa joint venture will provide research funds to the KAIST-Formosa Bio-Medical Research Center to be established in the future, secure the right to implement the intellectual property rights generated, and promote full-scale business.
The KAIST-Formosa Bio-Medical Research Center will establish a ‘brain organoid bank’ created by obtaining tissues from hundreds of patients with degenerative brain diseases, thereby securing high-dimensional data that will reveal the fundamental causes of aging and disease. It is expected that KAIST’s world-class artificial intelligence technology will analyze large-scale patient data to find the causes of aging and disease.
Through this business, it is expected that by 2030, five years from now, it will discover more than 10 types of intractable brain disease treatments and expand to more than 20 businesses, including human cell-centered diagnostics and preclinical businesses, and secure infrastructure and intellectual property rights that can create value worth approximately KRW 250 billion.
The Chang Gung Memorial Hospital in Taiwan has 10,000 beds and handles 35,000 patients per day, and systematically accumulates patient tissue and clinical data. Chang Gung Memorial Hospital will differentiate the tissues of patients with degenerative brain diseases and send them to the KAIST-Formosa Bio-Medical Research Center, which will then produce brain organoids to be used for disease research and new drug development. This will allow the world’s largest patient tissue data bank to be established.
Dean Daesoo Kim of the College of Life Science and Bioengineering at KAIST said, “This collaboration between KAIST and Formosa Group is a new research collaboration model that goes beyond joint research to establish a joint venture and global commercialization of developed technologies, and it is significant in that it can serve as an opportunity to promote biomedical research and development.”
With this agreement, KAIST, which has been promoting the KAIST Advanced Regenerative Medicine Engineering Center in Osong K-Bio Square, has secured a practical global partner.
< Representatives of the Formosa Group and KAIST >
KAIST’s Senior Vice President for Planning and Budget, Professor Kyung-Soo Kim emphasized, “KAIST has made great efforts to secure an edge in state-of-the-art biomedical fields such as stem cells and gene editing technology, by attracting the world’s best experts and discovering global cooperation partners, and these results can ultimately be linked to the Osong K-Bio Square project.”
SVP Kim then predicted, “In particular, the practical cooperation with Taiwan’s best Formosa Chang Gung Memorial Hospital, which has abundant clinical experience in stem cell treatment, will be an important axis of KAIST’s bio innovation strategy.”
Formosa Chairman Sandy Wang emphasized that this investment and cooperation is built on trust in KAIST’s R&D capabilities and the passion of its researchers. And added that through this, the Formosa Group will practice corporate social responsibility and take an important first step together with KAIST to protect the welfare and health of humanity. She also went on the say that she expects to see the cooperation expanded to various fields such as mobility and semiconductors based on the successes begotten from the cooperation in the bio field.
KAIST President Kwang-Hyung Lee said, “I evaluate this agreement as one of the most important events that will spearhead KAIST into overseas biotechnology stages,” and added, “I expect that this cooperation will be an opportunity for Taiwan and Korea, both of which have IT industry-centered structures, to create new growth engines in the bio industry.” Meanwhile, Formosa Group is a company founded by Chairman Sandy Wang’s father, Chairman Yung-Ching Wang. It is the world’s No. 1 plastic PVC producer and is leading core industries of the Taiwanese economy, including semiconductors, steel, heavy industry, bio, and batteries. Chairman Yung-Ching Wang was respected by the Taiwanese people for his exemplary return of wealth to society under the belief that the companies and assets he founded “belong to the people.”
2025.02.17 View 1874 -
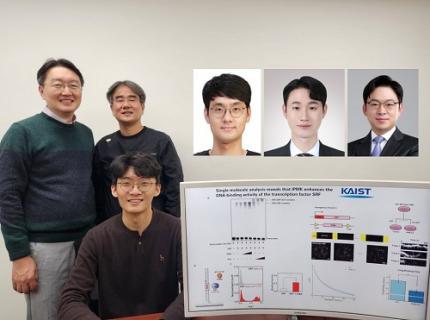 KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences>
A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for developing innovative therapeutic technologies.
On the 22nd of January, KAIST (represented by President Kwang Hyung Lee) announced that the joint research team led by Professors Seyun Kim, Gwangrog Lee, and Won-Ki Cho from the Department of Biological Sciences had uncovered essential mechanisms controlling gene expression in animal cells.
Inositol phosphate metabolites produced by inositol metabolism enzymes serve as vital secondary messengers in eukaryotic cell signaling systems and are broadly implicated in cancer, obesity, diabetes, and neurological disorders.
The research team demonstrated that the inositol polyphosphate multikinase (IPMK) enzyme, a key player in the inositol metabolism system, acts as a critical transcriptional activator within the core gene expression networks of animal cells. Notably, although IPMK was previously reported to play an important role in the transcription process governed by serum response factor (SRF), a representative transcription factor in animal cells, the precise mechanism of its action was unclear.
SRF is a transcription factor directly controlling the expression of at least 200–300 genes, regulating cell growth, proliferation, apoptosis, and motility, and is indispensable for organ development, such as in the heart.
The team discovered that IPMK binds directly to SRF, altering the three-dimensional structure of the SRF protein. This interaction facilitates the transcriptional activity of various genes through the SRF activated by IPMK, demonstrating that IPMK acts as a critical regulatory switch to enhance SRF's protein activity.
< Figure 1. The serum response factor (SRF) protein, a key transcription factor in animal cells, directly binds to inositol polyphosphate multikinase (IPMK) enzyme and undergoes structural change to acquire DNA binding ability, and precisely regulates growth and differentiation of animal cells through transcriptional activation. >
The team further verified that disruptions in the direct interaction between IPMK and SRF lead to the reduced functionality and activity of SRF, causing severe impairments in gene expression.
By highlighting the significance of the intrinsically disordered region (IDR) in SRF, the researchers underscored the biological importance of intrinsically disordered proteins (IDPs). Unlike most proteins that adopt distinct structures through folding, IDPs, including those with IDRs, do not exhibit specific structures but play crucial biological roles, attracting significant attention in the scientific community.
Professor Seyun Kim commented, "This study provides a vital mechanism proving that IPMK, a key enzyme in the inositol metabolism system, is a major transcriptional activator in the core gene expression network of animal cells. By understanding fundamental processes such as cancer development and metastasis, tissue differentiation from stem cells, and neural activation through SRF, we hope this discovery will lead to the broad application of innovative therapeutic technologies."
The findings were published on January 7th in the international journal Nucleic Acids Research (IF=16.7, top 1.8% in Biochemistry and Molecular Biology), under the title “Single-molecule analysis reveals that IPMK enhances the DNA-binding activity of the transcription factor SRF" (DOI: 10.1093/nar/gkae1281).
This research was supported by the National Research Foundation of Korea's Mid-career Research Program, Leading Research Center Program, and Global Research Laboratory Program, as well as by the Suh Kyungbae Science Foundation and the Samsung Future Technology Development Program.
2025.01.24 View 6932
KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences>
A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for developing innovative therapeutic technologies.
On the 22nd of January, KAIST (represented by President Kwang Hyung Lee) announced that the joint research team led by Professors Seyun Kim, Gwangrog Lee, and Won-Ki Cho from the Department of Biological Sciences had uncovered essential mechanisms controlling gene expression in animal cells.
Inositol phosphate metabolites produced by inositol metabolism enzymes serve as vital secondary messengers in eukaryotic cell signaling systems and are broadly implicated in cancer, obesity, diabetes, and neurological disorders.
The research team demonstrated that the inositol polyphosphate multikinase (IPMK) enzyme, a key player in the inositol metabolism system, acts as a critical transcriptional activator within the core gene expression networks of animal cells. Notably, although IPMK was previously reported to play an important role in the transcription process governed by serum response factor (SRF), a representative transcription factor in animal cells, the precise mechanism of its action was unclear.
SRF is a transcription factor directly controlling the expression of at least 200–300 genes, regulating cell growth, proliferation, apoptosis, and motility, and is indispensable for organ development, such as in the heart.
The team discovered that IPMK binds directly to SRF, altering the three-dimensional structure of the SRF protein. This interaction facilitates the transcriptional activity of various genes through the SRF activated by IPMK, demonstrating that IPMK acts as a critical regulatory switch to enhance SRF's protein activity.
< Figure 1. The serum response factor (SRF) protein, a key transcription factor in animal cells, directly binds to inositol polyphosphate multikinase (IPMK) enzyme and undergoes structural change to acquire DNA binding ability, and precisely regulates growth and differentiation of animal cells through transcriptional activation. >
The team further verified that disruptions in the direct interaction between IPMK and SRF lead to the reduced functionality and activity of SRF, causing severe impairments in gene expression.
By highlighting the significance of the intrinsically disordered region (IDR) in SRF, the researchers underscored the biological importance of intrinsically disordered proteins (IDPs). Unlike most proteins that adopt distinct structures through folding, IDPs, including those with IDRs, do not exhibit specific structures but play crucial biological roles, attracting significant attention in the scientific community.
Professor Seyun Kim commented, "This study provides a vital mechanism proving that IPMK, a key enzyme in the inositol metabolism system, is a major transcriptional activator in the core gene expression network of animal cells. By understanding fundamental processes such as cancer development and metastasis, tissue differentiation from stem cells, and neural activation through SRF, we hope this discovery will lead to the broad application of innovative therapeutic technologies."
The findings were published on January 7th in the international journal Nucleic Acids Research (IF=16.7, top 1.8% in Biochemistry and Molecular Biology), under the title “Single-molecule analysis reveals that IPMK enhances the DNA-binding activity of the transcription factor SRF" (DOI: 10.1093/nar/gkae1281).
This research was supported by the National Research Foundation of Korea's Mid-career Research Program, Leading Research Center Program, and Global Research Laboratory Program, as well as by the Suh Kyungbae Science Foundation and the Samsung Future Technology Development Program.
2025.01.24 View 6932 -
 A Way for Smartwatches to Detect Depression Risks Devised by KAIST and U of Michigan Researchers
- A international joint research team of KAIST and the University of Michigan developed a digital biomarker for predicting symptoms of depression based on data collected by smartwatches
- It has the potential to be used as a medical technology to replace the economically burdensome fMRI measurement test
- It is expected to expand the scope of digital health data analysis
The CORONA virus pandemic also brought about a pandemic of mental illness. Approximately one billion people worldwide suffer from various psychiatric conditions. Korea is one of more serious cases, with approximately 1.8 million patients exhibiting depression and anxiety disorders, and the total number of patients with clinical mental diseases has increased by 37% in five years to approximately 4.65 million. A joint research team from Korea and the US has developed a technology that uses biometric data collected through wearable devices to predict tomorrow's mood and, further, to predict the possibility of developing symptoms of depression.
