Disease+Molecule+Biochemistry+Lab
-
 Structural Insights into the Modulation of Synaptic Adhesion by MDGA for Synaptogenesis
Synapses connected by various synaptic adhesion molecules are communication spaces between neurons for transmitting information. Among various synaptic adhesion molecules, neuroligins are arguably the most widely studied class of postsynaptic adhesion molecules, which mainly interact with presynaptic neurexins to induce excitatory or inhibitory synapse development. Recently, the membrane-associated mucin (MAM) domain-containing GPI anchor protein 1 (MDGA1) has been characterized as a key suppressor of Neuroligin-2/Neurexin-1β-mediated inhibitory synapse development, but how it acts remains a mystery.
In a recent issue of Neuron, published on June 21, 2017, a research team led by Professor Ho Min Kim at the Graduate School of Medical Science and Engineering of KAIST reported the three-dimensional structure of MDGA1/Neuroligin-2 complex and mechanistic insights into how MDGAs negatively modulate synapse development governed by Neurexins/Neuroligins trans -synaptic adhesion complex.
MDGA1 consists of six Ig-like domains, fibronectin type III repeat domain, and MAM domain . The crystal structure of MDGA1/Neuroligin-2 complex reveals that they form the 2:2 hetero-tetrameric complex and only the Ig1-Ig2 domains of MDGA1 are involved in interactions with Neuroligin-2. The structural comparison between the MDGA1/Neuroligin-2 and Neurexin-1β/Neuroligin-1 complexes intriguingly indicates that the Neuroligin-2 region binding to MDGA1 largely overlaps with that of Neurexin-1β, but the interaction interface of the MDGA1/Neuroligin-2 complex is much larger than that of the Neurexin-1β/Neuroligin-1 complex. This explains why Neuroligin-2 binds stronger to MDGA1 than Neurexin-1β, and how the favored MDGA1 binding to Neuroligin-2 sterically blocks the interaction between Neuroligin-2 and Neurexin-1β, which is critical for the suppression of inhibitory synapse development.
“Although we found that MDGA Ig domains (Ig 1 and Ig 2) are sufficient to form a complex with NL2, other extracellular domains, including Ig 3–6, FN III, and MAM domains, may also contribute to stable cis-interactions between MDGA1 and Neuroligin-2 by providing conformational flexibility. Therefore, further structural analysis of full-length MDGA will be required,” Professor Kim said.
Neuroligin-2 specifically promotes the development of inhibitory synapses, whereas neuroligin-1 promotes the development of excitatory synapses. Recently, not only MDGA1, but also MDGA2 have emerged as synaptic regulators for the development of excitatory or inhibitory synapses. In vitro biochemical analysis in this research clearly demonstrates that Neuroligin-1 and Neuroligin-2 bind to both MDGA1 and MDGA2 with comparable affinity. However, pull-down assays using detergent-solubilized mouse brain membrane fractions show the specific interaction of MDGA1 with Neuroligin-2, but not with Neuroligin-1. “This suggests that unidentified processes may dictate the selective association of MDGA1 with Neuroligin-2 in vivo , ” explained Professor Jaewon Ko at the Daegu Gyeongbuk Institute of Science and Technology (DGIST).
A balance between excitatory and inhibitory synapses is crucial to healthy cognition and behavior. Mutations in neuroligins, neurexins, and MDGAs, which can disrupt the excitatory/inhibitory balance, are associated with neuropsychiatric diseases such as autism and schizophrenia. Jung A Kim at KAIST, first author in this study, said, “Our discovery from integrative investigations are an important first step both for a better understanding of Neuroligin/Neurexin synaptic adhesion pathways and MDGA-mediated regulation of synapse development as well as the development of potential new therapies for autism, schizophrenia, and epilepsy.”
2017.07.10 View 9695
Structural Insights into the Modulation of Synaptic Adhesion by MDGA for Synaptogenesis
Synapses connected by various synaptic adhesion molecules are communication spaces between neurons for transmitting information. Among various synaptic adhesion molecules, neuroligins are arguably the most widely studied class of postsynaptic adhesion molecules, which mainly interact with presynaptic neurexins to induce excitatory or inhibitory synapse development. Recently, the membrane-associated mucin (MAM) domain-containing GPI anchor protein 1 (MDGA1) has been characterized as a key suppressor of Neuroligin-2/Neurexin-1β-mediated inhibitory synapse development, but how it acts remains a mystery.
