Jae-Byum+Chang
-
 KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures
- Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS)
- Expected to have price competitiveness over bio-templating method as it expands the range of biological samples
- Expected to expand the range of application of nanostructure synthesis technology by utilizing various biological structures
< Photo 1. (From left) Professor Yeon Sik Jung, Ph.D. candidate Dae-Hyeon Song, Professor Jae-Byum Chang, and (from top right) Dr. Chang Woo Song and Dr. Seunghee H. Cho of the Department of Materials Science and Engineering >
Biological structures have complex characteristics that are difficult to replicate artificially, but biotemplating methods* that directly utilize these biological structures have been used in various fields of application. The KAIST research team succeeded in utilizing previously unusable biological structures and expanding the areas in which biotemplate methods can be applied.
*Biotemplating: A method of using biotemplates as a mold to create functional structural materials, utilizing the functions of these biological structures, from viruses to the tissues and organs that make up our bodies
KAIST (President Kwang Hyung Lee) announced on the 10th that a joint research team of Professors Jae-Byum Chang and Professor Yeon Sik Jung of the Department of Materials Science and Engineering developed a biotemplating method that utilizes specific intracellular proteins in biological samples and has high tunability.
Existing biotemplate methods mainly utilize only the external surface of biological samples or have limitations in utilizing the structure-function correlation of various biological structures due to limited dimensions and sample sizes, making it difficult to create functional nanostructures.
To solve this problem, the research team studied a way to utilize various biological structures within the cells while retaining high tunability.
< Figure 1. CamBio utilizing microtubules, a intracellular protein structure. The silver nanoparticle chains synthesized along the microtubules that span the entire cell interior can be observed through an electron microscope, and it is shown that this can be used as a successful SERS substrate. >
As a result of the research, the team developed the “Conversion to advanced materials via labeled Biostructure”, shortened as “CamBio”, which enables the selective synthesis of nanostructures with various characteristics and sizes from specific protein structures composed of diverse proteins within biological specimens.
The CamBio method secures high tunability of functional nanostructures that can be manufactured from biological samples by merging various manufacturing and biological technologies.
Through the technology of repeatedly attaching antibodies, arranging cells in a certain shape, and thinly slicing tissue, the functional nanostructures made with CamBio showed improved performance on the surface-enhanced Raman spectroscopy (SERS)* substrate used for material detection.
*Surface-enhanced Raman spectroscopy (SERS): A technology that can detect very small amounts of substances using light, based on the principle that specific substances react to light and amplifies signals on surfaces of metals such as gold or silver.
The research team found that the nanoparticle chains made using the intracellular protein structures through the process of repeated labeling with antibodies allowed easier control, and improved SERS performance by up to 230%.
In addition, the research team expanded from utilizing the structures inside cells to obtaining samples of muscle tissues inside meat using a cryostat and successfully producing a substrate with periodic bands made of metal particles by performing the CamBio process. This method of producing a substrate not only allows large-scale production using biological samples, but also shows that it is a cost-effective method.
< Figure 2. A method for securing tunability using CamBio at the cell level. Examples of controlling characteristics by integrating iterative labeling and cell pattering techniques with CamBio are shown. >
The CamBio developed by the research team is expected to be used as a way to solve problems faced by various research fields as it is to expand the range of bio-samples that can be produced for various usage.
The first author, Dae-Hyeon Song, a Ph.D. candidate of KAIST Department of Materials Science and Engineering said, “Through CamBio, we have comprehensively accumulated biotemplating methods that can utilize more diverse protein structures,” and “If combined with the state-of-the-art biological technologies such as gene editing and 3D bioprinting and new material synthesis technologies, biostructures can be utilized in various fields of application.”
< Figure 3. A method for securing tunability using CamBio at the tissue level. In order to utilize proteins inside muscle tissue, the frozen tissue sectioning technology is combined, and through this, a substrate with a periodic nanoparticle band pattern is successfully produced, and it is shown that large-area acquisition of samples and price competitiveness can be achieved. >
This study, in which the Ph.D. candidate Dae-Hyeon Song along with Dr. Chang Woo Song, and Dr. Seunghee H. Cho of the same department participated as the first authors, was published online in the international academic journal, Advanced Science, on November 13th, 2024.