< Figure 1. Schematic diagram of the research results. Based on the biometric data collected by a smartwatch, a mathematical algorithm that solves the inverse problem to estimate the brain's circadian phase and sleep stages has been developed. This algorithm can estimate the degrees of circadian disruption, and these estimates can be used as the digital biomarkers to predict depression risks. >
KAIST (President Kwang Hyung Lee) announced on the 15th of January that the research team under Professor Dae Wook Kim from the Department of Brain and Cognitive Sciences and the team under Professor Daniel B. Forger from the Department of Mathematics at the University of Michigan in the United States have developed a technology to predict symptoms of depression such as sleep disorders, depression, loss of appetite, overeating, and decreased concentration in shift workers from the activity and heart rate data collected from smartwatches.
According to WHO, a promising new treatment direction for mental illness focuses on the sleep and circadian timekeeping system located in the hypothalamus of the brain, which directly affect impulsivity, emotional responses, decision-making, and overall mood.
However, in order to measure endogenous circadian rhythms and sleep states, blood or saliva must be drawn every 30 minutes throughout the night to measure changes in the concentration of the melatonin hormone in our bodies and polysomnography (PSG) must be performed. As such treatments requires hospitalization and most psychiatric patients only visit for outpatient treatment, there has been no significant progress in developing treatment methods that take these two factors into account. In addition, the cost of the PSG test, which is approximately $1000, leaves mental health treatment considering sleep and circadian rhythms out of reach for the socially disadvantaged.
The solution to overcome these problems is to employ wearable devices for the easier collection of biometric data such as heart rate, body temperature, and activity level in real time without spatial constraints. However, current wearable devices have the limitation of providing only indirect information on biomarkers required by medical staff, such as the phase of the circadian clock.
The joint research team developed a filtering technology that accurately estimates the phase of the circadian clock, which changes daily, such as heart rate and activity time series data collected from a smartwatch. This is an implementation of a digital twin that precisely describes the circadian rhythm in the brain, and it can be used to estimate circadian rhythm disruption.
< Figure 2. The suprachiasmatic nucleus located in the hypothalamus of the brain is the central biological clock that regulates the 24-hour physiological rhythm and plays a key role in maintaining the body’s circadian rhythm. If the phase of this biological clock is disrupted, it affects various parts of the brain, which can cause psychiatric conditions such as depression. >
The possibility of using the digital twin of this circadian clock to predict the symptoms of depression was verified through collaboration with the research team of Professor Srijan Sen of the Michigan Neuroscience Institute and Professor Amy Bohnert of the Department of Psychiatry of the University of Michigan.
The collaborative research team conducted a large-scale prospective cohort study involving approximately 800 shift workers and showed that the circadian rhythm disruption digital biomarker estimated through the technology can predict tomorrow's mood as well as six symptoms, including sleep problems, appetite changes, decreased concentration, and suicidal thoughts, which are representative symptoms of depression.
< Figure 3. The circadian rhythm of hormones such as melatonin regulates various physiological functions and behaviors such as heart rate and activity level. These physiological and behavioral signals can be measured in daily life through wearable devices. In order to estimate the body’s circadian rhythm inversely based on the measured biometric signals, a mathematical algorithm is needed. This algorithm plays a key role in accurately identifying the characteristics of circadian rhythms by extracting hidden physiological patterns from biosignals. >
Professor Dae Wook Kim said, "It is very meaningful to be able to conduct research that provides a clue for ways to apply wearable biometric data using mathematics that have not previously been utilized for actual disease management." He added, "We expect that this research will be able to present continuous and non-invasive mental health monitoring technology. This is expected to present a new paradigm for mental health care. By resolving some of the major problems socially disadvantaged people may face in current treatment practices, they may be able to take more active steps when experiencing symptoms of depression, such as seeking counsel before things get out of hand."
< Figure 4. A mathematical algorithm was devised to circumvent the problems of estimating the phase of the brain's biological clock and sleep stages inversely from the biodata collected by a smartwatch. This algorithm can estimate the degree of daily circadian rhythm disruption, and this estimate can be used as a digital biomarker to predict depression symptoms. >
The results of this study, in which Professor Dae Wook Kim of the Department of Brain and Cognitive Sciences at KAIST participated as the joint first author and corresponding author, were published in the online version of the international academic journal npj Digital Medicine on December 5, 2024. (Paper title: The real-world association between digital markers of circadian disruption and mental health risks) DOI: 10.1038/s41746-024-01348-6
This study was conducted with the support of the KAIST's Research Support Program for New Faculty Members, the US National Science Foundation, the US National Institutes of Health, and the US Army Research Institute MURI Program.
2025.01.20 View 4063
A Way for Smartwatches to Detect Depression Risks Devised by KAIST and U of Michigan Researchers
- A international joint research team of KAIST and the University of Michigan developed a digital biomarker for predicting symptoms of depression based on data collected by smartwatches
- It has the potential to be used as a medical technology to replace the economically burdensome fMRI measurement test
- It is expected to expand the scope of digital health data analysis
The CORONA virus pandemic also brought about a pandemic of mental illness. Approximately one billion people worldwide suffer from various psychiatric conditions. Korea is one of more serious cases, with approximately 1.8 million patients exhibiting depression and anxiety disorders, and the total number of patients with clinical mental diseases has increased by 37% in five years to approximately 4.65 million. A joint research team from Korea and the US has developed a technology that uses biometric data collected through wearable devices to predict tomorrow's mood and, further, to predict the possibility of developing symptoms of depression.
< Figure 1. Schematic diagram of the research results. Based on the biometric data collected by a smartwatch, a mathematical algorithm that solves the inverse problem to estimate the brain's circadian phase and sleep stages has been developed. This algorithm can estimate the degrees of circadian disruption, and these estimates can be used as the digital biomarkers to predict depression risks. >
KAIST (President Kwang Hyung Lee) announced on the 15th of January that the research team under Professor Dae Wook Kim from the Department of Brain and Cognitive Sciences and the team under Professor Daniel B. Forger from the Department of Mathematics at the University of Michigan in the United States have developed a technology to predict symptoms of depression such as sleep disorders, depression, loss of appetite, overeating, and decreased concentration in shift workers from the activity and heart rate data collected from smartwatches.
According to WHO, a promising new treatment direction for mental illness focuses on the sleep and circadian timekeeping system located in the hypothalamus of the brain, which directly affect impulsivity, emotional responses, decision-making, and overall mood.
However, in order to measure endogenous circadian rhythms and sleep states, blood or saliva must be drawn every 30 minutes throughout the night to measure changes in the concentration of the melatonin hormone in our bodies and polysomnography (PSG) must be performed. As such treatments requires hospitalization and most psychiatric patients only visit for outpatient treatment, there has been no significant progress in developing treatment methods that take these two factors into account. In addition, the cost of the PSG test, which is approximately $1000, leaves mental health treatment considering sleep and circadian rhythms out of reach for the socially disadvantaged.
The solution to overcome these problems is to employ wearable devices for the easier collection of biometric data such as heart rate, body temperature, and activity level in real time without spatial constraints. However, current wearable devices have the limitation of providing only indirect information on biomarkers required by medical staff, such as the phase of the circadian clock.
The joint research team developed a filtering technology that accurately estimates the phase of the circadian clock, which changes daily, such as heart rate and activity time series data collected from a smartwatch. This is an implementation of a digital twin that precisely describes the circadian rhythm in the brain, and it can be used to estimate circadian rhythm disruption.
< Figure 2. The suprachiasmatic nucleus located in the hypothalamus of the brain is the central biological clock that regulates the 24-hour physiological rhythm and plays a key role in maintaining the body’s circadian rhythm. If the phase of this biological clock is disrupted, it affects various parts of the brain, which can cause psychiatric conditions such as depression. >
The possibility of using the digital twin of this circadian clock to predict the symptoms of depression was verified through collaboration with the research team of Professor Srijan Sen of the Michigan Neuroscience Institute and Professor Amy Bohnert of the Department of Psychiatry of the University of Michigan.
The collaborative research team conducted a large-scale prospective cohort study involving approximately 800 shift workers and showed that the circadian rhythm disruption digital biomarker estimated through the technology can predict tomorrow's mood as well as six symptoms, including sleep problems, appetite changes, decreased concentration, and suicidal thoughts, which are representative symptoms of depression.
< Figure 3. The circadian rhythm of hormones such as melatonin regulates various physiological functions and behaviors such as heart rate and activity level. These physiological and behavioral signals can be measured in daily life through wearable devices. In order to estimate the body’s circadian rhythm inversely based on the measured biometric signals, a mathematical algorithm is needed. This algorithm plays a key role in accurately identifying the characteristics of circadian rhythms by extracting hidden physiological patterns from biosignals. >
Professor Dae Wook Kim said, "It is very meaningful to be able to conduct research that provides a clue for ways to apply wearable biometric data using mathematics that have not previously been utilized for actual disease management." He added, "We expect that this research will be able to present continuous and non-invasive mental health monitoring technology. This is expected to present a new paradigm for mental health care. By resolving some of the major problems socially disadvantaged people may face in current treatment practices, they may be able to take more active steps when experiencing symptoms of depression, such as seeking counsel before things get out of hand."
< Figure 4. A mathematical algorithm was devised to circumvent the problems of estimating the phase of the brain's biological clock and sleep stages inversely from the biodata collected by a smartwatch. This algorithm can estimate the degree of daily circadian rhythm disruption, and this estimate can be used as a digital biomarker to predict depression symptoms. >
The results of this study, in which Professor Dae Wook Kim of the Department of Brain and Cognitive Sciences at KAIST participated as the joint first author and corresponding author, were published in the online version of the international academic journal npj Digital Medicine on December 5, 2024. (Paper title: The real-world association between digital markers of circadian disruption and mental health risks) DOI: 10.1038/s41746-024-01348-6
This study was conducted with the support of the KAIST's Research Support Program for New Faculty Members, the US National Science Foundation, the US National Institutes of Health, and the US Army Research Institute MURI Program.
2025.01.20 View 4063 -
 KAIST Proposes a New Way to Circumvent a Long-time Frustration in Neural Computing
The human brain begins learning through spontaneous random activities even before it receives sensory information from the external world. The technology developed by the KAIST research team enables much faster and more accurate learning when exposed to actual data by pre-learning random information in a brain-mimicking artificial neural network, and is expected to be a breakthrough in the development of brain-based artificial intelligence and neuromorphic computing technology in the future.