In a recent issue of Neuron, published on June 21, 2017, a research team led by Professor Ho Min Kim at the Graduate School of Medical Science and Engineering of KAIST reported the three-dimensional structure of MDGA1/Neuroligin-2 complex and mechanistic insights into how MDGAs negatively modulate synapse development governed by Neurexins/Neuroligins trans -synaptic adhesion complex.
MDGA1 consists of six Ig-like domains, fibronectin type III repeat domain, and MAM domain . The crystal structure of MDGA1/Neuroligin-2 complex reveals that they form the 2:2 hetero-tetrameric complex and only the Ig1-Ig2 domains of MDGA1 are involved in interactions with Neuroligin-2. The structural comparison between the MDGA1/Neuroligin-2 and Neurexin-1β/Neuroligin-1 complexes intriguingly indicates that the Neuroligin-2 region binding to MDGA1 largely overlaps with that of Neurexin-1β, but the interaction interface of the MDGA1/Neuroligin-2 complex is much larger than that of the Neurexin-1β/Neuroligin-1 complex. This explains why Neuroligin-2 binds stronger to MDGA1 than Neurexin-1β, and how the favored MDGA1 binding to Neuroligin-2 sterically blocks the interaction between Neuroligin-2 and Neurexin-1β, which is critical for the suppression of inhibitory synapse development.
“Although we found that MDGA Ig domains (Ig 1 and Ig 2) are sufficient to form a complex with NL2, other extracellular domains, including Ig 3–6, FN III, and MAM domains, may also contribute to stable cis-interactions between MDGA1 and Neuroligin-2 by providing conformational flexibility. Therefore, further structural analysis of full-length MDGA will be required,” Professor Kim said.
Neuroligin-2 specifically promotes the development of inhibitory synapses, whereas neuroligin-1 promotes the development of excitatory synapses. Recently, not only MDGA1, but also MDGA2 have emerged as synaptic regulators for the development of excitatory or inhibitory synapses. In vitro biochemical analysis in this research clearly demonstrates that Neuroligin-1 and Neuroligin-2 bind to both MDGA1 and MDGA2 with comparable affinity. However, pull-down assays using detergent-solubilized mouse brain membrane fractions show the specific interaction of MDGA1 with Neuroligin-2, but not with Neuroligin-1. “This suggests that unidentified processes may dictate the selective association of MDGA1 with Neuroligin-2 in vivo , ” explained Professor Jaewon Ko at the Daegu Gyeongbuk Institute of Science and Technology (DGIST).
A balance between excitatory and inhibitory synapses is crucial to healthy cognition and behavior. Mutations in neuroligins, neurexins, and MDGAs, which can disrupt the excitatory/inhibitory balance, are associated with neuropsychiatric diseases such as autism and schizophrenia. Jung A Kim at KAIST, first author in this study, said, “Our discovery from integrative investigations are an important first step both for a better understanding of Neuroligin/Neurexin synaptic adhesion pathways and MDGA-mediated regulation of synapse development as well as the development of potential new therapies for autism, schizophrenia, and epilepsy.”
2017.07.10 View 9695 -
 Complex responsible for protein breakdown in cells identified using Bio TEM
Professor Ho-Min Kim
- High resolution 3D structure analysis success using Bio Transmission Electron Microscopy (TEM), a giant step towards new anticancer treatment development
- Published in Nature on May 5th
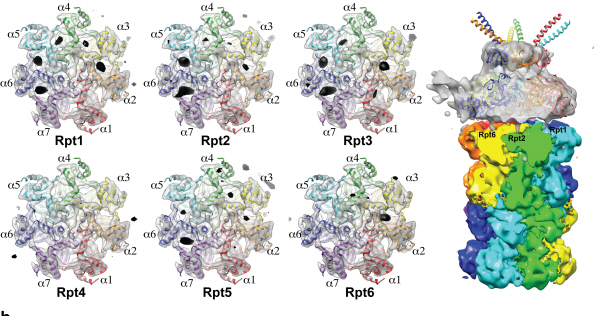
Using TEM to observe protein molecules and analysing its high resolution 3D structure is now possible. KAIST Biomedical Science and Engineering Department’s Professor Ho-Min Kim has identified the high resolution structure of proteasome complexes, which is responsible for protein breakdown in cells, using Bio TEM.