(Paper title: Highly Tunable, Nanomaterial-Functionalized Structural Templating of Intracellular Protein Structures Within Biological Species) https://doi.org/10.1002/advs.202406492
This study was conducted with a combination of support from various programs including the National Convergence Research of Scientific Challenges (National Research Foundation of Korea (NRF) 2024), Engineering Reseach Center (ERC) (Wearable Platform Materials Technology Center, NRF 2023), ERC (Global Bio-integrated Materials Center, NRF 2024), and the National Advanced Program for Biological Research Resources (Bioimaging Data Curation Center, NRF 2024) funded by Ministry of Science and ICT.
2025.01.10 View 6211
KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures
- Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS)
- Expected to have price competitiveness over bio-templating method as it expands the range of biological samples
- Expected to expand the range of application of nanostructure synthesis technology by utilizing various biological structures
< Photo 1. (From left) Professor Yeon Sik Jung, Ph.D. candidate Dae-Hyeon Song, Professor Jae-Byum Chang, and (from top right) Dr. Chang Woo Song and Dr. Seunghee H. Cho of the Department of Materials Science and Engineering >
Biological structures have complex characteristics that are difficult to replicate artificially, but biotemplating methods* that directly utilize these biological structures have been used in various fields of application. The KAIST research team succeeded in utilizing previously unusable biological structures and expanding the areas in which biotemplate methods can be applied.
*Biotemplating: A method of using biotemplates as a mold to create functional structural materials, utilizing the functions of these biological structures, from viruses to the tissues and organs that make up our bodies
KAIST (President Kwang Hyung Lee) announced on the 10th that a joint research team of Professors Jae-Byum Chang and Professor Yeon Sik Jung of the Department of Materials Science and Engineering developed a biotemplating method that utilizes specific intracellular proteins in biological samples and has high tunability.
Existing biotemplate methods mainly utilize only the external surface of biological samples or have limitations in utilizing the structure-function correlation of various biological structures due to limited dimensions and sample sizes, making it difficult to create functional nanostructures.
To solve this problem, the research team studied a way to utilize various biological structures within the cells while retaining high tunability.
< Figure 1. CamBio utilizing microtubules, a intracellular protein structure. The silver nanoparticle chains synthesized along the microtubules that span the entire cell interior can be observed through an electron microscope, and it is shown that this can be used as a successful SERS substrate. >
As a result of the research, the team developed the “Conversion to advanced materials via labeled Biostructure”, shortened as “CamBio”, which enables the selective synthesis of nanostructures with various characteristics and sizes from specific protein structures composed of diverse proteins within biological specimens.
The CamBio method secures high tunability of functional nanostructures that can be manufactured from biological samples by merging various manufacturing and biological technologies.
Through the technology of repeatedly attaching antibodies, arranging cells in a certain shape, and thinly slicing tissue, the functional nanostructures made with CamBio showed improved performance on the surface-enhanced Raman spectroscopy (SERS)* substrate used for material detection.
*Surface-enhanced Raman spectroscopy (SERS): A technology that can detect very small amounts of substances using light, based on the principle that specific substances react to light and amplifies signals on surfaces of metals such as gold or silver.
The research team found that the nanoparticle chains made using the intracellular protein structures through the process of repeated labeling with antibodies allowed easier control, and improved SERS performance by up to 230%.
In addition, the research team expanded from utilizing the structures inside cells to obtaining samples of muscle tissues inside meat using a cryostat and successfully producing a substrate with periodic bands made of metal particles by performing the CamBio process. This method of producing a substrate not only allows large-scale production using biological samples, but also shows that it is a cost-effective method.
< Figure 2. A method for securing tunability using CamBio at the cell level. Examples of controlling characteristics by integrating iterative labeling and cell pattering techniques with CamBio are shown. >
The CamBio developed by the research team is expected to be used as a way to solve problems faced by various research fields as it is to expand the range of bio-samples that can be produced for various usage.
The first author, Dae-Hyeon Song, a Ph.D. candidate of KAIST Department of Materials Science and Engineering said, “Through CamBio, we have comprehensively accumulated biotemplating methods that can utilize more diverse protein structures,” and “If combined with the state-of-the-art biological technologies such as gene editing and 3D bioprinting and new material synthesis technologies, biostructures can be utilized in various fields of application.”
< Figure 3. A method for securing tunability using CamBio at the tissue level. In order to utilize proteins inside muscle tissue, the frozen tissue sectioning technology is combined, and through this, a substrate with a periodic nanoparticle band pattern is successfully produced, and it is shown that large-area acquisition of samples and price competitiveness can be achieved. >
This study, in which the Ph.D. candidate Dae-Hyeon Song along with Dr. Chang Woo Song, and Dr. Seunghee H. Cho of the same department participated as the first authors, was published online in the international academic journal, Advanced Science, on November 13th, 2024.