KAIST (President Kwang-Hyung Lee) announced on the 16th of December that Professor Se-Bum Paik 's research team in the Department of Brain Cognitive Sciences solved the weight transport problem*, a long-standing challenge in neural network learning, and through this, explained the principles that enable resource-efficient learning in biological brain neural networks.
*Weight transport problem: This is the biggest obstacle to the development of artificial intelligence that mimics the biological brain. It is the fundamental reason why large-scale memory and computational work are required in the learning of general artificial neural networks, unlike biological brains.
Over the past several decades, the development of artificial intelligence has been based on error backpropagation learning proposed by Geoffery Hinton, who won the Nobel Prize in Physics this year. However, error backpropagation learning was thought to be impossible in biological brains because it requires the unrealistic assumption that individual neurons must know all the connected information across multiple layers in order to calculate the error signal for learning.
< Figure 1. Illustration depicting the method of random noise training and its effects >
This difficult problem, called the weight transport problem, was raised by Francis Crick, who won the Nobel Prize in Physiology or Medicine for the discovery of the structure of DNA, after the error backpropagation learning was proposed by Hinton in 1986. Since then, it has been considered the reason why the operating principles of natural neural networks and artificial neural networks will forever be fundamentally different.
At the borderline of artificial intelligence and neuroscience, researchers including Hinton have continued to attempt to create biologically plausible models that can implement the learning principles of the brain by solving the weight transport problem.
In 2016, a joint research team from Oxford University and DeepMind in the UK first proposed the concept of error backpropagation learning being possible without weight transport, drawing attention from the academic world. However, biologically plausible error backpropagation learning without weight transport was inefficient, with slow learning speeds and low accuracy, making it difficult to apply in reality.
KAIST research team noted that the biological brain begins learning through internal spontaneous random neural activity even before experiencing external sensory experiences. To mimic this, the research team pre-trained a biologically plausible neural network without weight transport with meaningless random information (random noise).
As a result, they showed that the symmetry of the forward and backward neural cell connections of the neural network, which is an essential condition for error backpropagation learning, can be created. In other words, learning without weight transport is possible through random pre-training.
< Figure 2. Illustration depicting the meta-learning effect of random noise training >
The research team revealed that learning random information before learning actual data has the property of meta-learning, which is ‘learning how to learn.’ It was shown that neural networks that pre-learned random noise perform much faster and more accurate learning when exposed to actual data, and can achieve high learning efficiency without weight transport.
< Figure 3. Illustration depicting research on understanding the brain's operating principles through artificial neural networks >
Professor Se-Bum Paik said, “It breaks the conventional understanding of existing machine learning that only data learning is important, and provides a new perspective that focuses on the neuroscience principles of creating appropriate conditions before learning,” and added, “It is significant in that it solves important problems in artificial neural network learning through clues from developmental neuroscience, and at the same time provides insight into the brain’s learning principles through artificial neural network models.”
This study, in which Jeonghwan Cheon, a Master’s candidate of KAIST Department of Brain and Cognitive Sciences participated as the first author and Professor Sang Wan Lee of the same department as a co-author, was presented at the 38th Neural Information Processing Systems (NeurIPS), the world's top artificial intelligence conference, on December 14th in Vancouver, Canada. (Paper title: Pretraining with random noise for fast and robust learning without weight transport)
This study was conducted with the support of the National Research Foundation of Korea's Basic Research Program in Science and Engineering, the Information and Communications Technology Planning and Evaluation Institute's Talent Development Program, and the KAIST Singularity Professor Program.
2024.12.16 View 4654
KAIST Proposes a New Way to Circumvent a Long-time Frustration in Neural Computing
The human brain begins learning through spontaneous random activities even before it receives sensory information from the external world. The technology developed by the KAIST research team enables much faster and more accurate learning when exposed to actual data by pre-learning random information in a brain-mimicking artificial neural network, and is expected to be a breakthrough in the development of brain-based artificial intelligence and neuromorphic computing technology in the future.
KAIST (President Kwang-Hyung Lee) announced on the 16th of December that Professor Se-Bum Paik 's research team in the Department of Brain Cognitive Sciences solved the weight transport problem*, a long-standing challenge in neural network learning, and through this, explained the principles that enable resource-efficient learning in biological brain neural networks.
*Weight transport problem: This is the biggest obstacle to the development of artificial intelligence that mimics the biological brain. It is the fundamental reason why large-scale memory and computational work are required in the learning of general artificial neural networks, unlike biological brains.
Over the past several decades, the development of artificial intelligence has been based on error backpropagation learning proposed by Geoffery Hinton, who won the Nobel Prize in Physics this year. However, error backpropagation learning was thought to be impossible in biological brains because it requires the unrealistic assumption that individual neurons must know all the connected information across multiple layers in order to calculate the error signal for learning.
< Figure 1. Illustration depicting the method of random noise training and its effects >
This difficult problem, called the weight transport problem, was raised by Francis Crick, who won the Nobel Prize in Physiology or Medicine for the discovery of the structure of DNA, after the error backpropagation learning was proposed by Hinton in 1986. Since then, it has been considered the reason why the operating principles of natural neural networks and artificial neural networks will forever be fundamentally different.
At the borderline of artificial intelligence and neuroscience, researchers including Hinton have continued to attempt to create biologically plausible models that can implement the learning principles of the brain by solving the weight transport problem.
In 2016, a joint research team from Oxford University and DeepMind in the UK first proposed the concept of error backpropagation learning being possible without weight transport, drawing attention from the academic world. However, biologically plausible error backpropagation learning without weight transport was inefficient, with slow learning speeds and low accuracy, making it difficult to apply in reality.
KAIST research team noted that the biological brain begins learning through internal spontaneous random neural activity even before experiencing external sensory experiences. To mimic this, the research team pre-trained a biologically plausible neural network without weight transport with meaningless random information (random noise).
As a result, they showed that the symmetry of the forward and backward neural cell connections of the neural network, which is an essential condition for error backpropagation learning, can be created. In other words, learning without weight transport is possible through random pre-training.
< Figure 2. Illustration depicting the meta-learning effect of random noise training >
The research team revealed that learning random information before learning actual data has the property of meta-learning, which is ‘learning how to learn.’ It was shown that neural networks that pre-learned random noise perform much faster and more accurate learning when exposed to actual data, and can achieve high learning efficiency without weight transport.
< Figure 3. Illustration depicting research on understanding the brain's operating principles through artificial neural networks >
Professor Se-Bum Paik said, “It breaks the conventional understanding of existing machine learning that only data learning is important, and provides a new perspective that focuses on the neuroscience principles of creating appropriate conditions before learning,” and added, “It is significant in that it solves important problems in artificial neural network learning through clues from developmental neuroscience, and at the same time provides insight into the brain’s learning principles through artificial neural network models.”
This study, in which Jeonghwan Cheon, a Master’s candidate of KAIST Department of Brain and Cognitive Sciences participated as the first author and Professor Sang Wan Lee of the same department as a co-author, was presented at the 38th Neural Information Processing Systems (NeurIPS), the world's top artificial intelligence conference, on December 14th in Vancouver, Canada. (Paper title: Pretraining with random noise for fast and robust learning without weight transport)
This study was conducted with the support of the National Research Foundation of Korea's Basic Research Program in Science and Engineering, the Information and Communications Technology Planning and Evaluation Institute's Talent Development Program, and the KAIST Singularity Professor Program.
2024.12.16 View 4654 -
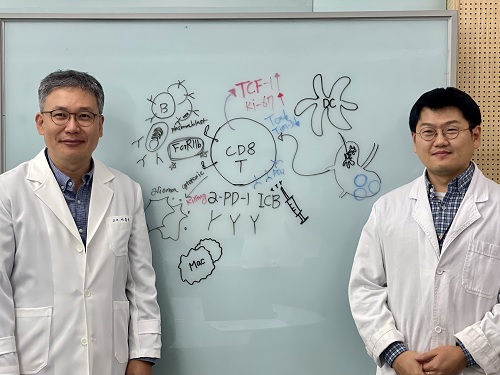 KAIST Unveils New Possibilities for Treating Intractable Brain Tumors
< Photo 1. (From left) Professor Heung Kyu Lee, KAIST Department of Biological Sciences, and Dr. Keun Bon Ku >
Immunotherapy, which enhances the immune system's T cell response to eliminate cancer cells, has emerged as a key approach in cancer treatment. However, in the case of glioblastoma, an aggressive and treatment-resistant brain tumor, numerous clinical trials have failed to confirm their efficacy. Korean researchers have recently analyzed the mechanisms that cause T cell exhaustion, which is characterized by a loss of function or a weakened response following prolonged exposure to antigens in such intractable cancers, identifying key control factors in T cell activation and clarifying the mechanisms that enhance therapeutic effectiveness.
KAIST (represented by President Kwang Hyung Lee) announced on the 6th of November that Professor Heung Kyu Lee’s team from the Department of Biological Sciences, in collaboration with the Korea Research Institute of Chemical Technology (represented by President Young Kuk Lee), has confirmed improved survival rates in a glioblastoma mouse model. By removing the inhibitory Fc gamma receptor (FcγRIIB), the research team was able to restore the responsiveness of cytotoxic T cells to immune checkpoint inhibitors, leading to enhanced anticancer activity.
The research team examined the effect of FcγRIIB, an inhibitory receptor recently found in cytotoxic T cells, on tumor-infiltrating T cells and the therapeutic effectiveness of the anti-PD-1 immune checkpoint inhibitor.
< Figure 1. Study results on improved survival rate due to increased antitumor activity of anti-PD-1 treatment in inhibitory Fc gamma receptor(Fcgr2b) ablation mice with murine glioblastoma. >
Their findings showed that deleting FcγRIIB induced the increase of tumor antigen-specific memory T cells, which helps to suppress exhaustion, enhances stem-like qualities, and reactivates T cell-mediated antitumor immunity, particularly in response to anti-PD-1 treatment. Furthermore, FcγRIIB deletion led to an increase in antigen-specific memory T cells that maintained continuous infiltration into the tumor tissue.
This study presents a new therapeutic target for tumors unresponsive to immune checkpoint inhibitors and demonstrates that combining FcγRIIB inhibition with anti-PD-1 treatment can produce synergistic effects, potentially improving therapeutic outcomes for tumors like glioblastoma, which typically show resistance to anti-PD-1 therapy.