This research has been published on the world"s most prestigious journal, Nature, online on May 5th. Our body controls many cellular processes through production and degradation of proteins to maintain homeostasis. A proteasome complex acts as a garbage disposal system and degrades cellular proteins when needed for regulation, which is one of the central roles of the body.
However, a mutation in proteasome complex leads to diseases such as cancer, degenerative brain diseases, and autoimmune diseases.
Currently, the anticancer drug Velcade is used to decrease proteasome function to treat Multiple Myeloma, a form of blood cancer. Research concerning proteasome complexes for more effective anticancer drugs and treatments with fewer side effects has been taking place for more than 20 years. There have been many difficulties in understanding proteasome function through 3D structure analysis since a proteasome complex, consisting of around 30 different proteins, has a great size and complexity.
The research team used Bio TEM instead of conventionally used protein crystallography technique. The protein sample was inserted into Bio TEM, hundreds of photographs were taken from various angles, and then a high–performance computer was used to analyse its structure. Bio TEM requires a smaller sample and can analyse the complexes of great size of proteins.
Professor Ho-Min Kim said, “Identifying proteasome complex assembly process and 3D structure will increase our understanding of cellular protein degradation process and hence assist in new drug development using this knowledge.” He added, “High resolution protein structure analysis using Bio TEM, used for the first time in Korea, will enable us to observe structure analysis of large protein complexes that were difficult to approach using protein crystallography.” Professor Kim continued, “If protein crystallography technology and Bio TEM could be used together to complement one another, it would bring a great synergetic effect to protein complex 3D structure analysis research in the future.”
Professor Ho-Min Kim has conducted this research since his post-doctorate at the University of California, San Francisco, under the advice of Professor Yifan Cheng; in co-operation with Harvard University and Colorado University.
Figure 1: A picture taken by Bio TEM of open state protein sample (proteasome complex)
Figure 2: Bio TEM image analysis showing protein 3D structure
2013.05.25 View 11913
Complex responsible for protein breakdown in cells identified using Bio TEM
Professor Ho-Min Kim
- High resolution 3D structure analysis success using Bio Transmission Electron Microscopy (TEM), a giant step towards new anticancer treatment development
- Published in Nature on May 5th
Using TEM to observe protein molecules and analysing its high resolution 3D structure is now possible. KAIST Biomedical Science and Engineering Department’s Professor Ho-Min Kim has identified the high resolution structure of proteasome complexes, which is responsible for protein breakdown in cells, using Bio TEM.
This research has been published on the world"s most prestigious journal, Nature, online on May 5th. Our body controls many cellular processes through production and degradation of proteins to maintain homeostasis. A proteasome complex acts as a garbage disposal system and degrades cellular proteins when needed for regulation, which is one of the central roles of the body.
However, a mutation in proteasome complex leads to diseases such as cancer, degenerative brain diseases, and autoimmune diseases.
Currently, the anticancer drug Velcade is used to decrease proteasome function to treat Multiple Myeloma, a form of blood cancer. Research concerning proteasome complexes for more effective anticancer drugs and treatments with fewer side effects has been taking place for more than 20 years. There have been many difficulties in understanding proteasome function through 3D structure analysis since a proteasome complex, consisting of around 30 different proteins, has a great size and complexity.
The research team used Bio TEM instead of conventionally used protein crystallography technique. The protein sample was inserted into Bio TEM, hundreds of photographs were taken from various angles, and then a high–performance computer was used to analyse its structure. Bio TEM requires a smaller sample and can analyse the complexes of great size of proteins.
Professor Ho-Min Kim said, “Identifying proteasome complex assembly process and 3D structure will increase our understanding of cellular protein degradation process and hence assist in new drug development using this knowledge.” He added, “High resolution protein structure analysis using Bio TEM, used for the first time in Korea, will enable us to observe structure analysis of large protein complexes that were difficult to approach using protein crystallography.” Professor Kim continued, “If protein crystallography technology and Bio TEM could be used together to complement one another, it would bring a great synergetic effect to protein complex 3D structure analysis research in the future.”
Professor Ho-Min Kim has conducted this research since his post-doctorate at the University of California, San Francisco, under the advice of Professor Yifan Cheng; in co-operation with Harvard University and Colorado University.
Figure 1: A picture taken by Bio TEM of open state protein sample (proteasome complex)
Figure 2: Bio TEM image analysis showing protein 3D structure
2013.05.25 View 11913