(Paper title: Highly Tunable, Nanomaterial-Functionalized Structural Templating of Intracellular Protein Structures Within Biological Species) https://doi.org/10.1002/advs.202406492
This study was conducted with a combination of support from various programs including the National Convergence Research of Scientific Challenges (National Research Foundation of Korea (NRF) 2024), Engineering Reseach Center (ERC) (Wearable Platform Materials Technology Center, NRF 2023), ERC (Global Bio-integrated Materials Center, NRF 2024), and the National Advanced Program for Biological Research Resources (Bioimaging Data Curation Center, NRF 2024) funded by Ministry of Science and ICT.
2025.01.10 View 6211 -
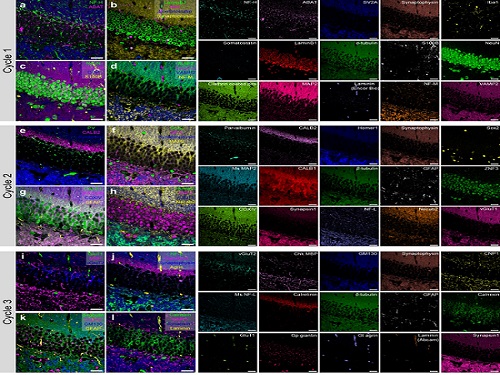 PICASSO Technique Drives Biological Molecules into Technicolor
The new imaging approach brings current imaging colors from four to more than 15 for mapping overlapping proteins
Pablo Picasso’s surreal cubist artistic style shifted common features into unrecognizable scenes, but a new imaging approach bearing his namesake may elucidate the most complicated subject: the brain. Employing artificial intelligence to clarify spectral color blending of tiny molecules used to stain specific proteins and other items of research interest, the PICASSO technique, allows researchers to use more than 15 colors to image and parse our overlapping proteins.
The PICASSO developers, based in Korea, published their approach on May 5 in Nature Communications.
Fluorophores — the staining molecules — emit specific colors when excited by a light, but if more than four fluorophores are used, their emitted colors overlap and blend. Researchers previously developed techniques to correct this spectral overlap by precisely defining the matrix of mixed and unmixed images. This measurement depends on reference spectra, found by identifying clear images of only one fluorophore-stained specimen or of multiple, identically prepared specimens that only contain a single fluorophore each.
“Such reference spectra measurement could be complicated to perform in highly heterogeneous specimens, such as the brain, due to the highly varied emission spectra of fluorophores depending on the subregions from which the spectra were measured,” said co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST. He explained that the subregions would each need their own spectra reference measurements, making for an inefficient, time-consuming process. “To address this problem, we developed an approach that does not require reference spectra measurements.”
The approach is the “Process of ultra-multiplexed Imaging of biomolecules viA the unmixing of the Signals of Spectrally Overlapping fluorophores,” also known as PICASSO. Ultra-multiplexed imaging refers to visualizing the numerous individual components of a unit. Like a cinema multiplex in which each theater plays a different movie, each protein in a cell has a different role. By staining with fluorophores, researchers can begin to understand those roles.
“We devised a strategy based on information theory; unmixing is performed by iteratively minimizing the mutual information between mixed images,” said co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering, KAIST. “This allows us to get away with the assumption that the spatial distribution of different proteins is mutually exclusive and enables accurate information unmixing.”
To demonstrate PICASSO’s capabilities, the researchers applied the technique to imaging a mouse brain. With a single round of staining, they performed 15-color multiplexed imaging of a mouse brain. Although small, mouse brains are still complex, multifaceted organs that can take significant resources to map. According to the researchers, PICASSO can improve the capabilities of other imaging techniques and allow for the use of even more fluorophore colors.
Using one such imaging technique in combination with PICASSO, the team achieved 45-color multiplexed imaging of the mouse brain in only three staining and imaging cycles, according to Yoon.
“PICASSO is a versatile tool for the multiplexed biomolecule imaging of cultured cells, tissue slices and clinical specimens,” Chang said. “We anticipate that PICASSO will be useful for a broad range of applications for which biomolecules’ spatial information is important. One such application the tool would be useful for is revealing the cellular heterogeneities of tumor microenvironments, especially the heterogeneous populations of immune cells, which are closely related to cancer prognoses and the efficacy of cancer therapies.”
The Samsung Research Funding & Incubation Center for Future Technology supported this work. Spectral imaging was performed at the Korea Basic Science Institute Western Seoul Center.