< Figure 2. Overview of the study on the enhanced response to anti-PD-1 therapy for glioblastoma brain tumors upon deletion of the inhibitory Fc gamma receptor (FcγRIIB) in tumor microenvironment. When the inhibitory Fc gamma receptor (FcγRIIB) of cytotoxic T cells is deleted, an increase in tumor-specific memory T cells (Ttsms) was observed. In addition, this T cell subset is identified as originating from the tumor-draining lymph nodes(TdLNs) and leads to persistent infiltration into the tumor tissue. Anti-PD-1 therapy leads to an increased anti-tumor immune response via Ttsms, which is confirmed by increased tumor cell toxicity and increased cell division and decreased cell de-migration indices. Ultimately, the increased cytotoxic T cell immune response leads to an increase in the survival rate of glioblastoma. >
Professor Heung Kyu Lee explained, "This study offers a way to overcome clinical failures in treating brain tumors with immune checkpoint therapy and opens possibilities for broader applications to other intractable cancers. It also highlights the potential of utilizing cytotoxic T cells for tumor cell therapy."
The study, led by Dr. Keun Bon Ku of KAIST (currently a senior researcher at the Korea Research Institute of Chemical Technology's Center for Infectious Disease Diagnosis and Prevention), along with Chae Won Kim, Yumin Kim, Byeong Hoon Kang, Jeongwoo La, In Kang, Won Hyung Park, Stephen Ahn, and Sung Ki Lee, was published online on October 26 in the Journal for ImmunoTherapy of Cancer, an international journal in tumor immunology and therapy from the Society for Immunotherapy of Cancer. (Paper title: “Inhibitory Fcγ receptor deletion enhances CD8 T cell stemness increasing anti-PD-1 therapy responsiveness against glioblastoma,” http://dx.doi.org/10.1136/jitc-2024-009449).
This research received support from the National Research Foundation of Korea, the Bio & Medical Technology Development Program, and the Samsung Science & Technology Foundation.
2024.11.15 View 3270
KAIST Unveils New Possibilities for Treating Intractable Brain Tumors
< Photo 1. (From left) Professor Heung Kyu Lee, KAIST Department of Biological Sciences, and Dr. Keun Bon Ku >
Immunotherapy, which enhances the immune system's T cell response to eliminate cancer cells, has emerged as a key approach in cancer treatment. However, in the case of glioblastoma, an aggressive and treatment-resistant brain tumor, numerous clinical trials have failed to confirm their efficacy. Korean researchers have recently analyzed the mechanisms that cause T cell exhaustion, which is characterized by a loss of function or a weakened response following prolonged exposure to antigens in such intractable cancers, identifying key control factors in T cell activation and clarifying the mechanisms that enhance therapeutic effectiveness.
KAIST (represented by President Kwang Hyung Lee) announced on the 6th of November that Professor Heung Kyu Lee’s team from the Department of Biological Sciences, in collaboration with the Korea Research Institute of Chemical Technology (represented by President Young Kuk Lee), has confirmed improved survival rates in a glioblastoma mouse model. By removing the inhibitory Fc gamma receptor (FcγRIIB), the research team was able to restore the responsiveness of cytotoxic T cells to immune checkpoint inhibitors, leading to enhanced anticancer activity.
The research team examined the effect of FcγRIIB, an inhibitory receptor recently found in cytotoxic T cells, on tumor-infiltrating T cells and the therapeutic effectiveness of the anti-PD-1 immune checkpoint inhibitor.
< Figure 1. Study results on improved survival rate due to increased antitumor activity of anti-PD-1 treatment in inhibitory Fc gamma receptor(Fcgr2b) ablation mice with murine glioblastoma. >
Their findings showed that deleting FcγRIIB induced the increase of tumor antigen-specific memory T cells, which helps to suppress exhaustion, enhances stem-like qualities, and reactivates T cell-mediated antitumor immunity, particularly in response to anti-PD-1 treatment. Furthermore, FcγRIIB deletion led to an increase in antigen-specific memory T cells that maintained continuous infiltration into the tumor tissue.
This study presents a new therapeutic target for tumors unresponsive to immune checkpoint inhibitors and demonstrates that combining FcγRIIB inhibition with anti-PD-1 treatment can produce synergistic effects, potentially improving therapeutic outcomes for tumors like glioblastoma, which typically show resistance to anti-PD-1 therapy.
< Figure 2. Overview of the study on the enhanced response to anti-PD-1 therapy for glioblastoma brain tumors upon deletion of the inhibitory Fc gamma receptor (FcγRIIB) in tumor microenvironment. When the inhibitory Fc gamma receptor (FcγRIIB) of cytotoxic T cells is deleted, an increase in tumor-specific memory T cells (Ttsms) was observed. In addition, this T cell subset is identified as originating from the tumor-draining lymph nodes(TdLNs) and leads to persistent infiltration into the tumor tissue. Anti-PD-1 therapy leads to an increased anti-tumor immune response via Ttsms, which is confirmed by increased tumor cell toxicity and increased cell division and decreased cell de-migration indices. Ultimately, the increased cytotoxic T cell immune response leads to an increase in the survival rate of glioblastoma. >
Professor Heung Kyu Lee explained, "This study offers a way to overcome clinical failures in treating brain tumors with immune checkpoint therapy and opens possibilities for broader applications to other intractable cancers. It also highlights the potential of utilizing cytotoxic T cells for tumor cell therapy."
The study, led by Dr. Keun Bon Ku of KAIST (currently a senior researcher at the Korea Research Institute of Chemical Technology's Center for Infectious Disease Diagnosis and Prevention), along with Chae Won Kim, Yumin Kim, Byeong Hoon Kang, Jeongwoo La, In Kang, Won Hyung Park, Stephen Ahn, and Sung Ki Lee, was published online on October 26 in the Journal for ImmunoTherapy of Cancer, an international journal in tumor immunology and therapy from the Society for Immunotherapy of Cancer. (Paper title: “Inhibitory Fcγ receptor deletion enhances CD8 T cell stemness increasing anti-PD-1 therapy responsiveness against glioblastoma,” http://dx.doi.org/10.1136/jitc-2024-009449).
This research received support from the National Research Foundation of Korea, the Bio & Medical Technology Development Program, and the Samsung Science & Technology Foundation.
2024.11.15 View 3270 -
 Unraveling Mitochondrial DNA Mutations in Human Cells
Throughout our lifetime, cells accumulate DNA mutations, which contribute to genetic diversity, or “mosaicism”, among cells. These genomic mutations are pivotal for the aging process and the onset of various diseases, including cancer. Mitochondria, essential cellular organelles involved in energy metabolism and apoptosis, possess their own DNA, which are susceptible to mutations. However, studies on mtDNA mutations and mosaicism have been limited due to a variety of technical challenges.
Genomic scientists from KAIST have revealed the genetic mosaicism characterized by variations in mitochondrial DNA (mtDNA) across normal human cells. This study provides fundamental insights into understanding human aging and disease onset mechanisms.
The study, “Mitochondrial DNA mosaicism in normal human somatic cells,” was published in Nature Genetics on July 22. It was led by graduate student Jisong An under the supervision of Professor Young Seok Ju from the Graduate School of Medical Science and Engineering.
Researchers from Seoul National University College of Medicine, Yonsei University College of Medicine, Korea University College of Medicine, Washington University School of Medicine National Cancer Center, Seoul National University Hospital, Gangnam Severance Hospital and KAIST faculty startup company Inocras Inc. also participated in this study.
< Figure 1. a. Flow of experiment. b. Schematic diagram illustrating the origin and dynamics of mtDNA alterations across a lifetime. >
The study involved a bioinformatic analysis of whole-genome sequences from 2,096 single cells obtained from normal human colorectal epithelial tissue, fibroblasts, and blood collected from 31 individuals. The study highlights an average of three significant mtDNA differences between cells, with approximately 6% of these variations confirmed to be inherited as heteroplasmy from the mother.
Moreover, mutations significantly increased during tumorigenesis, with some mutations contributing to instability in mitochondrial RNA. Based on these findings, the study illustrates a computational model that comprehensively elucidates the evolution of mitochondria from embryonic development to aging and tumorigenesis.
This study systematically reveals the mechanisms behind mitochondrial DNA mosaicism in normal human cells, establishing a crucial foundation for understanding the impact of mtDNA on aging and disease onset.
Professor Ju remarked, “By systematically utilizing whole-genome big data, we can illuminate previously unknown phenomena in life sciences.” He emphasized the significance of the study, adding, “For the first time, we have established a method to systematically understand mitochondrial DNA changes occurring during human embryonic development, aging, and cancer development.”
This work was supported by the National Research Foundation of Korea and the Suh Kyungbae Foundation.
2024.07.24 View 3912
Unraveling Mitochondrial DNA Mutations in Human Cells
Throughout our lifetime, cells accumulate DNA mutations, which contribute to genetic diversity, or “mosaicism”, among cells. These genomic mutations are pivotal for the aging process and the onset of various diseases, including cancer. Mitochondria, essential cellular organelles involved in energy metabolism and apoptosis, possess their own DNA, which are susceptible to mutations. However, studies on mtDNA mutations and mosaicism have been limited due to a variety of technical challenges.
Genomic scientists from KAIST have revealed the genetic mosaicism characterized by variations in mitochondrial DNA (mtDNA) across normal human cells. This study provides fundamental insights into understanding human aging and disease onset mechanisms.
The study, “Mitochondrial DNA mosaicism in normal human somatic cells,” was published in Nature Genetics on July 22. It was led by graduate student Jisong An under the supervision of Professor Young Seok Ju from the Graduate School of Medical Science and Engineering.
Researchers from Seoul National University College of Medicine, Yonsei University College of Medicine, Korea University College of Medicine, Washington University School of Medicine National Cancer Center, Seoul National University Hospital, Gangnam Severance Hospital and KAIST faculty startup company Inocras Inc. also participated in this study.
< Figure 1. a. Flow of experiment. b. Schematic diagram illustrating the origin and dynamics of mtDNA alterations across a lifetime. >
The study involved a bioinformatic analysis of whole-genome sequences from 2,096 single cells obtained from normal human colorectal epithelial tissue, fibroblasts, and blood collected from 31 individuals. The study highlights an average of three significant mtDNA differences between cells, with approximately 6% of these variations confirmed to be inherited as heteroplasmy from the mother.
Moreover, mutations significantly increased during tumorigenesis, with some mutations contributing to instability in mitochondrial RNA. Based on these findings, the study illustrates a computational model that comprehensively elucidates the evolution of mitochondria from embryonic development to aging and tumorigenesis.
This study systematically reveals the mechanisms behind mitochondrial DNA mosaicism in normal human cells, establishing a crucial foundation for understanding the impact of mtDNA on aging and disease onset.
Professor Ju remarked, “By systematically utilizing whole-genome big data, we can illuminate previously unknown phenomena in life sciences.” He emphasized the significance of the study, adding, “For the first time, we have established a method to systematically understand mitochondrial DNA changes occurring during human embryonic development, aging, and cancer development.”
This work was supported by the National Research Foundation of Korea and the Suh Kyungbae Foundation.
2024.07.24 View 3912 -
 KAIST begins full-scale cooperation with Taiwan’s Formosa Group
< (From left) Senior Vice President for Planning and Budget Kyung-Soo Kim, and Professor Minee Choi of the Department of Brain and Cognitive Sciences of KAIST along with Chairman of Formosa Group Sandy Wang and KAIST President Kwang-Hyung Lee, and Dean Daesoo Kim of KAIST College of Life Science and Bioengineering >
KAIST is pursuing cooperation in the fields of advanced biotechnology and eco-friendly energy with Formosa Plastics Group, one of Taiwan's three largest companies.
To this end, Chairman Sandy Wang, a member of Formosa Group's standing committee and leader of the group's bio and eco-friendly energy sector, will visit KAIST on the 13th of this month. This is the first time that the owner of Formosa Group has made an official visit to KAIST.
Cooperation between the two institutions began last March when our university signed a memorandum of understanding on comprehensive exchange and cooperation with Ming Chi University of Science and Technology (明志科技大學), Chang Gung University(長庚大學), and Chang Gung Memorial Hospital(長庚記念醫院), three of many institutions established and supported by Formosa Group.
Based on this, Chairman Sandy Wang, who visits our university to promote more exchanges and cooperation, talked about ‘the education of children and corporate social return and practice of his father, Chairman Yung-Ching Wang,’ through a special lecture for the school leadership as a part of the Monthly Lecture on KAIST’s Leadership Innovation Day.
She then visited KAIST's research and engineering facilities related to Taiwan's future industries, such as advanced biotechnology and eco-friendly energy, and discussed global industry-academic cooperation plans. In the future, the two organizations plan to appoint adjunct professors and promote practical global cooperation, including joint student guidance and research cooperation. We plan to pursue effective mid- to long-term cooperation, such as conducting battery application research with the KAIST Next-Generation ESS Research Center and opening a graduate program specialized in stem cell and gene editing technology in connection with Chang Gung University and Chang Gung Memorial Hospital. The newly established cooperative relationship will also promote Formosa Group's investment and cooperation with KAIST's outstanding venture companies related to bio and eco-friendly energy to lay the foundation for innovative industrial cooperation between Taiwan and Korea.
President Kwang-Hyung Lee said, “The Formosa Group has a global network, so we regard it to be a key partner that will position KAIST’s bio and engineering technology in the global stages.” He also said, “With Chairman Sandy Wang’s visit, Taiwan is emerging as a global economic powerhouse,” and added, “We expect to continue our close cooperative relationship with the company.”
Formosa Group is a company founded by the late Chairman Yung-Ching Wang, the father of Chairman Sandy Wang. As the world's No. 1 plastic PVC producer, it is leading the core industries of Taiwan's economy, including semiconductors, steel, heavy industry, bio, and batteries. Chairman Yung-Ching Wang was respected by the Taiwanese people by setting an example of returning his wealth to society under the belief that the companies and assets he built ‘belonged to the people.’ Chang Gung University, Chang Gung Memorial Hospital, and Ming Chi University of Technology, which are pursuing cooperation with our university, were also established as part of the social contribution promoted by Chairman Yung-Ching Wang and are receiving financial support from Formosa Group.
2024.05.09 View 4323
KAIST begins full-scale cooperation with Taiwan’s Formosa Group
< (From left) Senior Vice President for Planning and Budget Kyung-Soo Kim, and Professor Minee Choi of the Department of Brain and Cognitive Sciences of KAIST along with Chairman of Formosa Group Sandy Wang and KAIST President Kwang-Hyung Lee, and Dean Daesoo Kim of KAIST College of Life Science and Bioengineering >
KAIST is pursuing cooperation in the fields of advanced biotechnology and eco-friendly energy with Formosa Plastics Group, one of Taiwan's three largest companies.
To this end, Chairman Sandy Wang, a member of Formosa Group's standing committee and leader of the group's bio and eco-friendly energy sector, will visit KAIST on the 13th of this month. This is the first time that the owner of Formosa Group has made an official visit to KAIST.
Cooperation between the two institutions began last March when our university signed a memorandum of understanding on comprehensive exchange and cooperation with Ming Chi University of Science and Technology (明志科技大學), Chang Gung University(長庚大學), and Chang Gung Memorial Hospital(長庚記念醫院), three of many institutions established and supported by Formosa Group.
Based on this, Chairman Sandy Wang, who visits our university to promote more exchanges and cooperation, talked about ‘the education of children and corporate social return and practice of his father, Chairman Yung-Ching Wang,’ through a special lecture for the school leadership as a part of the Monthly Lecture on KAIST’s Leadership Innovation Day.
She then visited KAIST's research and engineering facilities related to Taiwan's future industries, such as advanced biotechnology and eco-friendly energy, and discussed global industry-academic cooperation plans. In the future, the two organizations plan to appoint adjunct professors and promote practical global cooperation, including joint student guidance and research cooperation. We plan to pursue effective mid- to long-term cooperation, such as conducting battery application research with the KAIST Next-Generation ESS Research Center and opening a graduate program specialized in stem cell and gene editing technology in connection with Chang Gung University and Chang Gung Memorial Hospital. The newly established cooperative relationship will also promote Formosa Group's investment and cooperation with KAIST's outstanding venture companies related to bio and eco-friendly energy to lay the foundation for innovative industrial cooperation between Taiwan and Korea.
President Kwang-Hyung Lee said, “The Formosa Group has a global network, so we regard it to be a key partner that will position KAIST’s bio and engineering technology in the global stages.” He also said, “With Chairman Sandy Wang’s visit, Taiwan is emerging as a global economic powerhouse,” and added, “We expect to continue our close cooperative relationship with the company.”
Formosa Group is a company founded by the late Chairman Yung-Ching Wang, the father of Chairman Sandy Wang. As the world's No. 1 plastic PVC producer, it is leading the core industries of Taiwan's economy, including semiconductors, steel, heavy industry, bio, and batteries. Chairman Yung-Ching Wang was respected by the Taiwanese people by setting an example of returning his wealth to society under the belief that the companies and assets he built ‘belonged to the people.’ Chang Gung University, Chang Gung Memorial Hospital, and Ming Chi University of Technology, which are pursuing cooperation with our university, were also established as part of the social contribution promoted by Chairman Yung-Ching Wang and are receiving financial support from Formosa Group.
2024.05.09 View 4323 -
 A Korean research team develops a new clinical candidate for fatty liver disease
A team of Korean researchers have succeeded in developing a new drug candidate for the treatment of non-alcoholic fatty liver disease (NAFLD) acting on peripheral tissues. To date, there has not been an optimal treatment for non-alcoholic steatohepatitis (NASH), and this discovery is expected to set the grounds for the development of new drugs that can safely suppress both liver fat accumulation and liver fibrosis at the same time.
A joint research team led by Professor Jin Hee Ahn from Gwangju Institute of Science and Technology (GIST) and Professor Hail Kim from the KAIST Graduate School of Medical Science and Engineering developed a new chemical that can suppress disease-specific protein (HTR2A) through years of basic research. The team also revealed to have verified its efficacy and safety through preclinical tests (animal tests) at JD Bioscience Inc., a start-up company founded by Professor Ahn.
Although NAFLD has a prevalence rate as high as 20-30%, and about 5% of the global adult population suffers from NASH, there are no commercial drugs targeting them to date. NAFLD is a chronic disease that starts from the fatty liver and progresses into steatohepatitis, fibrosis, cirrhosis, and liver cancer. The mortality rate of patients increases with accompanied cardiovascular diseases and liver-related complications, and appropriate treatment in the early stage is hence necessary.
< Figure 1. Strategy and history of 5HT2A antagonists. Library and rational design for the development of compound 11c as a potent 5HT2A antagonist. Previous research efforts were discontinued due to limited oral absorption and safety. A therapeutic candidate to overcome this problem was identified and phase 1 clinical trials are currently in progress. >
The new synthetic chemical developed by the joint GIST-KAIST research is an innovative drug candidate that shows therapeutic effects on NASH based on a dual action mechanism that inhibits the accumulation of fat in the liver and liver fibrosis by suppressing the serotonin receptor protein 5HT2A.
The research team confirmed its therapeutic effects in animal models for NAFLD and NASH, in which hepatic steatosis and liver fibrosis* caused by fat accumulation in the liver were suppressed simultaneously by 50-70%.
*fibrosis: stiffening of parts of the liver, also used as a major indicator to track the prognosis of steatosis
The research team explained that the material was designed with optimal polarity and lipid affinity to minimize its permeability across the blood-brain barrier. It therefore does not affect the brain, and causes little side effects in the central nervous system (CNS) such as depression and suicidal ideations, while demonstrating excellent inhibition on its target protein present in tissues outside brain (IC50* = 14 nM). The team also demonstrated its superior efficacy in improving liver fibrosis when compared to similar drugs in the phase 3 clinical trial.
*IC50 (half maximal inhibitory concentration): the concentration at which a chemical suppresses 50% of a particular biological function
< Figure 2. GM-60106 (11c)'s effect on obesity: When GM-60106 was administered to an obese animal model (mice) for 2 months, body weight, body fat mass, and blood sugar were significantly reduced (a-d). In addition, the steatohepatitis level (NAFLD Activity Score) and the expression of genes of the treated mice involved in adipogenesis along with blood/liver fat decreased (e-h) >
Based on the pharmacological data obtained through preclinical trials, the team evaluated the effects of the drug on 88 healthy adults as part of their phase 1 clinical trial, where the side effects and the safe dosage of a drug are tested against healthy adults. Results showed no serious side effects and a good level of drug safety.
In addition, a preliminary efficacy evaluation on eight adults with steatohepatitis is currently underway.
Professor Jin Hee Ahn said, “The aim of this research is to develop a treatment for NASH with little side effects and guaranteed safety by developing a new target. The developed chemical is currently going through phase 1 of the global clinical trial in Australia through JD Bioscience Inc., a bio venture company for innovative drug development.” he added, “The candidate material the research team is currently developing shows not only a high level of safety and preventative effects by suppressing fat accumulation in the liver, but also a direct therapeutic effect on liver fibrosis. This is a strength that distinguishes our material from other competing drugs.”