-PublicationJunyoung Seo, Yeonbo Sim, Jeewon Kim, Hyunwoo Kim, In Cho, Hoyeon Nam, Yong-Gyu Yoon, Jae-Byum Chang, “PICASSO allows ultra-multiplexed fluorescence imaging of spatiallyoverlapping proteins without reference spectra measurements,” May 5, Nature Communications (doi.org/10.1038/s41467-022-30168-z)
-ProfileProfessor Jae-Byum ChangDepartment of Materials Science and EngineeringCollege of EngineeringKAIST
Professor Young-Gyu YoonSchool of Electrical EngineeringCollege of EngineeringKAIST
2022.06.22 View 12396
PICASSO Technique Drives Biological Molecules into Technicolor
The new imaging approach brings current imaging colors from four to more than 15 for mapping overlapping proteins
Pablo Picasso’s surreal cubist artistic style shifted common features into unrecognizable scenes, but a new imaging approach bearing his namesake may elucidate the most complicated subject: the brain. Employing artificial intelligence to clarify spectral color blending of tiny molecules used to stain specific proteins and other items of research interest, the PICASSO technique, allows researchers to use more than 15 colors to image and parse our overlapping proteins.
The PICASSO developers, based in Korea, published their approach on May 5 in Nature Communications.
Fluorophores — the staining molecules — emit specific colors when excited by a light, but if more than four fluorophores are used, their emitted colors overlap and blend. Researchers previously developed techniques to correct this spectral overlap by precisely defining the matrix of mixed and unmixed images. This measurement depends on reference spectra, found by identifying clear images of only one fluorophore-stained specimen or of multiple, identically prepared specimens that only contain a single fluorophore each.
“Such reference spectra measurement could be complicated to perform in highly heterogeneous specimens, such as the brain, due to the highly varied emission spectra of fluorophores depending on the subregions from which the spectra were measured,” said co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST. He explained that the subregions would each need their own spectra reference measurements, making for an inefficient, time-consuming process. “To address this problem, we developed an approach that does not require reference spectra measurements.”
The approach is the “Process of ultra-multiplexed Imaging of biomolecules viA the unmixing of the Signals of Spectrally Overlapping fluorophores,” also known as PICASSO. Ultra-multiplexed imaging refers to visualizing the numerous individual components of a unit. Like a cinema multiplex in which each theater plays a different movie, each protein in a cell has a different role. By staining with fluorophores, researchers can begin to understand those roles.
“We devised a strategy based on information theory; unmixing is performed by iteratively minimizing the mutual information between mixed images,” said co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering, KAIST. “This allows us to get away with the assumption that the spatial distribution of different proteins is mutually exclusive and enables accurate information unmixing.”
To demonstrate PICASSO’s capabilities, the researchers applied the technique to imaging a mouse brain. With a single round of staining, they performed 15-color multiplexed imaging of a mouse brain. Although small, mouse brains are still complex, multifaceted organs that can take significant resources to map. According to the researchers, PICASSO can improve the capabilities of other imaging techniques and allow for the use of even more fluorophore colors.
Using one such imaging technique in combination with PICASSO, the team achieved 45-color multiplexed imaging of the mouse brain in only three staining and imaging cycles, according to Yoon.
“PICASSO is a versatile tool for the multiplexed biomolecule imaging of cultured cells, tissue slices and clinical specimens,” Chang said. “We anticipate that PICASSO will be useful for a broad range of applications for which biomolecules’ spatial information is important. One such application the tool would be useful for is revealing the cellular heterogeneities of tumor microenvironments, especially the heterogeneous populations of immune cells, which are closely related to cancer prognoses and the efficacy of cancer therapies.”
The Samsung Research Funding & Incubation Center for Future Technology supported this work. Spectral imaging was performed at the Korea Basic Science Institute Western Seoul Center.
-PublicationJunyoung Seo, Yeonbo Sim, Jeewon Kim, Hyunwoo Kim, In Cho, Hoyeon Nam, Yong-Gyu Yoon, Jae-Byum Chang, “PICASSO allows ultra-multiplexed fluorescence imaging of spatiallyoverlapping proteins without reference spectra measurements,” May 5, Nature Communications (doi.org/10.1038/s41467-022-30168-z)
-ProfileProfessor Jae-Byum ChangDepartment of Materials Science and EngineeringCollege of EngineeringKAIST
Professor Young-Gyu YoonSchool of Electrical EngineeringCollege of EngineeringKAIST
2022.06.22 View 12396