< Figure 3. Efficacy of GM-60106 (11c) on liver fibrosis: When GM-60106 was administered to a steatohepatitis model (mice) for 3 months, the expression of genes associated with tissue fibrosis was significantly reduced (b-c). As a result of a detailed analysis of the tissues of the animal model, it was confirmed that the rate of tissue fibrosis was reduced and the expression rate of genes related to tissue fibrosis and inflammation was also significantly reduced (e-h). >
Professor Hail Kim from KAIST said, “Until now, this disease did not have a method of treatment other than weight control, and there has been no attempt to develop a drug that can be used for non-obese patients.” He added, “Through this research, we look forward to the development of various treatment techniques targeting a range of metabolic diseases including NASH that do not affect the weight of the patient.”
This study, conducted together by the research teams led by Professor Ahn from GIST and Professor Kim from KAIST, as well as the research team from JD Bioscience Inc., was supported by the Ministry of Science and ICT, and the National New Drug Development Project. The results of this research were published by Nature Communications on January 20.
The team also presented the results of their clinical study on the candidate material coded GM-60106 targeting metabolic abnormality-related MASH* at NASH-TAG Conference 2024, which was held in Utah for three days starting on January 4, which was selected as an excellent abstract.
*MASH (Metabolic Dysfunction-Associated Steatohepatitis): new replacement term for NASH
2024.02.21 View 7372
A Korean research team develops a new clinical candidate for fatty liver disease
A team of Korean researchers have succeeded in developing a new drug candidate for the treatment of non-alcoholic fatty liver disease (NAFLD) acting on peripheral tissues. To date, there has not been an optimal treatment for non-alcoholic steatohepatitis (NASH), and this discovery is expected to set the grounds for the development of new drugs that can safely suppress both liver fat accumulation and liver fibrosis at the same time.
A joint research team led by Professor Jin Hee Ahn from Gwangju Institute of Science and Technology (GIST) and Professor Hail Kim from the KAIST Graduate School of Medical Science and Engineering developed a new chemical that can suppress disease-specific protein (HTR2A) through years of basic research. The team also revealed to have verified its efficacy and safety through preclinical tests (animal tests) at JD Bioscience Inc., a start-up company founded by Professor Ahn.
Although NAFLD has a prevalence rate as high as 20-30%, and about 5% of the global adult population suffers from NASH, there are no commercial drugs targeting them to date. NAFLD is a chronic disease that starts from the fatty liver and progresses into steatohepatitis, fibrosis, cirrhosis, and liver cancer. The mortality rate of patients increases with accompanied cardiovascular diseases and liver-related complications, and appropriate treatment in the early stage is hence necessary.
< Figure 1. Strategy and history of 5HT2A antagonists. Library and rational design for the development of compound 11c as a potent 5HT2A antagonist. Previous research efforts were discontinued due to limited oral absorption and safety. A therapeutic candidate to overcome this problem was identified and phase 1 clinical trials are currently in progress. >
The new synthetic chemical developed by the joint GIST-KAIST research is an innovative drug candidate that shows therapeutic effects on NASH based on a dual action mechanism that inhibits the accumulation of fat in the liver and liver fibrosis by suppressing the serotonin receptor protein 5HT2A.
The research team confirmed its therapeutic effects in animal models for NAFLD and NASH, in which hepatic steatosis and liver fibrosis* caused by fat accumulation in the liver were suppressed simultaneously by 50-70%.
*fibrosis: stiffening of parts of the liver, also used as a major indicator to track the prognosis of steatosis
The research team explained that the material was designed with optimal polarity and lipid affinity to minimize its permeability across the blood-brain barrier. It therefore does not affect the brain, and causes little side effects in the central nervous system (CNS) such as depression and suicidal ideations, while demonstrating excellent inhibition on its target protein present in tissues outside brain (IC50* = 14 nM). The team also demonstrated its superior efficacy in improving liver fibrosis when compared to similar drugs in the phase 3 clinical trial.
*IC50 (half maximal inhibitory concentration): the concentration at which a chemical suppresses 50% of a particular biological function
< Figure 2. GM-60106 (11c)'s effect on obesity: When GM-60106 was administered to an obese animal model (mice) for 2 months, body weight, body fat mass, and blood sugar were significantly reduced (a-d). In addition, the steatohepatitis level (NAFLD Activity Score) and the expression of genes of the treated mice involved in adipogenesis along with blood/liver fat decreased (e-h) >
Based on the pharmacological data obtained through preclinical trials, the team evaluated the effects of the drug on 88 healthy adults as part of their phase 1 clinical trial, where the side effects and the safe dosage of a drug are tested against healthy adults. Results showed no serious side effects and a good level of drug safety.
In addition, a preliminary efficacy evaluation on eight adults with steatohepatitis is currently underway.
Professor Jin Hee Ahn said, “The aim of this research is to develop a treatment for NASH with little side effects and guaranteed safety by developing a new target. The developed chemical is currently going through phase 1 of the global clinical trial in Australia through JD Bioscience Inc., a bio venture company for innovative drug development.” he added, “The candidate material the research team is currently developing shows not only a high level of safety and preventative effects by suppressing fat accumulation in the liver, but also a direct therapeutic effect on liver fibrosis. This is a strength that distinguishes our material from other competing drugs.”
< Figure 3. Efficacy of GM-60106 (11c) on liver fibrosis: When GM-60106 was administered to a steatohepatitis model (mice) for 3 months, the expression of genes associated with tissue fibrosis was significantly reduced (b-c). As a result of a detailed analysis of the tissues of the animal model, it was confirmed that the rate of tissue fibrosis was reduced and the expression rate of genes related to tissue fibrosis and inflammation was also significantly reduced (e-h). >
Professor Hail Kim from KAIST said, “Until now, this disease did not have a method of treatment other than weight control, and there has been no attempt to develop a drug that can be used for non-obese patients.” He added, “Through this research, we look forward to the development of various treatment techniques targeting a range of metabolic diseases including NASH that do not affect the weight of the patient.”
This study, conducted together by the research teams led by Professor Ahn from GIST and Professor Kim from KAIST, as well as the research team from JD Bioscience Inc., was supported by the Ministry of Science and ICT, and the National New Drug Development Project. The results of this research were published by Nature Communications on January 20.
The team also presented the results of their clinical study on the candidate material coded GM-60106 targeting metabolic abnormality-related MASH* at NASH-TAG Conference 2024, which was held in Utah for three days starting on January 4, which was selected as an excellent abstract.
*MASH (Metabolic Dysfunction-Associated Steatohepatitis): new replacement term for NASH
2024.02.21 View 7372 -
 A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time.
On January 9, a joint research team led by Professor Won Do Heo from the KAIST Department of Biological Sciences, Professor Hyung-Bae Kwon from Johns Hopkins School of Medicine, and Professor Sangkyu Lee from the Institute for Basic Science (IBS) revealed that they have developed the world’s first technique to allow a real-time observation of synapse formation, extinction, and alterations.
Professor Heo’s team conjugated dimerization-dependent fluorescent proteins (ddFP) to synapses in order to observe the process in which synapses create connections between neurons in real time. The team named this technique SynapShot, by combining the words ‘synapse’ and snapshot’, and successfully tracked and observed the live formation and extinction processes of synapses as well as their dynamic changes.
< Figure 1. To observe dynamically changing synapses, dimerization-dependent fluorescent protein (ddFP) was expressed to observe flourescent signals upon synapse formation as ddFP enables fluorescence detection through reversible binding to pre- and postsynaptic terminals. >
Through a joint research project, the teams led by Professor Heo and Professor Sangkyu Lee at IBS together designed a SynapShot with green and red fluorescence, and were able to easily distinguish the synapse connecting two different neurons. Additionally, by combining an optogenetic technique that can control the function of a molecule using light, the team was able to observe the changes in the synapses while simultaneously inducing certain functions of the neurons using light.
Through more joint research with the team led by Professor Hyung-Bae Kwon at the Johns Hopkins School of Medicine, Professor Heo’s team induced several situations on live mice, including visual discrimination training, exercise, and anaesthesia, and used SynapShot to observe the changes in the synapses during each situation in real time. The observations revealed that each synapse could change fairly quickly and dynamically. This was the first-ever case in which the changes in synapses were observed in a live mammal.
< Figure 2. Microscopic photos observed through changes of the flourescence of the synapse sensor (SynapShot) by cultivating the neurons of an experimental rat and expressing the SynapShot. The changes in the synapse that is created when the pre- and post-synaptic terminals come into contact and the synapse that disappears after a certain period of time are measured by the fluorescence of the SynapShot. >
Professor Heo said, “Our group developed SynapShot through a collaboration with domestic and international research teams, and have opened up the possibility for first-hand live observations of the quick and dynamic changes of synapses, which was previously difficult to do. We expect this technique to revolutionize research methodology in the neurological field, and play an important role in brightening the future of brain science.”
This research, conducted by co-first authors Seungkyu Son (Ph.D. candidate), Jinsu Lee (Ph.D. candidate) and Dr. Kanghoon Jung from Johns Hopkins, was published in the online edition of Nature Methods on January 8 under the title “Real-time visualization of structural dynamics of synapses in live cells in vivo”, and will be printed in the February volume.
< Figure 3. Simultaneous use of green-SynapShot and red-SynapShot to distinguish and observe synapses with one post-terminal and different pre-terminals. >
< Figure 4. Dimer-dependent fluorescent protein (ddFP) exists as a green fluorescent protein as well as a red fluorescent protein, and can be applied together with blue light-activated optogenetic technology. After activating Tropomyosin receptor kinase B (TrkB) by blue light using optogenetic technology, the strengthening of synaptic connections through signals of brain-derived neurotrophic factor is observed using red-SynapShot. >
< Figure 5. Micrographs showing real-time changing synapses in the visual cortex of mice trained through visual training using in vivo imaging techniques such as two-photon microscopy as well as at the cellular level. >
This research was supported by Mid-Sized Research Funds and the Singularity Project from KAIST, and by IBS.
2024.01.18 View 5570
A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time.
On January 9, a joint research team led by Professor Won Do Heo from the KAIST Department of Biological Sciences, Professor Hyung-Bae Kwon from Johns Hopkins School of Medicine, and Professor Sangkyu Lee from the Institute for Basic Science (IBS) revealed that they have developed the world’s first technique to allow a real-time observation of synapse formation, extinction, and alterations.
Professor Heo’s team conjugated dimerization-dependent fluorescent proteins (ddFP) to synapses in order to observe the process in which synapses create connections between neurons in real time. The team named this technique SynapShot, by combining the words ‘synapse’ and snapshot’, and successfully tracked and observed the live formation and extinction processes of synapses as well as their dynamic changes.
< Figure 1. To observe dynamically changing synapses, dimerization-dependent fluorescent protein (ddFP) was expressed to observe flourescent signals upon synapse formation as ddFP enables fluorescence detection through reversible binding to pre- and postsynaptic terminals. >
Through a joint research project, the teams led by Professor Heo and Professor Sangkyu Lee at IBS together designed a SynapShot with green and red fluorescence, and were able to easily distinguish the synapse connecting two different neurons. Additionally, by combining an optogenetic technique that can control the function of a molecule using light, the team was able to observe the changes in the synapses while simultaneously inducing certain functions of the neurons using light.
Through more joint research with the team led by Professor Hyung-Bae Kwon at the Johns Hopkins School of Medicine, Professor Heo’s team induced several situations on live mice, including visual discrimination training, exercise, and anaesthesia, and used SynapShot to observe the changes in the synapses during each situation in real time. The observations revealed that each synapse could change fairly quickly and dynamically. This was the first-ever case in which the changes in synapses were observed in a live mammal.
< Figure 2. Microscopic photos observed through changes of the flourescence of the synapse sensor (SynapShot) by cultivating the neurons of an experimental rat and expressing the SynapShot. The changes in the synapse that is created when the pre- and post-synaptic terminals come into contact and the synapse that disappears after a certain period of time are measured by the fluorescence of the SynapShot. >
Professor Heo said, “Our group developed SynapShot through a collaboration with domestic and international research teams, and have opened up the possibility for first-hand live observations of the quick and dynamic changes of synapses, which was previously difficult to do. We expect this technique to revolutionize research methodology in the neurological field, and play an important role in brightening the future of brain science.”
This research, conducted by co-first authors Seungkyu Son (Ph.D. candidate), Jinsu Lee (Ph.D. candidate) and Dr. Kanghoon Jung from Johns Hopkins, was published in the online edition of Nature Methods on January 8 under the title “Real-time visualization of structural dynamics of synapses in live cells in vivo”, and will be printed in the February volume.
< Figure 3. Simultaneous use of green-SynapShot and red-SynapShot to distinguish and observe synapses with one post-terminal and different pre-terminals. >
< Figure 4. Dimer-dependent fluorescent protein (ddFP) exists as a green fluorescent protein as well as a red fluorescent protein, and can be applied together with blue light-activated optogenetic technology. After activating Tropomyosin receptor kinase B (TrkB) by blue light using optogenetic technology, the strengthening of synaptic connections through signals of brain-derived neurotrophic factor is observed using red-SynapShot. >
< Figure 5. Micrographs showing real-time changing synapses in the visual cortex of mice trained through visual training using in vivo imaging techniques such as two-photon microscopy as well as at the cellular level. >
This research was supported by Mid-Sized Research Funds and the Singularity Project from KAIST, and by IBS.
2024.01.18 View 5570 -
 KAIST-UCSD researchers build an enzyme discovering AI
- A joint research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Bernhard Palsson of UCSD developed ‘DeepECtransformer’, an artificial intelligence that can predict Enzyme Commission (EC) number of proteins.
- The AI is tasked to discover new enzymes that have not been discovered yet, which would allow prediction for a total of 5,360 types of Enzyme Commission (EC) numbers
- It is expected to be used in the development of microbial cell factories that produce environmentally friendly chemicals as a core technology for analyzing the metabolic network of a genome.
While E. coli is one of the most studied organisms, the function of 30% of proteins that make up E. coli has not yet been clearly revealed. For this, an artificial intelligence was used to discover 464 types of enzymes from the proteins that were unknown, and the researchers went on to verify the predictions of 3 types of proteins were successfully identified through in vitro enzyme assay.
KAIST (President Kwang-Hyung Lee) announced on the 24th that a joint research team comprised of Gi Bae Kim, Ji Yeon Kim, Dr. Jong An Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Dr. Charles J. Norsigian and Professor Bernhard O. Palsson of the Department of Bioengineering at UCSD has developed DeepECtransformer, an artificial intelligence that can predict the enzyme functions from the protein sequence, and has established a prediction system by utilizing the AI to quickly and accurately identify the enzyme function.
Enzymes are proteins that catalyze biological reactions, and identifying the function of each enzyme is essential to understanding the various chemical reactions that exist in living organisms and the metabolic characteristics of those organisms. Enzyme Commission (EC) number is an enzyme function classification system designed by the International Union of Biochemistry and Molecular Biology, and in order to understand the metabolic characteristics of various organisms, it is necessary to develop a technology that can quickly analyze enzymes and EC numbers of the enzymes present in the genome.
Various methodologies based on deep learning have been developed to analyze the features of biological sequences, including protein function prediction, but most of them have a problem of a black box, where the inference process of AI cannot be interpreted. Various prediction systems that utilize AI for enzyme function prediction have also been reported, but they do not solve this black box problem, or cannot interpret the reasoning process in fine-grained level (e.g., the level of amino acid residues in the enzyme sequence).
The joint team developed DeepECtransformer, an AI that utilizes deep learning and a protein homology analysis module to predict the enzyme function of a given protein sequence. To better understand the features of protein sequences, the transformer architecture, which is commonly used in natural language processing, was additionally used to extract important features about enzyme functions in the context of the entire protein sequence, which enabled the team to accurately predict the EC number of the enzyme. The developed DeepECtransformer can predict a total of 5360 EC numbers.
The joint team further analyzed the transformer architecture to understand the inference process of DeepECtransformer, and found that in the inference process, the AI utilizes information on catalytic active sites and/or the cofactor binding sites which are important for enzyme function. By analyzing the black box of DeepECtransformer, it was confirmed that the AI was able to identify the features that are important for enzyme function on its own during the learning process.
"By utilizing the prediction system we developed, we were able to predict the functions of enzymes that had not yet been identified and verify them experimentally," said Gi Bae Kim, the first author of the paper. "By using DeepECtransformer to identify previously unknown enzymes in living organisms, we will be able to more accurately analyze various facets involved in the metabolic processes of organisms, such as the enzymes needed to biosynthesize various useful compounds or the enzymes needed to biodegrade plastics." he added.
"DeepECtransformer, which quickly and accurately predicts enzyme functions, is a key technology in functional genomics, enabling us to analyze the function of entire enzymes at the systems level," said Professor Sang Yup Lee. He added, “We will be able to use it to develop eco-friendly microbial factories based on comprehensive genome-scale metabolic models, potentially minimizing missing information of metabolism.”
The joint team’s work on DeepECtransformer is described in the paper titled "Functional annotation of enzyme-encoding genes using deep learning with transformer layers" written by Gi Bae Kim, Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST and their colleagues. The paper was published via peer-review on the 14th of November on “Nature Communications”.
This research was conducted with the support by “the Development of next-generation biorefinery platform technologies for leading bio-based chemicals industry project (2022M3J5A1056072)” and by “Development of platform technologies of microbial cell factories for the next-generation biorefineries project (2022M3J5A1056117)” from National Research Foundation supported by the Korean Ministry of Science and ICT (Project Leader: Distinguished Professor Sang Yup Lee, KAIST).
< Figure 1. The structure of DeepECtransformer's artificial neural network >
2023.11.24 View 4984
KAIST-UCSD researchers build an enzyme discovering AI
- A joint research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Bernhard Palsson of UCSD developed ‘DeepECtransformer’, an artificial intelligence that can predict Enzyme Commission (EC) number of proteins.
- The AI is tasked to discover new enzymes that have not been discovered yet, which would allow prediction for a total of 5,360 types of Enzyme Commission (EC) numbers
- It is expected to be used in the development of microbial cell factories that produce environmentally friendly chemicals as a core technology for analyzing the metabolic network of a genome.
While E. coli is one of the most studied organisms, the function of 30% of proteins that make up E. coli has not yet been clearly revealed. For this, an artificial intelligence was used to discover 464 types of enzymes from the proteins that were unknown, and the researchers went on to verify the predictions of 3 types of proteins were successfully identified through in vitro enzyme assay.
KAIST (President Kwang-Hyung Lee) announced on the 24th that a joint research team comprised of Gi Bae Kim, Ji Yeon Kim, Dr. Jong An Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Dr. Charles J. Norsigian and Professor Bernhard O. Palsson of the Department of Bioengineering at UCSD has developed DeepECtransformer, an artificial intelligence that can predict the enzyme functions from the protein sequence, and has established a prediction system by utilizing the AI to quickly and accurately identify the enzyme function.
Enzymes are proteins that catalyze biological reactions, and identifying the function of each enzyme is essential to understanding the various chemical reactions that exist in living organisms and the metabolic characteristics of those organisms. Enzyme Commission (EC) number is an enzyme function classification system designed by the International Union of Biochemistry and Molecular Biology, and in order to understand the metabolic characteristics of various organisms, it is necessary to develop a technology that can quickly analyze enzymes and EC numbers of the enzymes present in the genome.
Various methodologies based on deep learning have been developed to analyze the features of biological sequences, including protein function prediction, but most of them have a problem of a black box, where the inference process of AI cannot be interpreted. Various prediction systems that utilize AI for enzyme function prediction have also been reported, but they do not solve this black box problem, or cannot interpret the reasoning process in fine-grained level (e.g., the level of amino acid residues in the enzyme sequence).
The joint team developed DeepECtransformer, an AI that utilizes deep learning and a protein homology analysis module to predict the enzyme function of a given protein sequence. To better understand the features of protein sequences, the transformer architecture, which is commonly used in natural language processing, was additionally used to extract important features about enzyme functions in the context of the entire protein sequence, which enabled the team to accurately predict the EC number of the enzyme. The developed DeepECtransformer can predict a total of 5360 EC numbers.
The joint team further analyzed the transformer architecture to understand the inference process of DeepECtransformer, and found that in the inference process, the AI utilizes information on catalytic active sites and/or the cofactor binding sites which are important for enzyme function. By analyzing the black box of DeepECtransformer, it was confirmed that the AI was able to identify the features that are important for enzyme function on its own during the learning process.
"By utilizing the prediction system we developed, we were able to predict the functions of enzymes that had not yet been identified and verify them experimentally," said Gi Bae Kim, the first author of the paper. "By using DeepECtransformer to identify previously unknown enzymes in living organisms, we will be able to more accurately analyze various facets involved in the metabolic processes of organisms, such as the enzymes needed to biosynthesize various useful compounds or the enzymes needed to biodegrade plastics." he added.
"DeepECtransformer, which quickly and accurately predicts enzyme functions, is a key technology in functional genomics, enabling us to analyze the function of entire enzymes at the systems level," said Professor Sang Yup Lee. He added, “We will be able to use it to develop eco-friendly microbial factories based on comprehensive genome-scale metabolic models, potentially minimizing missing information of metabolism.”
The joint team’s work on DeepECtransformer is described in the paper titled "Functional annotation of enzyme-encoding genes using deep learning with transformer layers" written by Gi Bae Kim, Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST and their colleagues. The paper was published via peer-review on the 14th of November on “Nature Communications”.
This research was conducted with the support by “the Development of next-generation biorefinery platform technologies for leading bio-based chemicals industry project (2022M3J5A1056072)” and by “Development of platform technologies of microbial cell factories for the next-generation biorefineries project (2022M3J5A1056117)” from National Research Foundation supported by the Korean Ministry of Science and ICT (Project Leader: Distinguished Professor Sang Yup Lee, KAIST).
< Figure 1. The structure of DeepECtransformer's artificial neural network >
2023.11.24 View 4984 -
 KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 7678
KAIST presents a microbial cell factory as a source of eco-friendly food and cosmetic coloring
Despite decades of global population growth, global food crisis seems to be at hand yet again because the food productivity is cut severely due to prolonged presence of abnormal weather from intensifying climate change and global food supply chain is deteriorated due to international conflicts such as wars exacerbating food shortages and nutritional inequality around the globe. At the same time, however, as awareness of the environment and sustainability rises, an increase in demand for more eco-friendly and high-quality food and beauty products is being observed not without a sense of irony. At a time like this, microorganisms are attracting attention as a key that can handle this couple of seemingly distant problems.
KAIST (President Kwang-Hyung Lee) announced on the 26th that Kyeong Rok Choi, a research professor of the Bioprocess Research Center and Sang Yup Lee, a Distinguished Professor of the Department of Chemical and Biomolecular Engineering, published a paper titled “Metabolic Engineering of Microorganisms for Food and Cosmetics Production” upon invitation by “Nature Reviews Bioengineering” to be published online published by Nature after peer review.
※ Paper title: Systems metabolic engineering of microorganisms for food and cosmetics production
※ Author information: Kyeong Rok Choi (first author) and Sang Yup Lee (corresponding author)
Systems metabolic engineering is a research field founded by Distinguished Professor Sang Yup Lee of KAIST to more effectively develop microbial cell factories, the core factor of the next-generation bio industry to replace the existing chemical industry that relies heavily on petroleum. By applying a systemic metabolic engineering strategy, the researchers have developed a number of high-performance microbial cell factories that produce a variety of food and cosmetic compounds including natural substances like heme and zinc protoporphyrin IX compounds which can improve the flavor and color of synthetic meat, lycopene and β-carotene which are functional natural pigments that can be widely used in food and cosmetics, and methyl anthranilate, a grape-derived compound widely used to impart grape flavor in food and beverage manufacturing.
In this paper written upon invitation by Nature, the research team covered remarkable cases of microbial cell factory that can produce amino acids, proteins, fats and fatty acids, vitamins, flavors, pigments, alcohols, functional compounds and other food additives used in various foods and cosmetics and the companies that have successfully commercialized these microbial-derived materials Furthermore, the paper organized and presents systems metabolic engineering strategies that can spur the development of industrial microbial cell factories that can produce more diverse food and cosmetic compounds in an eco-friendly way with economic feasibility.
< Figure 1. Examples of production of food and cosmetic compounds using microbial cell factories >
For example, by producing proteins or amino acids with high nutritional value through non-edible biomass used as animal feed or fertilizer through the microbial fermentation process, it will contribute to the increase in production and stable supply of food around the world. Furthermore, by contributing to developing more viable alternative meat, further reducing dependence on animal protein, it can also contribute to reducing greenhouse gases and environmental pollution generated through livestock breeding or fish farming.
In addition, vanillin or methyl anthranilate, which give off vanilla or grape flavor, are widely added to various foods, but natural products isolated and refined from plants are low in production and high in production cost, so in most cases, petrochemicals substances derived from vanillin and methylanthranilic acid are added to food. These materials can also be produced through an eco-friendly and human-friendly method by borrowing the power of microorganisms.
Ethical and resource problems that arise in producing compounds like Calmin (cochineal pigment), a coloring added to various cosmetics and foods such as red lipstick and strawberry-flavored milk, which must be extracted from cochineal insects that live only in certain cacti. and Hyaluronic acid, which is widely consumed as a health supplement, but is only present in omega-3 fatty acids extracted from shark or fish livers, can also be resolved when they can be produced in an eco-friendly way using microorganisms.
KAIST Research Professor Kyeong Rok Choi, the first author of this paper, said, “In addition to traditional fermented foods such as kimchi and yogurt, foods produced with the help of microorganisms like cocoa butter, a base ingredient for chocolate that can only be obtained from fermented cacao beans, and monosodium glutamate, a seasoning produced through microbial fermentation are already familiar to us”. “In the future, we will be able to acquire a wider variety of foods and cosmetics even more easily produced in an eco-friendly and sustainable way in our daily lives through microbial cell factories.” he added.
< Figure 2. Systems metabolic engineering strategy to improve metabolic flow in microbial cell factories >
Distinguished Professor Sang Yup Lee said, “It is engineers’ mission to make the world a better place utilizing science and technology.” and added, “Continuous advancement and active use of systems metabolic engineering will contribute greatly to easing and resolving the problems arising from both the food crisis and the climate change."
This research was carried out as a part of the “Development of Protein Production Technology from Inorganic Substances through Control of Microbial Metabolism System Project” (Project Leader: Kyeong Rok Choi, KAIST Research Professor) of the the Center for Agricultural Microorganism and Enzyme (Director Pahn-Shick Chang) supported by the Rural Development Administration and the “Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project” (Project Leader: Sang Yup Lee, KAIST Distinguished Professor) of the Petroleum-Substitute Eco-friendly Chemical Technology Development Program supported by the Ministry of Science and ICT.
2023.07.28 View 7678 -
 KAIST researchers discovers the neural circuit that reacts to alarm clock
KAIST (President Kwang Hyung Lee) announced on the 20th that a research team led by Professor Daesoo Kim of the Department of Brain and Cognitive Sciences and Dr. Jeongjin Kim 's team from the Korea Institute of Science and Technology (KIST) have identified the principle of awakening animals by responding to sounds even while sleeping.
Sleep is a very important physiological process that organizes brain activity and maintains health. During sleep, the function of sensory nerves is blocked, so the ability to detect danger in the proximity is reduced. However, many animals detect approaching predators and respond even while sleeping. Scientists thought that animals ready for danger by alternating between deep sleep and light sleep.
A research team led by Professor Daesoo Kim at KAIST discovered that animals have neural circuits that respond to sounds even during deep sleep. While awake, the medial geniculate thalamus responds to sounds, but during deep sleep, or Non-REM sleep, the Mediodorsal thalamus responds to sounds to wake up the brain.
As a result of the study, when the rats fell into deep sleep, the nerves of the medial geniculate thalamus were also sleeping, but the nerves of mediodorsal thalamus were awake and responded immediately to sounds. In addition, it was observed that when mediodorsal thalamus was inhibited, the rats could not wake up even when a sound was heard, and when the mediodorsal thalamus was stimulated, the rats woke up within a few seconds without sound.
This is the first study to show that sleep and wakefulness can transmit auditory signals through different neural circuits, and was reported in the international journal, Current Biology on February 7, and was highlighted by Nature. (https://www.nature.com/articles/d41586-023-00354-0)
Professor Daesoo Kim explained, “The findings of this study can used in developing digital healthcare technologies to be used to improve understanding of disorders of senses and wakefulness seen in various brain diseases and to control the senses in the future.”
This research was carried out with the support from the National Research Foundation of Korea's Mid-Career Research Foundation Program.
Figure 1. Traditionally, sound signals were thought to be propagated from the auditory nerve to the auditory thalamus. However, while in slow-wave sleep, the auditory nerve sends sound signals to the mediodorsal thalamic neurons via the brainstem nerve to induce arousal in the brain.
Figure 2. GRIK4 dorsomedial nerve in response to sound stimulation. The awakening effect is induced as the activity of the GRIK4 dorsal medial nerve increases based on the time when sound stimulation is given.
2023.03.03 View 5389
KAIST researchers discovers the neural circuit that reacts to alarm clock
KAIST (President Kwang Hyung Lee) announced on the 20th that a research team led by Professor Daesoo Kim of the Department of Brain and Cognitive Sciences and Dr. Jeongjin Kim 's team from the Korea Institute of Science and Technology (KIST) have identified the principle of awakening animals by responding to sounds even while sleeping.
Sleep is a very important physiological process that organizes brain activity and maintains health. During sleep, the function of sensory nerves is blocked, so the ability to detect danger in the proximity is reduced. However, many animals detect approaching predators and respond even while sleeping. Scientists thought that animals ready for danger by alternating between deep sleep and light sleep.
A research team led by Professor Daesoo Kim at KAIST discovered that animals have neural circuits that respond to sounds even during deep sleep. While awake, the medial geniculate thalamus responds to sounds, but during deep sleep, or Non-REM sleep, the Mediodorsal thalamus responds to sounds to wake up the brain.
As a result of the study, when the rats fell into deep sleep, the nerves of the medial geniculate thalamus were also sleeping, but the nerves of mediodorsal thalamus were awake and responded immediately to sounds. In addition, it was observed that when mediodorsal thalamus was inhibited, the rats could not wake up even when a sound was heard, and when the mediodorsal thalamus was stimulated, the rats woke up within a few seconds without sound.
This is the first study to show that sleep and wakefulness can transmit auditory signals through different neural circuits, and was reported in the international journal, Current Biology on February 7, and was highlighted by Nature. (https://www.nature.com/articles/d41586-023-00354-0)
Professor Daesoo Kim explained, “The findings of this study can used in developing digital healthcare technologies to be used to improve understanding of disorders of senses and wakefulness seen in various brain diseases and to control the senses in the future.”
This research was carried out with the support from the National Research Foundation of Korea's Mid-Career Research Foundation Program.
Figure 1. Traditionally, sound signals were thought to be propagated from the auditory nerve to the auditory thalamus. However, while in slow-wave sleep, the auditory nerve sends sound signals to the mediodorsal thalamic neurons via the brainstem nerve to induce arousal in the brain.
Figure 2. GRIK4 dorsomedial nerve in response to sound stimulation. The awakening effect is induced as the activity of the GRIK4 dorsal medial nerve increases based on the time when sound stimulation is given.
2023.03.03 View 5389